Abstract
DNA methylation is a key epigenetic mechanism responsible for gene regulation, chromatin remodeling, and genome stability, playing a fundamental role during embryonic development. The aim of this study was to determine if these epigenetic marks are associated with chromosomal aneuploidy in human blastocysts. Surplus, cryopreserved blastocysts that were donated to research with IRB consent were chosen with varying chromosomal aneuploidies and respective implantation potential: monosomies and trisomies 7, 11, 15, 21, and 22. DNA methylation analysis was performed using the Illumina Infinium HumanMethylation450 BeadChip (~485,000 CpG sites). The methylation profiles of these human blastocysts were found to be similar across all samples, independent of chromosome constitution; however, more detailed examination identified significant hypomethylation in the chromosome involved in the monosomy. Real-time PCR was also performed to determine if downstream messenger RNA (mRNA) was affected for genes on the monosomy chromosome. Gene dysregulation was observed for monosomy blastocysts within significant regions of hypo-methylation (AVEN, CYFIP1, FAM189A1, MYO9A, ADM2, PACSIN2, PARVB, and PIWIL3) (P < 0.05). Additional analysis was performed to examine the gene expression profiles of associated methylation regulators including: DNA methyltransferases (DNMT1, DNMT3A, DNMT3B, DNMT3L), chromatin modifying regulators (CSNK1E, KDM1, PRKCA), and a post-translational modifier (PRMT5). Decreased RNA transcription was confirmed for each DNMT, and the regulators that impact DNMT activity, for only monosomy blastocysts (P < 0.05). In summary, monosomy blastocysts displayed hypomethylation for the chromosome involved in the error, as well as transcription alterations of associated developmental genes. Together, these modifications may be contributing to genetic instability and therefore be responsible for the limited implantation potential observed for full monosomy blastocysts.
Introduction
During reproduction, an embryo receives one set of chromosomes from the sperm, and one set from the oocyte, resulting in a complete set of 23 chromosome pairs. Errors during meiotic or mitotic cell division can lead to extra or missing chromosomes, termed aneuploidy, which is the leading cause of miscarriage, stillbirth, and congenital birth defects [1]. The most significant risk factor for an aneuploid conception is advanced maternal age. In fact, 35–50% of oocytes from women aged 35–39 will have chromosomal aneuploidies and this will climb to over 80% once a woman reaches 45 years of age [2]. Aneuploidy can occur for any chromosome with the highest proportion observed in conception belonging to the smaller sized chromosomes (15–22) [3]. Only a fraction of full aneuploidies, specifically trisomies 13, 18, 21, XXY, and XYY, will develop past the first trimester and may even result in a live birth [4]. Nevertheless, even the vast majority (>95%) of these trisomies will perish in utero. This is in contrast to full monosomies which almost never implant or result in an ongoing pregnancy. Turner Syndrome (XO) is the only exception and the only full monosomy known to reach term. Partial fetal autosomal monosomies are observed during clinical pregnancy, where only a portion of a chromosome is missing, and these imbalances can lead to various phenotypes depending on the chromosome involved, the size of the absent chromosome, and which genes are impacted [5].
DNA methylation is a biochemical process that plays an important role in regulating gene expression without altering the underlying DNA sequence and involves the addition of a methyl group to a cytosine in a CpG dinucleotide. This is established either de novo, by DNA methyltransferases DNMT3A, DNMT3B, and DNMT3L, or during replication by the maintenance methyltransferase DNMT1 [6]. Appropriate methylation is essential for both normal cell differentiation and development [7, 8]. Methylation is involved in chromatin structure which is responsible for proper chromosome segregation during cell division as well as regulating gene expression [9]. Global epigenetic reprogramming begins in the early embryo with DNA demethylation occurring post fertilization through to the blastocyst stage. This process is essential for the establishment of embryonic gene expression patterns during re-methylation which is required for implantation and ongoing fetal development [8]. Only imprinted genes escape demethylation to preserve their exclusive parent-of-origin-specific gene expression profiles [10]. DNA methylation is also shown to be vital in the maintenance of X chromosome inactivation which is crucial for female embryos due to the presence of two X chromosomes [11]. It is well known that disturbances during these methylation processes can result in developmental delays and/or embryo death. Loss of Dnmt1 activity results in significantly lower DNA methylation levels, as well as impaired implantation and embryo development in mice [12]. Dnmt3a mutant mice develop to term but are runted and die at around 4 weeks of age while Dnmt3b mutant mice have no viable births as their embryos are found to have multiple developmental defects [13]. Dnmt3l interacts with Dnmt3a and 3b and has been shown in mice to play an important role in the regulation of genomic imprinting and embryonic development [14].
Given the importance of DNA methylation and chromosome constitution to healthy fetal/embryonic development, the aim of this study was to investigate the association between methylation, the molecular processes involved in establishing methylation, and chromosomal aneuploidies. Results revealed that trisomy blastocysts had similar methylation profiles to their euploid counterparts. In contrast, monosomy blastocysts were hypomethylated for the chromosome involved in the error and displayed altered expression of developmental genes and DNMTs, which could be contributing to their overall compromised implantation potential.
Methods and Materials
Blastocysts
Surplus, cryopreserved blastocysts (n = 316) from the Colorado Center for Reproductive Medicine were donated for research with written IRB consent, including blastocysts donated from donor oocyte cycles. This study was approved by HCA-HealthONE (study #231587) and Western Institutional Review Board (study #1145350). All blastocysts were viable and morphologically similar, graded as high quality expanded blastocysts (≥ 3BB) on day 5 of embryonic development using the Gardner and Schoolcraft system [15]. Blastocysts underwent trophectoderm biopsy for comprehensive chromosome screening prior to vitrification using the cryotop method as previously described [16]. The control group consisted of euploid, day 5 blastocysts produced from donor oocyte IVF cycles with no male factor infertility. Specific aneuploidies were chosen based on their differing implantation potential and included chromosomes 7, 11, 15, 21, and 22. Trisomies 7 and 11 are most likely to result in implantation failure; trisomies 15 and 22 are able to implant however will always result in miscarriage; and trisomy 21 embryos will implant but result in either miscarriage, still birth, or live birth.
DNA Lysis and Methylation Analysis
After warming, blastocysts (n = 230) were lysed using the EZ DNA Methylation-Direct™ Kit (Zymo Research, Irvine CA). Briefly, pools of 10 re-expanded blastocysts, with 2–3 biological replicates per group, were washed through a series of PBS washes before being lysed in a digestion buffer containing 20ug Proteinase K in a 20ul final volume. Samples were incubated at 50°C for 20 minutes and then stored at -80°C. All 20ul of each sample were bisulfite converted by adding 130ul of CT Conversion Reagent and incubated at 98°C for 8 minutes and 64°C for 3.5 hours. Samples were then purified on the Zymo-Spin™ IC Column according to manufacturer’s protocol and eluted in 10ul of M-Elution Buffer. 500ng of each sample were amplified, fragmented, and hybridized to the Infinium HumanMethylation450K BeadChip (Illumina, San Diego CA). GenomeStudio Methylation Module 1.0 software (Illumina) was used for image processing and to perform normalization and differential methylation analysis. Normalization was performed using both normalization control probes as well as background subtraction. Methylation beta values were then determined for each sample which estimate the methylation level of the CpG locus using the ratio intensities between methylated and unmethylated alleles. A value of “0” represents no methylation and a value of “1” indicates full methylation. DiffScore was calculated using the Illumina Custom Model to determine significance at P < 0.05. Variance was estimated across replicate samples.
RNA Isolation, Reverse Transcription, and Real-Time PCR
RNA was either isolated using the PicoPure RNA Isolation Kit (Life Technologies, Grand Island, NY) or lysed and deoxyribonuclease treated using the Taqman® Gene Expression Cells-to-Ct™ Kit (Life Technologies). For primer-based assays, warmed blastocysts (n = 50) were washed through ice-cold phosphate buffered saline (PBS) containing bovine serum albumin (BSA) prior to being transferred into 10ul of Extraction Buffer. RNA was then purified from individual blastocysts (PicoPure) according to manufacturer’s protocol with minor modifications [17]. RNA quantity and quality were assessed using the NanoDrop® Spectrophotometer ND-1000 (Thermo Scientific, Wilmington DE) before being reverse transcribed using the High Capacity cDNA Archive Kit (Life Technologies) where 20ul of a master mix was combined with all 20ul of the purified RNA and incubated according to protocol.
For Taqman® assays, warmed blastocysts (n = 36) were washed as previously mentioned prior to being individually transferred into 10ul of Lysis Solution containing DNase I (Cells-to-Ct™) and incubated at room temperature for 8 minutes. 1ul of Stop Solution was added to each sample and incubated at room temperature for 2 minutes. Samples were reverse transcribed with 30ul of master mix and 10ul of RNA lysate. cDNA was then amplified by using 37.5ul of Taqman® PreAmp Master Mix containing 0.05X of each Taqman® probe with 12.5ul of the cDNA under the following thermal cycling conditions: 95°C for 10 minutes and 12 cycles at 95°C for 15 seconds and 60°C for 4 minutes.
Primer-based quantitative real-time PCR (qPCR) was performed using the ABI 7300 Real-Time PCR System (Life Technologies) by combining 5ul of diluted cDNA (1:4) with 7ul water, 12.5ul SYBR Green PCR Master Mix (Life Technologies) and 0.5ul of 5uM primer pool. After a 10 minute incubation at 95°C, amplification occurred for 40 cycles at 95°C for 15 seconds and 60°C for 1 minute, followed by a dissociation stage. Standard curves were calculated for each gene by performing 10-fold serial dilutions of reference RNA (Agilent, Santa Clara CA). Expression of 8 genes of interest were analyzed in duplicates (AVEN, CYFIP1, FAM189A1, MYO9A, ADM2, PACSIN2, PARVB, and PIWIL3) relative to an internal house-keeping gene, PPIA, which had the most consistent expression across all samples (Table 1). Negative controls were performed for each gene and all remained unamplified with Ct values at 40.
Table 1. Primer information and qPCR efficiencies for chromosomes 15, 22, and housekeeping genes.
| Gene | Accession # | Slope | Amplicon GC | Chromosome | Primer Sequence (3'-5') |
|---|---|---|---|---|---|
| Intercept | Amplicon Length | ||||
| R2 | |||||
| AVEN | NM_020371 | -3.29 | 46% | 15 | F: AAGAGCTGGAAGACTGGTTGGA |
| 28.37 | 98 | R: TATGCCCACCTGCCGTTAG | |||
| 0.99 | |||||
| CYFIP1 | NM_014608 | -3.51 | 51% | 15 | F: ACGACCACTCAGCGTACAAGAG |
| 25.71 | 78 | R: TCTGCGATTCCTGGATGGA | |||
| 0.99 | |||||
| FAM189A1 | NM_015307 | -3.07 | 62% | 15 | F: GGGACACCCAGGATGATCTG |
| 31.47 | 97 | R: GGAAATGCAATCCCCAAAGAG | |||
| 0.99 | |||||
| MYO9A | NM_006901 | -3.26 | 45% | 15 | F: CAATACACTGGAACGCCTCATC |
| 27.11 | 92 | R: ACACAATGGCCAAAGCATTAGC | |||
| 0.99 | |||||
| ADM2 | NM_001253845 | -2.97 | 59% | 22 | F: GAGCCTAAACACCCTGAAATTGTG |
| 28.92 | 88 | R: TCTCTGAAGCGCTTAGCATCTG | |||
| 0.99 | |||||
| PACSIN2 | NM_001184970 | -3.57 | 54% | 22 | F: AAGCCCTGGGCCAAGAAG |
| 26.99 | 59 | R: GCTGCATGGTGGGCTTTC | |||
| 0.99 | |||||
| PARVB | NM_001003828 | -3.45 | 62% | 22 | F: TCTCTGGCCATGCACTTCAG |
| 25.31 | 65 | R: ACCACCACCTGCACCGTTAC | |||
| 0.99 | |||||
| PIWIL3 | NM_001008496 | -1.51 | 44% | 22 | F: AAAGAGCGGAGAGTGGAATGG |
| 32.88 | 91 | R: ACGTGGGCGTGAGTTCTTTG | |||
| 0.95 | |||||
| PPIA | NM_021130 | -4.81 | 51% | 7 | F: GCTTTGGGTCCAGGAATGG |
| 21.52 | 59 | R: TTGTCCACAGTCAGCAATGG | |||
| 0.96 |
Taqman® qPCR was performed by combining 4ul of diluted pre-amplified product (1:5) with 5ul nuclease-free water, 10ul Taqman® Gene Expression Master Mix, and 1ul Taqman® probe. This was run on the ABI7900HT Fast Real-Time PCR System (Life Technologies) at 95°C for 10 minutes, and 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. Standard curves were calculated as previously described and expression of 8 genes of interest were analyzed in duplicates (DNMT1, DNMT3A, DNMT3B, DNMT3L, CSNK1E, KDM1, PRKCA, and PRMT5) relative to an internal house-keeping gene, RPL19, which had the most consistent expression across all samples (Table 2). Negative controls were also performed for each Taqman® assay and all were found to be unamplified.
Table 2. Genes involved in DNA methylation processes (including two housekeeping genes) and qPCR efficiency information (Taqman® assays; Life Technologies).
| Gene | Entrez ID / Catalog # | Slope | Chromosome | Function |
|---|---|---|---|---|
| Intercept | ||||
| R2 | ||||
| DNMT1 | 1786 / Hs00154749_m1 | -3.46 | 19 | Maintanence methyltransferase |
| 27.92 | ||||
| 0.99 | ||||
| DNMT3A | 1788 / Hs01027166_m1 | -3.48 | 2 | de novo methlytransferase |
| 31.02 | ||||
| 0.99 | ||||
| DNMT3B | 1789 / Hs00171876_m1 | -3.35 | 20 | de novo methlytransferase |
| 29.97 | ||||
| 0.99 | ||||
| DNMT3L | 29947 / Hs01081364_m1 | -2.73 | 21 | In-active methyltransferase essential for the |
| 34.00 | function of DNMT3A and DNMT3B | |||
| 0.95 | ||||
| CSNK1E | 1454 / Hs00266431_m1 | -3.95 | 22 | Post-translational regulation |
| 28.02 | ||||
| 0.99 | ||||
| KDM1 | 23028 / Hs01002741_m1 | -3.76 | 1 | Post-translational regulation |
| 29.90 | ||||
| 0.99 | ||||
| PRKCA | 5578 / Hs00925193_m1 | -4.13 | 17 | Post-translational regulation |
| 30.52 | ||||
| 0.97 | ||||
| PRMT5 | 10419 / Hs01047356_m1 | -3.77 | 14 | Chromatin modifying protein |
| 29.17 | ||||
| 0.99 | ||||
| PPIA | 5478 / Hs04194521_s1 | -3.71 | 7 | Housekeeping |
| 24.43 | ||||
| 0.99 | ||||
| RPL19 | 6143 / Hs01577060_gH | -3.53 | 17 | Housekeeping |
| 25.18 | ||||
| 0.99 |
Data normalization and analysis were performed using REST 2009 software (Qiagen, Valencia CA). REST software uses the correction for exact PCR efficiencies with mean crossing point deviations between sample and control groups to determine an expression ratio that is tested for significance by a Pair Wise Fixed Reallocation Randomization Test. Significance was defined as P < 0.05.
Results
Global Methylation Analysis
Analysis of the blastocyst methylome for monosomies 7, 11, 15, 21, and 22, as well as trisomies 7, 11, 15, 21, and 22, compared to control blastocysts, was performed using the Illumina Infinium HumanMethylation450K BeadChip. To avoid bias, groups were blinded and Illumina GenomeStudio Software was used for normalization, beta value calculations, and DiffScore determination. When analyzing the overall methylation profiles of any blastocyst group, no significant differences were observed regardless of chromosome constitution. The average beta value (0 = no methylation, 1 = full methylation) for each group was similar, ranging from 0.20 to 0.21, representing an overall hypomethylated state (Table 3). For comparison, a typical somatic cell has a beta value of around 0.5 [18]. Further examination of the methylome of each individual chromosome revealed all trisomy blastocysts, independent of which chromosome had a third copy (7, 11, 15, 21, or 22), were similar to the diploid state (Table 4). For example, the beta value of chromosome 11 in trisomy 11 blastocysts was 0.21 (Table 4, column D) and the beta value of chromosome 11 in control blastocysts was 0.22 (Table 4, column A). In contrast, all monsomy blastocysts showed a decreased methylated state for the specific missing chromosome in comparison to controls. In this case, monosomy 11 blastocysts displayed significant hypomethylation of chromosome 11 with a beta value of 0.17 (Table 4, columnE, P < 0.05) compared to chromosome 11 in either trisomy or control blastocysts which had beta values of 0.21 and 0.22 respectively. All other correctly-paired chromosomes from these aneuploid blastocysts displayed a methylation profile similar to control blastocysts (Table 4).
Table 3. Methylome profiles of pooled human blastocysts (n = 10 each pool with 2–3 replicates per group).
Beta value reflects the level of global methylation with no variation observed in association with blastocyst chromosome constitution. (no statistical significance). Standard deviation (STDEV) was calculated for each group, reflecting low biological variability between replicates.
| Group | Beta Value | STDEV |
|---|---|---|
| (Avg) | ||
| Diploid (Euploid) | 0.21 | 0.01 |
| Trisomy 7 | 0.21 | 0.01 |
| Trisomy 11 | 0.20 | 0.01 |
| Trisomy 15 | 0.20 | 0.01 |
| Trisomy 21 | 0.20 | 0.01 |
| Trisomy 22 | 0.20 | 0.01 |
| Monosomy 7 | 0.20 | 0.01 |
| Monosomy 11 | 0.20 | 0.06 |
| Monosomy 15 | 0.20 | 0.00 |
| Monosomy 21 | 0.20 | 0.01 |
| Monosomy 22 | 0.21 | 0.00 |
Table 4. Methylation profiles of individual chromosomes for blastocysts with a specific chromosome constitution: A) Diploid Control, B) Trisomy 7, C) Monosomy 7, D) Trisomy 11, E) Monosomy 11, F) Trisomy 15, G) Monosomy 15, H) Trisomy 21, I) Monosomy 21, (J) Trisomy 22, and (K) Monosomy 22 (*P < 0.05).
| A | B | C | D | E | F | G | H | I | J | K | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Control | +7 | -7 | +11 | -11 | +15 | -15 | +21 | -21 | +22 | -22 |
| 1 | 0.19 | 0.19 | 0.19 | 0.18 | 0.18 | 0.18 | 0.18 | 0.18 | 0.18 | 0.18 | 0.19 |
| 2 | 0.22 | 0.22 | 0.22 | 0.20 | 0.20 | 0.21 | 0.22 | 0.21 | 0.22 | 0.22 | 0.23 |
| 3 | 0.22 | 0.22 | 0.22 | 0.21 | 0.21 | 0.20 | 0.21 | 0.21 | 0.21 | 0.21 | 0.22 |
| 4 | 0.23 | 0.23 | 0.23 | 0.22 | 0.23 | 0.22 | 0.22 | 0.21 | 0.23 | 0.22 | 0.23 |
| 5 | 0.21 | 0.21 | 0.21 | 0.20 | 0.21 | 0.20 | 0.21 | 0.20 | 0.21 | 0.20 | 0.21 |
| 6 | 0.20 | 0.20 | 0.20 | 0.19 | 0.19 | 0.19 | 0.19 | 0.19 | 0.19 | 0.19 | 0.20 |
| 7 | 0.25 | 0.26 | 0.23* | 0.24 | 0.24 | 0.24 | 0.24 | 0.24 | 0.24 | 0.24 | 0.25 |
| 8 | 0.24 | 0.24 | 0.23 | 0.23 | 0.23 | 0.22 | 0.23 | 0.22 | 0.23 | 0.23 | 0.24 |
| 9 | 0.21 | 0.21 | 0.20 | 0.20 | 0.20 | 0.20 | 0.20 | 0.19 | 0.20 | 0.20 | 0.21 |
| 10 | 0.23 | 0.22 | 0.22 | 0.21 | 0.22 | 0.21 | 0.22 | 0.21 | 0.22 | 0.22 | 0.22 |
| 11 | 0.22 | 0.21 | 0.21 | 0.21 | 0.17* | 0.21 | 0.21 | 0.20 | 0.21 | 0.21 | 0.22 |
| 12 | 0.22 | 0.21 | 0.21 | 0.20 | 0.21 | 0.20 | 0.21 | 0.20 | 0.21 | 0.21 | 0.21 |
| 13 | 0.24 | 0.24 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.24 |
| 14 | 0.21 | 0.20 | 0.20 | 0.20 | 0.20 | 0.19 | 0.20 | 0.19 | 0.20 | 0.20 | 0.21 |
| 15 | 0.22 | 0.22 | 0.22 | 0.21 | 0.22 | 0.22 | 0.18* | 0.21 | 0.22 | 0.22 | 0.22 |
| 16 | 0.23 | 0.22 | 0.22 | 0.22 | 0.21 | 0.22 | 0.22 | 0.21 | 0.22 | 0.22 | 0.23 |
| 17 | 0.20 | 0.20 | 0.19 | 0.19 | 0.19 | 0.19 | 0.19 | 0.19 | 0.19 | 0.19 | 0.20 |
| 18 | 0.18 | 0.18 | 0.18 | 0.18 | 0.18 | 0.17 | 0.18 | 0.17 | 0.18 | 0.18 | 0.18 |
| 19 | 0.18 | 0.18 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.18 |
| 20 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.16 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 |
| 21 | 0.23 | 0.22 | 0.22 | 0.21 | 0.22 | 0.21 | 0.22 | 0.22 | 0.18* | 0.22 | 0.23 |
| 22 | 0.18 | 0.18 | 0.18 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.18 | 0.14* |
| X | 0.16 | 0.17 | 0.16 | 0.15 | 0.16 | 0.15 | 0.16 | 0.16 | 0.16 | 0.16 | 0.16 |
| Y | 0.16 | 0.17 | 0.18 | 0.18 | 0.16 | 0.15 | 0.14 | 0.16 | 0.17 | 0.16 | 0.15 |
Blastocyst Gene Expression
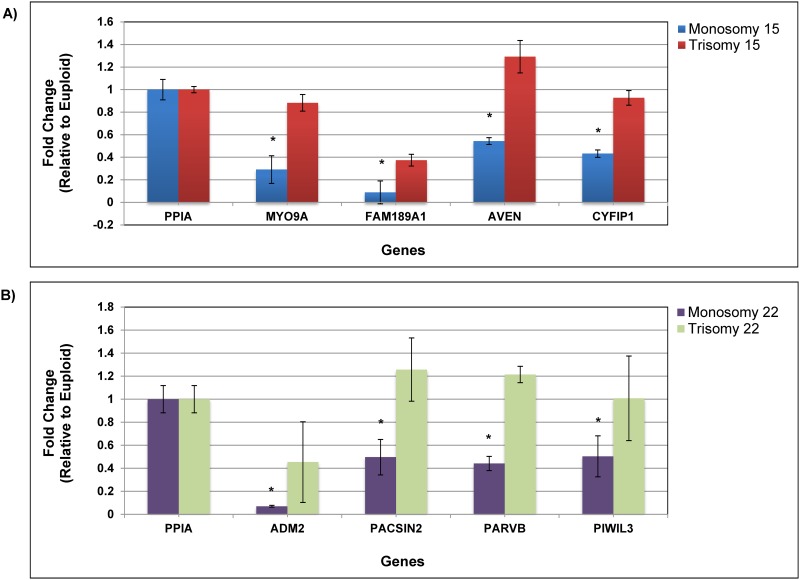
mRNA analysis was performed on individual blastocysts that were monsomy or trisomy for chromosome 15, monosomy or trisomy for chromosome 22, and controls for key developmental genes located in cytoband regions with significantly altered methylation. The chromosome 15 genes AVEN (15q13.1), CYFIP1 (15q11), FAM189A1 (15q13.1) and MYO9A (15q22-q23) were all determined to have reduced expression in monosomy 15 blastocysts compared to control blastocysts (P < 0.05; Fig 1A). In contrast, trisomy 15 blastocysts had similar expression profiles to controls (ns). Chromosome 22 genes ADM2 (22q13.33), PACSIN2 (22q13.2-q13.33), PARVB (22q13.2-q13.33), and PIWIL3 (22q11.23) were all shown to have significantly lower expression levels in monosomy 22 blastocysts compared to controls (P < 0.05; Fig 1B) with trisomy 22 blastocysts displaying no significant differences. All samples were normalized to the housekeeping gene, PPIA, which had stable expression within all sample groups.
Fig 1. Developmental gene expression in individual human blastocysts (n = 10 replicates for each group) was performed by qPCR.
Ct values were normalized to PPIA, an internal, constant housekeeping gene. Fold change was determined using the ΔΔCt method on the average of technical duplicates. Error bars represent standard error and the y-axis denotes fold change between euploid and aneuploidy. A) Signigicant decreased expression of chromosome 15 genes in monosomy 15 blastocysts compared to controls (*P < 0.05). B) Significant decreased expression of chromosome 22 genes in monosomy 22 blastocysts compared to controls (*P < 0.05).
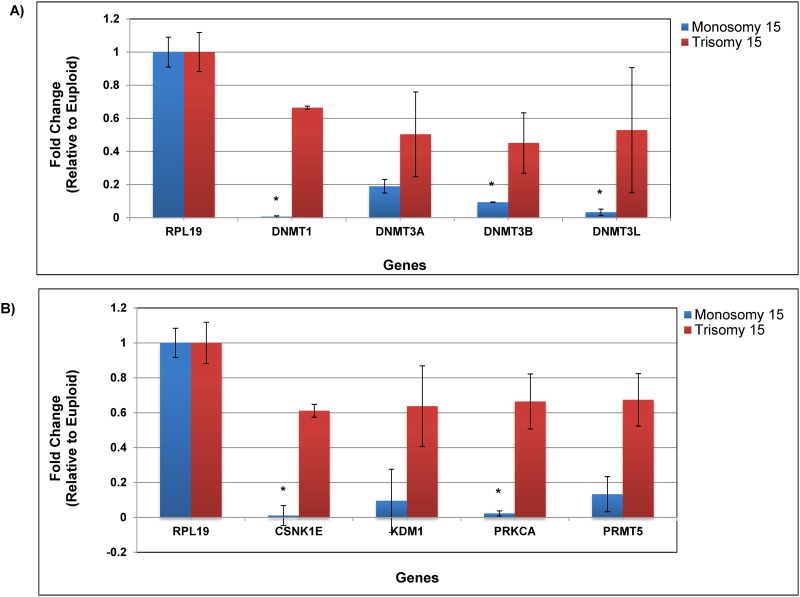
Gene expression analysis was also examined on additional, individual blastocysts for DNA methyltransferases and regulatory genes associated with establishing methylation. Monosomy 15 and trisomy 15 blastocysts were analyzed alongside controls for the following genes: DNMT1, DNMT3A, DNMT3B, DNMT3L, CSNK1E, KDM1, PRKCA, and PRMT5. It is important to note that none of these genes are located on chromosome 15 to avoid expression bias in these aneuploid samples. RPL19 was used as the internal housekeeping gene and had constant, stable expression in all sample groups. All 4 DNA methyltransferases showed decreased expression in monsomy 15 blastocysts compared to controls (Fig 2A) with DNMT1, DNMT3B, and DNMT3L being statistically significant (P < 0.05). DNMT1 showed the largest difference with a nearly 10-fold decrease in expression observed in the monosomy 15 blastocysts. Post-translational regulatory genes responsible for the regulation of DNMT1 gene expression (CSNK1E, KDM1, and PRKCA) revealed reduced expression in monosomy 15 blastocysts compared to controls (Fig 2B) with CSNK1E, PRKCA, and showing statistical significance (P < 0.05). The chromatin modifying protein, PRMT5, also displayed significantly decreased expression in monosomy 15 blastocysts (P < 0.05; Fig 2B). No expression differences were calculated to be statistically significant for any of the 8 genes examined between trisomy 15 and control blastocysts.
Fig 2. Epigenetic regulator expression in individual human blastocysts by qPCR (n = 12 replicates for each group).
Ct values were normalized to RPL19, an internal, constant housekeeping gene. Fold change was determined using the ΔΔCt method on the average of technical duplicates. Error bars represent standard error and the y-axis denotes fold change between euploid and aneuploidy. A) The expression of DNA methylatransferases was analyzed in euploid, trisomy 15, and monosomy 15 blastocysts (*P < 0.05). B) The expression of post-translational regulators and the chromatin modifying protein, PRMT5 (*P < 0.05).
Discussion
Chromosome segregation errors during maternal or paternal meiosis that lead to aneuploidy in the resulting embryo are well documented in human reproduction. While a handful of trisomy embryos (chromosomes 13, 18, 21, X and Y) can result in ongoing clinical pregnancies, monosomy embryos are rarely observed post-implantation, with Turner Syndrome being the only exception [19]. The bias against implantation of autosomal monosomies indicates that the lack of an autosomal chromosome is critical for development. This study investigated the relationship between chromosome aneuploidy epigenetic mechanisms and gene transcription as possible mechanisms to explain the low implantation potential of monosomy embryos.
Mammalian embryos undergo active and passive global demethylation, following fertilization, which reaches minimum levels at the morula/blastocyst stage. Therefore, unlike somatic cells with 50% methylation, the methylation status of human a blastocyst is significantly reduced [20]. Our results revealed a similar hypomethylated state of human blastocysts, independent of the blastocysts’ chromosome constitution (monosomy, trisomy, or diploid). Closer examination of the methylation profile of each individual chromosome revealed reduced methylation on the chromosome involved in the error for monosomy blastocysts. This could be reflective of the presence of only a single chromosome from the pair of chromosomes. In contrast, no methylation differences were observed between trisomy blastocysts and diploid controls, including for the extra chromosome involved in the aneuploidy. This observation could reflect dosage alterations of the trisomy blastocyst to normalize its transcriptome in order to offset the presence of the third chromosome [21]. In fact, evidence of this has been reported in studies of Down Syndrome that have shown tissue specific differences in the transcript levels of chromosome 21 genes [22, 23]. While there were genes that displayed an expected 50% increase in transcription, others exhibited no expression differences, and in some cases, even decreased expression was observed [22]. Transcriptional regulation in response to gene copy number for specific cell types could be the mechanism responsible for the observed gene dosage compensation [22]. Specifically, it has been suggested that stimulating mRNA degradation could be the active mechanism that allows for post-transcriptional buffering of aneuploidy in trisomy cells [24].
To determine if the methylation changes observed on the chromosome associated with the error were disrupting gene transcription, mRNA analysis was performed on key developmental genes. AVEN, CYFIP1, FAM189A1, and MYO9A are located within specific cytogenic regions of chromosome 15 that displayed the most substantial levels of hypomethylation in monosomy 15 blastocysts. Transcriptional analysis revealed significant reduction in expression compared to controls for these developmental genes. AVEN plays an important role in male and female germ cell development and has been shown to induce apoptosis in cells that have large amounts of DNA damage [25]. Reduced expression would diminish the apoptotic activity required to prevent abnormal cells from further development, thereby allowing these monosomy embryos to progress further than they should, forcing their demise prior to implantation. CYFIP1 is involved in mRNA translation and knockout mouse embryos have been shown to be significantly reduced in size, developmentally delayed, and do not survive past the blastocyst stage [26, 27]. This has important implications for monosomy embryos. Although they can grow to the blastocyst stage and appear to be of good quality, suitable expression levels of CYFIP1 are essential for further embryonic development and proper implantation. FAM189A1 is a CD20-like multi-pass transmembrane protein that is required for cell signaling [28]. These proteins are expressed on the surface of B-cells which are important for antibody response. With pregnancy being a pro-inflammatory state, proper expression of these proteins would be required for successful implantation to occur. MYO9A mutations are known to cause several diseases in humans [29]. This gene is a class IX myosin molecule that is important for epithelial formation and downregulation of MYO9A has been shown to affect cell morphology and differentiation [30]. Complete knockdown disrupts the formation and stabilization of cell-to-cell contacts during early development. Reduced expression of MYO9A could be greatly impacting the ability of monosomy blastocysts to have functional interactions with the uterus, thereby reducing the ability to implant and develop into a viable pregnancy.
ADM2, PACSIN2, PARVB, and PIWIL3 are located within highly significant hypo-methylated cytogenic regions of chromosome 22 and were all found to have significantly lower expression in monosomy 22 blastocysts compared to controls. ADM2 is an invasion promoting peptide that regulates placental mucin 1 (MUC1) and plays an important role in embryo implantation by promoting placental growth and inhibiting MUC1 expression in order to assist in trophoblast invasion [31]. PACSIN2 plays a role in endocytosis [32] and cell migration [33]. Decreases in PACSIN2 expression have been postulated to result in unregulated activation of α5β1 integrin which would reduce the ability of mesodermal cells to migrate [34]. This would have a very severe impact on the ability of a monosomy embryo to implant. PARVB is involved in cell adhesion and survival and also plays an important role in angiogenesis which promotes tumor growth in cancers [35–37]. The biology of tumor development and progression is similar to that of trophoblast invasion required for implantation. Reduced expression would prevent these cells from sufficiently being able to invade the maternal uterus. Likewise, PIWI genes are mainly expressed in germ cells and their proteins participate in germ cell differentiation with overexpression leading to malignancy [37]. PIWIL3, specifically, is required for early mammalian oogenesis and embryogenesis [38] and the under expression observed in monosomy blastocysts, again, could prevent trophoblast invasion leading to failed implantation.
Each of these developmental genes on chromosomes 15 and 22 displayed, roughly, a 0.5-fold expression decrease in monosomy blastocysts and could be contributing to their overall reduced competence and lack of implantation potential. Gene dosage is likely a contributing factor for this reduced expression, with the presence of only a single chromosome from the pair of chromosomes. In contrast, the transcription levels for each of the developmental genes in trisomy 15 and trisomy 22 blastocysts remained unchanged compared to controls. This indicates transcriptional compensation by trisomy embryos, away from the expected 1.5-fold increase, which could explain their future implantation potential.
Additional mRNA analysis was performed to determine if the processes involved in establishing methylation are impacted in monosomy blastocysts. DNA methyltransferases are the enzymes responsible for DNA methylation acquisition and maintenance during embryogenesis. DNMT1 is the maintenance methyltransferase that replicates methylation patterns on daughter DNA strands during mitosis [39]. DNMT3A, 3B, and 3L are de novo methyltransferases that set up DNA methylation patterns early in embryonic development, initiating at the blastocyst stage, and are also required for establishing maternal genomic imprints in gametes [14]. DNMT1, DNMT3B, and DNMT3L all displayed significantly reduced expression in monosomy blastocysts compared to either controls or trisomy blastocysts.
Reduced transcription was also observed in monosomy blastocysts for two post-translational regulatory genes, CSNK1E and PRKCA, which are required for DNMT1 activity. These two genes showed no differences when comparing expression profiles between trisomy blastocysts and controls. Furthermore, reduced gene expression was confirmed only in monosomy blastocysts for a chromatin modifying protein, PRMT5, which is recruited along with the DNMTs, to remodel histones through arginine methylation, resulting in the silencing of genes [40]. PRMT5 has been shown to be required throughout the resetting of the epigenome, during preimplantation development [41]. These combined mRNA expression data in monosomy blastocysts compared to trisomy or controls suggest that a decrease in the functionality of DNMT machinery may result during cell division and DNA replication due to the presence of only a single chromosome from the pair, thereby compromising further development.
In conclusion, this novel study revealed hypomethylation of the chromosome involved in the error for monosomy blastocysts, alongside decreased expression of developmental genes located on the chromosome of error and altered transcription of DNA methylation processes. Taken together, the altered methylation and disrupted downstream transcription could be directly impacting the developmental and implantation potential of monosomy blastocysts as it is well known that the autosomal monosomy state of a whole chromosome is not well tolerated during the window of implantation. In contrast, the trisomy blastocyst displays transcriptional dosage compensatory mechanisms for the presence of an additional chromosome, revealing similar methylation and gene expression to controls, and thereby giving an explanation for the difference in the implantation potential between trisomy and monosomy embryos. Future studies investigating epigenetic mechanisms associated with chromosome constitution may further expand our knowledge of human chromosomal aneuploidy and increase our understanding of its origins and impact during the window of implantation.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Human molecular genetics. 2007;16 Spec No. 2:R203–8. 10.1093/hmg/ddm243 . [DOI] [PubMed] [Google Scholar]
- 2.http://www.advancedfertility.com/age-eggs-chromosomes.htm.
- 3.Fragouli E, Alfarawati S, Goodall NN, Sanchez-Garcia JF, Colls P, Wells D. The cytogenetics of polar bodies: insights into female meiosis and the diagnosis of aneuploidy. Molecular human reproduction. 2011;17(5):286–95. 10.1093/molehr/gar024 . [DOI] [PubMed] [Google Scholar]
- 4.Sanchez JM, Franzi L, Collia F, De Diaz SL, Panal M, Dubner M. Cytogenetic study of spontaneous abortions by transabdominal villus sampling and direct analysis of villi. Prenatal diagnosis. 1999;19(7):601–3. . [PubMed] [Google Scholar]
- 5.Luthardt FW, Keitges E. Chromosomal Syndromes and Genetic Disease: Nature Publishing Group; 2001.
- 6.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harbor perspectives in biology. 2014;6(2). 10.1101/cshperspect.a018382 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–10. 10.1038/nature13544 . [DOI] [PubMed] [Google Scholar]
- 8.Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes & development. 2014;28(8):812–28. 10.1101/gad.234294.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301(5634):798–802. 10.1126/science.1086887 . [DOI] [PubMed] [Google Scholar]
- 10.Jacob S, Moley KH. Gametes and embryo epigenetic reprogramming affect developmental outcome: implication for assisted reproductive technologies. Pediatric research. 2005;58(3):437–46. 10.1203/01.PDR.0000179401.17161.D3 . [DOI] [PubMed] [Google Scholar]
- 11.Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, et al. DNA methylation profiles of human active and inactive X chromosomes. Genome research. 2011;21(10):1592–600. 10.1101/gr.112680.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin LJ, Zhang Y, Lv PP, He WH, Wu YT, Liu AX, et al. Insufficient maintenance DNA methylation is associated with abnormal embryonic development. BMC medicine. 2012;10:26 10.1186/1741-7015-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. . [DOI] [PubMed] [Google Scholar]
- 14.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–93. . [DOI] [PubMed] [Google Scholar]
- 15.Gardner D, Schoolcraft W. In-vitro culture of human blastocysts In: Jansen R, Mortimer D, editors. Towards Reproductive Certainty: Fertility and Genetics Beyond 1999. Parthenon Press: Carnforth; 1999. p. 378–88. [Google Scholar]
- 16.Treff NR, Scott RT Jr. Four-hour quantitative real-time polymerase chain reaction-based comprehensive chromosome screening and accumulating evidence of accuracy, safety, predictive value, and clinical efficacy. Fertility and sterility. 2013;99(4):1049–53. 10.1016/j.fertnstert.2012.11.007 . [DOI] [PubMed] [Google Scholar]
- 17.Parks JC, McCallie BR, Janesch AM, Schoolcraft WB, Katz-Jaffe MG. Blastocyst gene expression correlates with implantation potential. Fertility and sterility. 2011;95(4):1367–72. 10.1016/j.fertnstert.2010.08.009 . [DOI] [PubMed] [Google Scholar]
- 18.Ludwig G, Nejman D, Hecht M, Orlanski S, Abu-Remaileh M, Yanuka O, et al. Aberrant DNA methylation in ES cells. PloS one. 2014;9(5):e96090 10.1371/journal.pone.0096090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robberecht C, Vanneste E, Pexsters A, D'Hooghe T, Voet T, Vermeesch JR. Somatic genomic variations in early human prenatal development. Current genomics. 2010;11(6):397–401. 10.2174/138920210793175967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511(7511):611–5. 10.1038/nature13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179(2):737–46. 10.1534/genetics.108.090878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahlem P, Sultan M, Herwig R, Steinfath M, Balzereit D, Eppens B, et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome research. 2004;14(7):1258–67. 10.1101/gr.1951304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ait Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. American journal of human genetics. 2007;81(3):475–91. 10.1086/520000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veitia RA, Potier MC. Gene dosage imbalances: action, reaction, and models. Trends Biochem Sci. 2015;40(6):309–17. 10.1016/j.tibs.2015.03.011 . [DOI] [PubMed] [Google Scholar]
- 25.O'Shea LC, Hensey C, Fair T. Progesterone regulation of AVEN protects bovine oocytes from apoptosis during meiotic maturation. Biology of reproduction. 2013;89(6):146 10.1095/biolreprod.113.111880 . [DOI] [PubMed] [Google Scholar]
- 26.Pathania M, Davenport EC, Muir J, Sheehan DF, Lopez-Domenech G, Kittler JT. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Translational psychiatry. 2014;4:e374 10.1038/tp.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134(6):1042–54. 10.1016/j.cell.2008.07.031 . [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Bartenhagen C, Gombert M, Okpanyi V, Binder V, Rottgers S, et al. Next-generation-sequencing-based risk stratification and identification of new genes involved in structural and sequence variations in near haploid lymphoblastic leukemia. Genes, chromosomes & cancer. 2013;52(6):564–79. 10.1002/gcc.22054 . [DOI] [PubMed] [Google Scholar]
- 29.Gorman SW, Haider NB, Grieshammer U, Swiderski RE, Kim E, Welch JW, et al. The cloning and developmental expression of unconventional myosin IXA (MYO9A) a gene in the Bardet-Biedl syndrome (BBS4) region at chromosome 15q22-q23. Genomics. 1999;59(2):150–60. 10.1006/geno.1999.5867 . [DOI] [PubMed] [Google Scholar]
- 30.Abouhamed M, Grobe K, San IV, Thelen S, Honnert U, Balda MS, et al. Myosin IXa regulates epithelial differentiation and its deficiency results in hydrocephalus. Mol Biol Cell. 2009;20(24):5074–85. 10.1091/mbc.E09-04-0291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chauhan ML, Balakrishnan M, Chan R, Yallampalli C. Adrenomedullin 2 (ADM2) Regulates Mucin-1 at the Maternal-Fetal Interface in Human Pregnancy. Biology of reproduction. 2015. 10.1095/biolreprod.115.134296 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. Journal of cell science. 2000;113 Pt 24:4511–21. . [DOI] [PubMed] [Google Scholar]
- 33.Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, et al. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol. 2001;11(12):918–30. . [DOI] [PubMed] [Google Scholar]
- 34.Cousin H, Desimone DW, Alfandari D. PACSIN2 regulates cell adhesion during gastrulation in Xenopus laevis. Developmental biology. 2008;319(1):86–99. 10.1016/j.ydbio.2008.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Javerzat S, Franco M, Herbert J, Platonova N, Peille AL, Pantesco V, et al. Correlating global gene regulation to angiogenesis in the developing chick extra-embryonic vascular system. PloS one. 2009;4(11):e7856 10.1371/journal.pone.0007856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura M, Murakami T, Kizaka-Kondoh S, Itoh M, Yamamoto K, Hojo Y, et al. Functional molecular imaging of ILK-mediated Akt/PKB signaling cascades and the associated role of beta-parvin. Journal of cell science. 2010;123(Pt 5):747–55. 10.1242/jcs.052498 . [DOI] [PubMed] [Google Scholar]
- 37.Li L, Yu C, Gao H, Li Y. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer. 2010;10:38 10.1186/1471-2407-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenkranz D, Han CT, Roovers EF, Zischler H, Ketting RF. Piwi proteins and piRNAs in mammalian oocytes and early embryos: From sample to sequence. Genom Data. 2015;5:309–13. 10.1016/j.gdata.2015.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes & cancer. 2011;2(6):607–17. Epub 2011/09/24. 10.1177/1947601910393957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature reviews Genetics. 2002;3(9):662–73. 10.1038/nrg887 . [DOI] [PubMed] [Google Scholar]
- 41.Kim S, Gunesdogan U, Zylicz JJ, Hackett JA, Cougot D, Bao S, et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Molecular cell. 2014;56(4):564–79. 10.1016/j.molcel.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.