Abstract
Many bacterial species coexist in the same niche as heterogeneous clones with different phenotypes; however, understanding of infectious diseases by polyphenotypic bacteria is still limited. In the present study, encapsulation in isolates of the porcine pathogen Streptococcus suis from persistent endocarditis lesions was examined. Coexistence of both encapsulated and unencapsulated S. suis isolates was found in 26 out of 59 endocarditis samples. The isolates were serotype 2, and belonged to two different sequence types (STs), ST1 and ST28. The genomes of each of the 26 pairs of encapsulated and unencapsulated isolates from the 26 samples were sequenced. The data showed that each pair of isolates had one or more unique nonsynonymous mutations in the cps gene, and the encapsulated and unencapsulated isolates from the same samples were closest to each other. Pairwise comparisons of the sequences of cps genes in 7 pairs of encapsulated and unencapsulated isolates identified insertion/deletions (indels) ranging from one to 104 bp in different cps genes of unencapsulated isolates. Capsule expression was restored in a subset of unencapsulated isolates by complementation in trans with cps expression vectors. Examination of gene content common to isolates indicated that mutation frequency was higher in ST28 pairs than in ST1 pairs. Genes within mobile genetic elements were mutation hot spots among ST28 isolates. Taken all together, our results demonstrate the coexistence of dual phenotype (encapsulated and unencapsulated) bacterial clones and suggest that the dual phenotypes arose independently in each farm by means of spontaneous mutations in cps genes.
Introduction
Bacteria can adapt to environmental changes through various adaptive strategies. One example is coexistence of different phenotypes of the same organism at specific niches [1]. Polyphenotypic infections by a single species have been reported in many bacterial species, including Pseudomonas aeruginosa, Helicobacter pylori, Escherichia coli, and Staphylococcus epidermidis [1–4]. The ecologies of these infections have shown that the composition of polyphenotypic clones permit adaptation to various environmental conditions. Coexistence by polyphenotypic clones is thus thought to be a barrier to treatment and prophylaxis [2, 4–7]. On the other hand, the compositions of coexistable bacteria are affected by antimicrobials and host immunity [8, 9]. Therefore, bacterial population dynamics in response to environmental changes play an important role in evasion from host immune system [10–12].
Variation of surface structures in bacteria is important for adaptation and survival in variable environments. Although capsules are major bacterial extracellular components and show antiphagocytic effect, the loss of capsules can also be beneficial [13–16]. Hanage et al. reported that one or more capsule-related genes were not detected by PCR in isolates of Streptococcus pneumoniae from nasopharyngeal swabs and middle ear fluid [10]. This finding suggested that unencapsulation afforded advantages to capsule-less clones in some habitats. In Streptococcus pyogenes, unencapsulated clones isolated from pharyngitis and invasive diseases are known to be invasive and cause diseases [16, 17]. Similarly, unencapsulated clones of the zoonotic pathogen Streptococcus suis [18–20] were isolated from porcine endocarditis lesions [21]. The role of these phenotypic changes has, however, not yet been established. In addition, although S. suis isolates from different worldwide locations are known to be genetically highly diverse, there is no information about the genetic diversity of isolates recovered from a single lesion [22].
The S. suis capsule is composed of capsular polysaccharides (CPs), whose biosynthesis is dependent on the concerted action of enzymes encoded by genes located in the so-called capsular polysaccharide synthesis (cps) locus [23–27]. Different antigenicities of the S. suis CPs are the basis of S. suis serotyping [27, 28]. S. suis CPs have been shown to play a major role in protection against host phagocytes [29–31], and unencapsulated S. suis clones have been described as low virulent or completely avirulent, leading to the notion that loss of capsule is unfavorable for S. suis virulence. However, the ability to form biofilms and to adhere to epithelial cells and platelets was previously reported to be stronger in unencapsulated clones than in encapsulated ones [32–34], as unencapsulated clones exclusively invaded into epithelial cells [32, 34, 35]. Therefore, from an ecological perspective, it is plausible that unencapsulated and encapsulated S. suis clones coexist in the same lesions, and that the presence of both encapsulated and unencapsulated cells originated from genetically identical clones facilitate adherence and invasion to host cells, thereby permitting efficient evasion from the immune system and persistence of the clonal population in a particular habitat or niche.
In the present study, to determine whether encapsulated and unencapsulated, or dual phenotypic S. suis clones coexist in porcine endocarditis lesions, we collected multiple S. suis isolates from each endocarditis lesion and confirm encapsulation status of the isolates. Finally, comparative genome analyses allowed us to precisely define the close relationships between encapsulated and unencapsulated isolates from the same endocarditis lesion. Our findings suggest that mutations from encapsulated cells to unencapsulated ones occurred independently in each pig or farm.
Materials and Methods
Bacteria, plasmids and growth condition
Fifty-nine heart valve vegetations of porcine endocarditis from 24 farms (Fig 1A) were collected in meat inspection centers in Japan between 2013 and 2014. These samples were stamped onto Todd-Hewitt (TH, Becton Dickinson, MD, U.S.) agar plates containing 5% horse blood, followed by incubation at 37°C for 16 hours under a 5% CO2 atmosphere. Twenty-four colonies were picked from each sample and purified twice by single colony isolation. Amplification of the S. suis recN gene, which serves to confirm species [36] was performed by PCR, and the positive isolates were further examined by PCR permitting serotype estimation [37, 38]. All isolates were stored in Luria-Bertani (LB, Becton Dickinson) broth containing 30% glycerol at -80°C. For S. suis isolates, encapsulation was determined using a co-agglutination test [21]. One pair of encapsulated and unencapsulated S. suis isolates was selected from each of 26 endocarditis samples that yielded both encapsulation and unencapsulated phenotypes (a total of 52 isolates, Table 1). Escherichia coli strains and plasmids used in this study are listed in S1 Table [39, 40]. E. coli strains were grown on LB broth at 37°C for 16 hours. When necessary, media were supplemented with spectinomycin (50 μg/ml or 100 μg/ml for E. coli and S. suis, respectively).
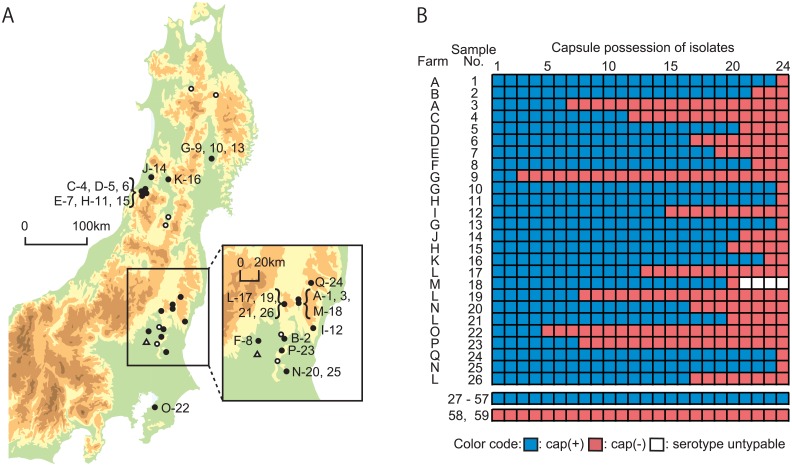
Fig 1. Geographic location of porcine farms examined and results of encapsulated and unencapsulated S. suis isolates.
(A) The map shows the northern east area of the main island of Japan from which S. suis was isolated in the present study. The farms are indicated by the following symbols: blank circles and blank triangles: encapsulated and unencapsulated S. suis, respectively, were exclusively isolated, and filled circles: encapsulated and unencapsulated S. suis were both isolated. The farm identifiers (alphabetical characters) and sample Nos. of endocarditis samples are indicated along the above symbols. The map was publicly available from the Geospatial Information Authority in Japan. (B) The heat map shows the encapsulation traits of 24 S. suis isolates in each of 59 endocarditis samples, with the following colors: blue for encapsulated, and red for unencapsulated isolates. Untypeable isolates in serotype-specific PCR are indicated by blanks. Encapsulated and unencapsulated isolates were exclusively from 31 (Nos. 27–57) and 2 (Nos. 58 and 59) endocarditis samples, respectively. Sample Nos. 1 to 26 were hereafter used as the No. of the pair of encapsulated and unencapsulated isolates.
Table 1. S. suis isolates from Japan used in this study.
| Pair No. | Isolate No. | Farm | Location | Serotype | MLST | Accession No. of DRA* | ||
|---|---|---|---|---|---|---|---|---|
| Encapsulated | Unencapsulated | Encapsulated | Unencapsulated | |||||
| 1 | SUT709 | SUT708 | A | Nasu-gun, Tochigi | 2 | ST28 | DRX045114 | DRX045113 |
| 2 | SUT780 | SUT785 | B | Utsunomiya-shi, Tochigi | 2 | ST28 | DRX045115 | DRX045116 |
| 3 | SUT806 | SUT804 | A | Nasu-gun, Tochigi | 2 | ST28 | DRX045118 | DRX045117 |
| 4 | SUT877 | SUT876 | C | Tsuruoka-shi, Yamagata | 2 | ST1 | DRX045120 | DRX045119 |
| 5 | SUT906 | SUT907 | D | Tsuruoka-shi, Yamagata | 2 | ST1 | DRX045121 | DRX045122 |
| 6 | SUT926 | SUT927 | D | Tsuruoka-shi, Yamagata | 2 | ST1 | DRX045123 | DRX045124 |
| 7 | SUT1006 | SUT1007 | E | Tsuruoka-shi, Yamagata | 2 | ST28 | DRX045125 | DRX045126 |
| 8 | SUT1020 | SUT1024 | F | Shimotsuga-gun, Tochigi | 2 | ST1 | DRX045127 | DRX045128 |
| 9 | SUT1046 | SUT1044 | G | Tanzawa-gun, Iwate | 2 | ST28 | DRX045130 | DRX045129 |
| 10 | SUT1189 | SUT1190 | G | Tanzawa-gun, Iwate | 2 | ST28 | DRX045131 | DRX045132 |
| 11 | SUT1330 | SUT1329 | H | Tsuruoka-shi, Yamagata | 2 | ST28 | DRX045134 | DRX045133 |
| 12 | SUT1337 | SUT1334 | I | Hitachiomiya-shi, Ibaraki | 2 | ST28 | DRX045136 | DRX045135 |
| 13 | SUT1468 | SUT1467 | G | Tanzawa-gun, Iwate | 2 | ST28 | DRX045138 | DRX045137 |
| 14 | SUT1582 | SUT1583 | J | Shonai-shi, Yamagata | 2 | ST28 | DRX045139 | DRX045140 |
| 15 | SUT1596 | SUT1607 | H | Tsuruoka-shi, Yamagata | 2 | ST28 | DRX045141 | DRX045142 |
| 16 | SUT1678 | SUT1675 | K | Mogami-gun, Yamagata | 2 | ST28 | DRX045144 | DRX045143 |
| 17 | SUT1789 | SUT1797 | L | Nasukarasuyama-shi, Tochigi | 2 | ST28 | DRX045145 | DRX045146 |
| 18 | SUT1865 | SUT1882 | M | Nasu-machi, Nasu-gun, Tochigi | 2 | ST28 | DRX045147 | DRX045148 |
| 19 | SUT1914 | SUT1908 | L | Nasukarasuyama-shi, Tochigi | 2 | ST1 | DRX045150 | DRX045149 |
| 20 | SUT2052 | SUT2055 | N | Ishioka-shi, Ibaraki | 2 | ST28 | DRX045151 | DRX045152 |
| 21 | SUT2083 | SUT2080 | L | Nasukarasuyama-shi, Tochigi | 2 | ST28 | DRX045154 | DRX045153 |
| 22 | SUT2148 | SUT2154 | O | Ichihara-shi, Chiba | 2 | ST28 | DRX045155 | DRX045156 |
| 23 | SUT2188 | SUT2189 | P | Moka-shi, Tochigi | 2 | ST28 | DRX045157 | DRX045158 |
| 24 | SUT2199 | SUT2210 | Q | Higashishirakawa-gun, Fukushima | 2 | ST1 | DRX045159 | DRX045160 |
| 25 | SUT2222 | SUT2241 | N | Ishioka-shi, Ibaraki | 2 | ST28 | DRX045161 | DRX045162 |
| 26 | SUT2256 | SUT2250 | L | Nasukarasuyama-shi, Tochigi | 2 | ST28 | DRX045164 | DRX045163 |
* DRA, the DNA Data Bank of Japan (DDBJ) Sequence Read Archive
Genomic DNA extraction
Genomic DNA was extracted from the 52 S. suis isolates as described previously [41]. DNA concentrations were determined using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies Corporation, CA, U.S.), and quality was checked using a NanoDrop 1000 instrument (Thermo Fisher Scientific, DE, U.S.).
Multilocus sequence typing (MLST)
The 52 S. suis isolates were examined by MLST, as described previously [42]. The sequence type (ST) was determined by comparing the nucleotide sequences of 7 loci with the data deposited in the S. suis MLST database (http://ssuis.mlst.net).
Whole-genome sequencing
Genomic libraries were prepared using Nextera XT DNA kits (Illumina Inc., CA, U.S.). Paired-end sequencing was performed using MiSeq Reagent Kit v3 (600-cycles) in the Illumina MiSeq platform. Quality trimming and filtering of the obtained sequence reads were performed using CLC Genomics Workbench v8.0.1 (CLC bio, Aarhus, Denmark) with the following parameters: Quality Limit = 0.01, Adapters Trimming = Yes, Remove 5′ Terminal Nucleotides = Yes, Number of 5′ Terminal Nucleotides to Remove = 20, Remove 3′ Terminal Nucleotides = Yes, Number of 3′ Terminal Nucleotides to Remove = 5, and Discard Reads below Length = 50.
Construction of phylogenetic trees
The preprocessed reads were de novo assembled using A5-miseq, as of May 22th, 2015, with default parameters [43]. A maximum parsimony phylogenetic tree for the 52 S. suis isolates was constructed using kSNP3 v3.0 with a k-mer length of 19 [44]. kSNP3 was also used to construct maximum parsimony phylogenetic trees for 12 ST1 isolates with k-mer 19 and for 40 ST28 isolates with k-mer 21. The Kchooser program in kSNP3 was used to estimate these optimum k-mer values. Trees were visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree).
Sequence determination of cps genes by Sanger sequencing
The nucleotide sequences of the cps genes of 7 pairs of encapsulated and unencapsulated isolates (pair no. 1, 2, 3, 9, 10, 11 and 15) were determined as described previously [21]. Sequencher v4.8 and Molecular Evolutionary Genetics Analysis (MEGA) v5 [45] were used for sequence alignment and detection of mutations, respectively.
Complementation analysis
In order to construct cps2E and cps2H expression vectors, we individually amplified the cps2E and cps2H genes by PCR using genomic DNA of encapsulated S. suis isolates as template, primers listed in S2 Table, and PrimeSTAR Max DNA Polymerase (TaKaRa Bio, Shiga, Japan). Expression plasmid pMX1 [40] was digested with EcoRΙ (TaKaRa Bio), and then fused with either cps2E or cps2H fragments using the In-Fusion HD Cloning Kit (TaKaRa Bio). Insert orientation and nucleotide sequences of cloned genes were verified by PCR and Sanger sequencing. The resultant expression vectors, pCps2E and pCps2H, were introduced into unencapsulated S. suis isolates by electroporation. Restoration of capsule expression was examined by the co-agglutination test as described previously [21, 46].
Detection of nucleotide mutations
Contigs assembled by A5-miseq for the 52 isolates were annotated using Prokka v1.11, run with default parameters [47]. The pan-genomes analysis pipeline (PGAP) v1.02 [48], run with default parameters, was used for the identification of single-copy common genes that were present in the genomes of all isolates. Single-copy common genes were identified independently for ST1 and ST28. For identification of mobile genetic elements (MGEs), the positions of MGEs in the genome of reference strain NSUI002 [49] were used. PHAST [50] and IslandViewer 3 [51], run with default parameters, were also used to assess MGE contents. The preprocessed (as described above) short-read genome data of 12 ST1 and 40 ST28 isolates were mapped against the P1/7 (ST1) and NSUI002 (ST28) complete genome sequences, respectively, using CLC Genomics Workbench. Mapping was performed with the following parameters: Mismatch Cost = 2, Affine Gap Cost = Yes, Insertion Open Cost = 6, Insertion Extend Cost = 1, Deletion Open Cost = 6, Deletion Extend Cost = 1, Length Fraction = 0.5, Similarity Fraction = 0.8, Auto-Detect Paired Distance = Yes, and Non-Specific Match Handling = Map Randomly. Mutation calling was performed using the Basic Variant Detection tool in the CLC Genomics Workbench with the following parameters: Ignore Broken Pairs = Yes, Minimum Coverage = 10, Minimum Count = 10, Minimum Frequency (%) = 80, Base Quality Filter = Yes, Neighborhood Radius = 5, Minimum Central Quality = 30, Minimum Neighborhood Quality = 25, Relative Read Direction Filter = Yes, and Significance = 1.0% [52, 53]. In each pair of encapsulated and unencapsulated isolates, mutations with nonsynonymous amino acid substitutions in the cps genes and other single-copy common genes were identified and were visualized by bar graphs with genome structure of the ST28 reference strain using R v3.2.2. The cps gene cluster was included in this comparison even though cps genes had been excluded from pool of common genes used in precious analysis due to presence of the indels spanning over vast nucleotide stretches. The mutation frequency for each single-copy common gene in each ST was calculated as the rate of the number of nucleotides with a mutation divided by the total number.
Data access
The genome sequence data obtained in this study were deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive under accession number DRA004206.
Results
Coexistence of encapsulated and unencapsulated S. suis in 26 endocarditis samples
A total of 1,416 isolates from 59 endocarditis samples were identified to be S. suis. In 58 of the endocarditis samples, PCR genotyping showed that 1,388 isolates were serotype 2, and 4 isolates were untypeable, whereas the 24 isolates from the remaining endocarditis sample were serotype 16. On the basis of the co-agglutination test, 31 (53%) and 2 (3%) of the 59 samples, contained encapsulated only, or unencapsulated only isolates, respectively. On the other hand, both encapsulated and unencapsulated isolates were found in 26 samples (44%). Encapsulated and unencapsulated phenotypes were observed in 17 out of the 24 farms examined (70.8%) (Fig 1A). The ratio of encapsulated to unencapsulated isolates varied (Fig 1B).
Phylogenetic relationships of encapsulated and unencapsulated S. suis isolates
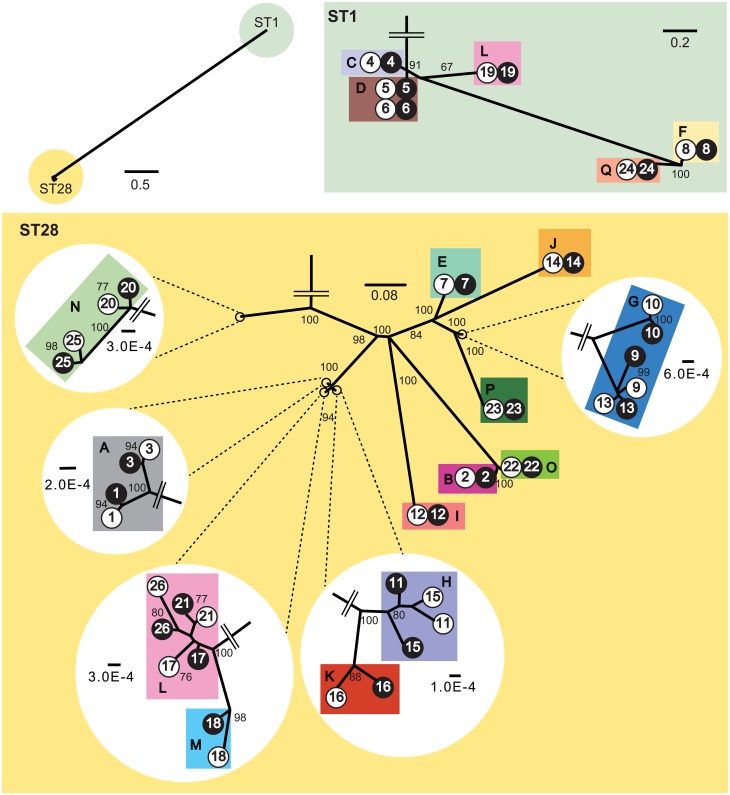
We selected a pair of encapsulated and unencapsulated S. suis isolates from each of the 26 endocarditis samples in order to examine their phylogenetic relationships. The STs of encapsulated and unencapsulated isolates were identical in each of 20 pairs of ST28, and 6 pairs of ST1. Whole-genome short-read data was assembled using A5-miseq (S3 Table presents statistics of the assemblies). Maximum parsimony trees were constructed using genome-wide single-nucleotide polymorphisms (SNPs) compared with the reference stains. In the tree of all 52 isolates, ST1 and ST28 isolates were separately clustered (Fig 2). In each pair of ST1 isolates, the encapsulated and unencapsulated isolates belonged to the same lineage and were phylogenetically distinct from other pairs, except for pairs 5 and 6, which were isolated from the same farm D (Fig 2). In each pair of ST28 isolates, the two were phylogenetically closely related to each other, except for pairs 11 and 15 (Fig 2).
Fig 2. A phylogenetic tree for 26 encapsulated and 26 unencapsulated S. suis isolates.
The tree was constructed using the maximum parsimony method for genome-wide SNPs in 52 S. suis isolates. The 52 isolates analyzed were composed of 26 encapsulated and unencapsulated isolate pairs from 17 porcine farms, indicated by the following symbols: blank circles for encapsulated, and filled circles for unencapsulated isolates, with pair Nos. and farm identifiers (colored rectangles) along with the symbols. The isolates were separately clustered into two STs, ST1 and ST28; a detailed tree structure in each ST is shown as an enlarged tree. Several ends of the enlarged tree in ST28 are further enlarged. Seventeen colors are used to show distribution of farms in the tree. Only the bootstrap values >50% are indicated on the branches.
Mutations in a cps gene cluster and complementation with cps expression vectors
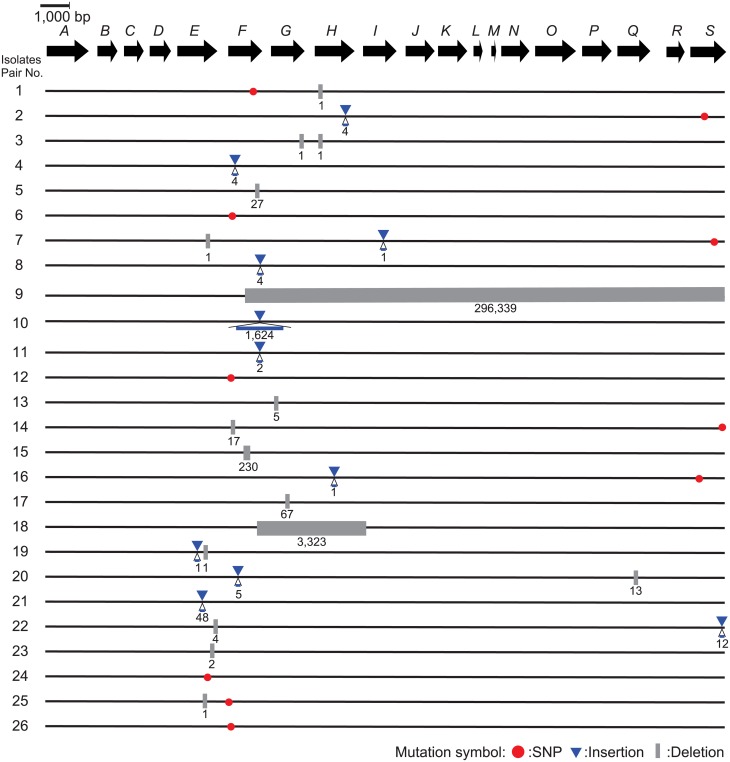
The nucleotide sequences of the cps gene cluster in pairs no. 1, 2, 3, 9, 10, 11, and 15 were determined by Sanger sequencing. The length of indels resulting in frameshift mutations ranged from 1 to 4 nucleotides in 4 pairs (pairs no. 1, 2, 3, and 11), whereas indels with sizes spanning from 102- to 104-bp were found in the remaining 3 pairs (pairs no. 9, 10, and 15) (Fig 3). We next complemented in trans the unencapsulated isolates of pairs no. 1, 2, 3 and 11 with expression vectors of cps genes. Construct pCps2H was used for those of the pairs no. 1, 2, and 3, which had indels in the cps2H gene, while construct pCps2E was used to complement pair no. 11, which had mutations in the cps2E gene. All transformants showed a positive reaction in the co-agglutination test using anti-serotype 2 serum, indicating restoration of capsule expression.
Fig 3. Nonsynonymous mutations and indels in the cps gene cluster between encapsulated and unencapsulated S. suis isolates in 26 pairs.
The genetic organization of the cps gene cluster (from cps2A to cps2S) is shown as a sequence of the arrows located by transcriptional directions with the identifiers such as “A” for “cps2A”. The mutations between encapsulated and unencapsulated isolates in each pair are shown along with the genetic map, with the following symbols: red circles for nonsynonymous mutations, blue triangles for insertions, and gray rectangles for deletions along with the nucleotide length of the mutations.
Occurrence of mutations in single-copy common genes, in the cps gene cluster, and in MGEs
Mapping results of 52 isolates using CLC Genomics Workbench are listed in S3 Table. The nucleotide sequences of single-copy common genes and the cps gene cluster were compared within each pair. The analysis was verified by the consistency of the mutation profiles in the Sanger and MiSeq-based methods. No mutation was found in the ccpA gene that regulated the production of CPs, and nonsynonymous mutations found between the pairs were depicted in Fig 3 [54].
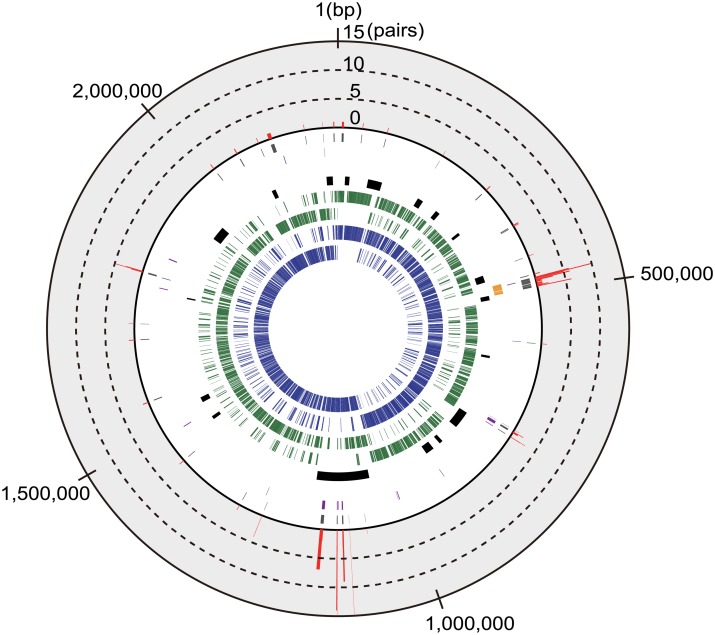
There were differences between ST1 and ST28 pairs in the number of genome-wide mutations identified. Only a few nonsynonymous mutations were identified in the ST1 pairs, whereas the ST28 pairs had a large number of unique nonsynonymous mutations (Fig 4, S4 and S5 Tables). Indeed, the mean mutation frequencies in all single-copy common genes between ST1 and ST28 were calculated to be 2.70 × 10−7 and 3.25 × 10−5, respectively. At the nucleotide level, most of the mutations were unique for each pair. All the pairs showed SNPs within the cps gene cluster. In addition, SNPs within a particular MGE (position 1,054,531–1,178,032 bp of the reference genome NSUI002) were found in many pairs. This MGE contains genes related to conjugative transposons (Fig 4, S6 Table).
Fig 4. The genome structure of the ST28 S. suis strain NSUI002.
The following information on the NSUI002 genome is shown from the innermost circle: protein-coding genes transcribed clockwise (1st track, blue) and counterclockwise (2nd, blue), common genes that were shared among NSUI002 and all ST28 S. suis isolates, transcribed clockwise (3rd, green) and counterclockwise (4th, green), MGEs (5th, black), and genes in the cps gene cluster (6th, yellow). The common genes in which synonymous and nonsynonymous mutations were detected are shown in the 7th (purple) and 8th (gray) tracks, respectively. The outermost bar graph for the common genes in the 9th track (red) shows the number of pairs in which nonsynonymous mutations were detected. No common gene exhibited a number of pairs more than 15, the maximum value of the graph.
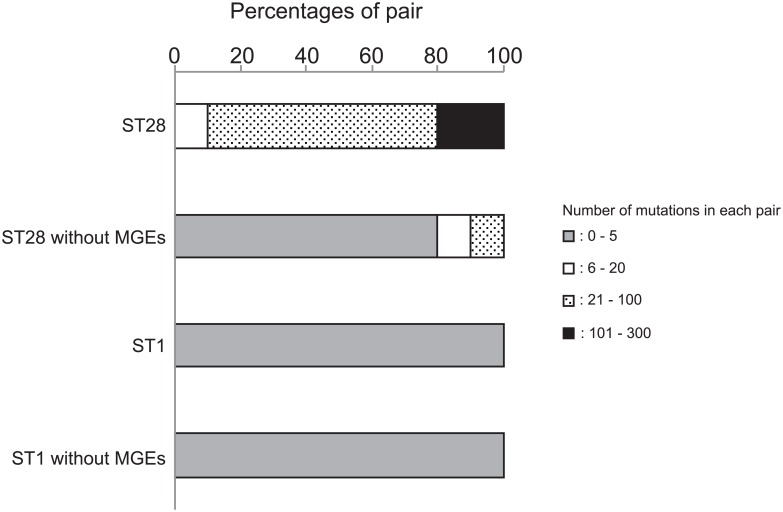
The maximum numbers of nonsynonymous mutations per gene were 3 and 15 in the ST1 and ST28 pairs, respectively. Among ST28 pairs, most genes with mutations were part of MGEs (Fig 4). Therefore, we excluded the single-copy common genes in MGEs and examined the profiles of mutations. The number of mutations in each pair ranged between 0 to 5, except for the genes within MGEs, which ranged between 21 to 302 (Fig 5). Thus, the mutation frequencies were amended to be 2.95 × 10−7 in ST1, and 3.25 × 10−6 in ST28.
Fig 5. The proportion of each pair classified by number of mutations.
The number of mutations was calculated in each of 26 pairs, and its distribution is shown in ST1 and ST28 isolates as a percentage of pairs, as follows: gray bars for pairs with 0–5 mutations, blank bars for 6–20, dotted bars for 21–100, and filled bars for 101–300. Percentages are shown in ST1 and ST28 clones when genes on MGEs are excluded from the calculation.
Discussion
In the present study, we demonstrated the coexistence of encapsulated and unencapsulated S. suis in the same endocarditis lesion. The capsule protects S. suis cells from the host immune response [24, 30, 55, 56]. S. suis is known, however, to escape the vaccine-immunity by means of capsule loss [57–59], such as reported previously for the capsule loss of Haemophilus influenzae [11, 12]. Unencapsulation can also afford S. suis cells some advantages, such as high adhesion and invasion of host cells [32–35]. This led us to hypothesize that encapsulated and unencapsulated S. suis “cooperate” by expressing both advantageous characteristics at lesions, and that each of these phenotypically distinct, but genetically very similar clonal subpopulations provides an advantage that results in persistence of the global population of the clone. Taken all together, our results seem to support the abovementioned hypothesis. However, we cannot disregard the alternative hypothesis that the unencapsulated cells in the endocarditis lesions may not have the capacity to participate in any significant way in inducing heart lesions, but they simply arise by their faster growth in the particular niche.
We obtained only unencapsulated isolates from 2 samples. Moreover, the ratio of both phenotypes varied among 26 samples. It is conceivable that the encapsulated and unencapsulated cells did not form a completely mixed jumble within the lesions but instead formed different layers of either encapsulated or unencapsulated cells. Assuming that the layer of unencapsulated cells was cut out and stamped on an agar medium, we can isolate only unencapsulated cells from the sample. Indeed, we always succeeded in isolating both phenotypes from the same lesion by cutting the samples in many pieces for stamp (our unpublished observation). We collected endocarditis samples from many geographically separated farms, and many of these unrelated samples harbored both encapsulated and unencapsulated phenotypes of the same clone of S. suis (Fig 1A). The high prevalence may suggest that occurrence of unencapsulated isolates does not require specific conditions and occur frequently in every farm.
We only detected ST1 and ST28 S. suis isolates in our sample (Fig 1B). Both ST1 and ST28 are prevalent in Japan [60]. ST1 is also prevalent worldwide and the isolates belonging to ST1 generally express a hemolysin known as suilysin [61]. ST28 has become prevalent in several countries, including the United States of America, Canada, and China [49, 62] and exhibits a broad range of virulence in porcine groups [49]. The very close genetic relationship of the isolates within each ST (Fig 2) suggests transmission of each lineage across distant farms by porcine transportations, similar to the transmission of Salmonella spp. from slaughterhouses to farms [63]. However, we were unable to find any relationships between the geographical location of the farms and STs of the isolates (Fig 1).
Some bacterial species have been known to cause polyphenotypic infections. Previous studies reported the coexistence of multiple P. aeruginosa clones that exhibit different antimicrobial resistance and colony formation. Because such phenotypic differences were observed after the establishment of infection and lesion formation, it was suggested that they differentiate in situ at lung lesions [6, 64, 65]. Since encapsulated and unencapsulated S. suis isolates in most pairs were phylogenetically closest to each other (Fig 2), the encapsulated and unencapsulated lineages may have differentiated in the farm, i.e., environment or host body. Bacteria can change their phenotype to adapt to the surrounding environment, such as genetic change in P. aeruginosa, or transcriptional regulation in Streptococcus equi [66]. In the present analyses of the cps gene cluster, all pairs of encapsulated and unencapsulated isolates had mutations that included single-nucleotide polymorphisms and indels (Fig 3). As reported in a previous study, in which capsule production by unencapsulated S. suis isolates was complemented with the intact cps genes of reference strains P1/7 and 89–1591 [21, 46], we also demonstrated here the complementation of capsule production in unencapsulated isolates with cps expression vectors. This result suggests that capsule expression by S. suis in endocarditis lesions was affected by genetic changes in the cps gene cluster.
The expression of capsule is generally affected by regulatory genes, such as ccpA. In the present study, we did not find any modification in the ccpA gene. Although there may be the other regulatory genes that affect expression of capsule, spontaneous mutations in the cps genes cluster are likely to be the main mechanism leading to unencapsulation. Indeed, all cps mutants examined to date had nucleotide changes in structural genes of enzymes for capsule production [21, 46]. Among the mechanisms that generate nucleotide changes, spontaneous mutations are a major cause that are frequently observed in various events such as errors in DNA replication [67]. In the mutation profiles of all single-copy common genes, the spontaneous mutations unique for a single pair were predominant throughout the genomes (Fig 4); however, there were mutations in the cps gene cluster that occurred in multiple pairs. This may have been due to the limitation of the examined isolates and a bias for the selection of the phenotypes. Mutation frequencies differed between the ST1 and ST28 pairs; however, the difference may be explained by the large number of mutations concentrated within a MGE in ST28 pairs (approximately 90% of mutations were within a MGE (Fig 4, S6 Table)). After the exclusion of the single-copy common genes within MGEs, the mutation frequency was almost the same to those in other Streptococcus spp. such as S. equi (5.22 × 10−7) [68], S. pneuomoniae (1.57 × 10−6) [69] and, S. pyogenes (1.1 × 10−6) [70]. Collectively, these findings suggest that mutations may spontaneously arise in the cps gene cluster; therefore, these mutations may alter the phenotypic change through capsule expression. On the other hand, the MGE found in ST28 contained two genomic regions that exhibited high nucleotide sequence similarity with attL and attR, parts of a conjugative element, as well as genes related to its transfer [71]. Although precise cause why the MGE carried more mutations than other genome regions remains unknown, we assume the region may be related to the cause of endocarditis, which will be revealed by genome-wide association study in the future [72].
In summary, we herein demonstrate that different phenotypes of genetically very closely related S. suis isolates were from the same endocarditis lesion. S. suis may adapt to its surrounding environment through the loss of capsule expression, and endocarditis lesion may be developed by dual phenotypes of a single clones. Although unencapsulated S. suis isolates are, in general, easily phagocytized by immune cells, it is possible that they persist and proliferate in the host by the assistance of encapsulated cells. In addition, a comparison of ST1 and ST28 suggested that the isolates of different STs employed various mechanisms by which they adapt to the surrounding environment. Future studies about coexistence of different phenotypes, which may enable them to persist in endocarditis lesions, and experimental reproduction of endocarditis with both phenotypes, will provide insights into the pathogenicity, ecology, diversity and evolution of S. suis.
Supporting Information
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We are grateful to Daisuke Takamatsu, Makoto Osaki and Masatoshi Okura, National Institute of Animal Health, National Agriculture and Food Research Organization for helpful discussion and advice. We appreciate Hiroyuki Nakayama, Laboratory of Veterinary Pathology, The University of Tokyo for valuable comments. We thank meat inspection centers of Utsunomiya-City, Yamagata-Shonai, and Ibaraki-Kensei in Japan for help with collecting porcine endocarditis samples.
Data Availability
All relevant data are within the paper and its Supporting Information files. All genome sequence data files are available from the DNA Data Bank of Japan Sequence Read Archive under accession number DRA004206.
Funding Statement
This work was supported by JSPS KAKENHI Grant Number 15K15675, 25713060, 24117508 (to FM), 25305013 (to IN), 15J10486 (to SA), 26861544, 16K08015 (to TW), 26660226, 15H02651 and 16K15037 (to TS).
References
- 1.Levert M, Zamfir O, Clermont O, Bouvet O, Lespinats S, Hipeaux MC, et al. Molecular and evolutionary bases of within-patient genotypic and phenotypic diversity in Escherichia coli extraintestinal infections. PLoS Pathog. 2010;6(9):e1001125 Epub 2010/10/14. 10.1371/journal.ppat.1001125 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Eldere J, Peetermans WE, Struelens M, Deplano A, Bobbaers H. Polyclonal Staphylococcal endocarditis caused by genetic variability. Clin Infect Dis. 2000;31(1):24–30. Epub 2000/07/29. 10.1086/313915 . [DOI] [PubMed] [Google Scholar]
- 3.Romling U, Fiedler B, Bosshammer J, Grothues D, Greipel J, von der Hardt H, et al. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis. 1994;170(6):1616–1621. Epub 1994/12/01. . [DOI] [PubMed] [Google Scholar]
- 4.Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S, et al. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci U S A. 2001;98(25):14625–14630. Epub 2001/11/29. 10.1073/pnas.251551698 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser MJ, Berg DE. Helicobacter pylori genetic diversity and risk of human disease. J Clin Invest. 2001;107(7):767–773. Epub 2001/04/04. 10.1172/jci12672 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashish A, Paterson S, Mowat E, Fothergill JL, Walshaw MJ, Winstanley C. Extensive diversification is a common feature of Pseudomonas aeruginosa populations during respiratory infections in cystic fibrosis. J Cyst Fibros. 2013;12(6):790–793. Epub 2013/05/07. 10.1016/j.jcf.2013.04.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Romero MA, Casadesus J. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc Natl Acad Sci U S A. 2014;111(1):355–360. Epub 2013/12/20. 10.1073/pnas.1316084111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Temime L, Boelle PY, Opatowski L, Guillemot D. Impact of capsular switch on invasive pneumococcal disease incidence in a vaccinated population. PLoS One. 2008;3(9):e3244 Epub 2008/09/20. 10.1371/journal.pone.0003244 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki S, Horinouchi T, Furusawa C. Prediction of antibiotic resistance by gene expression profiles. Nat Commun. 2014;5:5792 Epub 2014/12/18. 10.1038/ncomms6792 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanage WP, Kaijalainen T, Saukkoriipi A, Rickcord JL, Spratt BG. A successful, diverse disease-associated lineage of nontypeable pneumococci that has lost the capsular biosynthesis locus. J Clin Microbiol. 2006;44(3):743–749. Epub 2006/03/07. 10.1128/jcm.44.3.743-749.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dworkin MS, Park L, Borchardt SM. The changing epidemiology of invasive Haemophilus influenzae disease, especially in persons > or = 65 years old. Clin Infect Dis. 2007;44(6):810–816. Epub 2007/02/17. 10.1086/511861 . [DOI] [PubMed] [Google Scholar]
- 12.Farley MM, Stephens DS, Brachman PS Jr, Harvey RC, Smith JD, Wenger JD. Invasive Haemophilus influenzae disease in adults. A prospective, population-based surveillance. CDC Meningitis Surveillance Group. Ann Intern Med. 1992;116(10):806–812. Epub 1992/05/25. . [DOI] [PubMed] [Google Scholar]
- 13.Hilty M, Wuthrich D, Salter SJ, Engel H, Campbell S, Sa-Leao R, et al. Global phylogenomic analysis of nonencapsulated Streptococcus pneumoniae reveals a deep-branching classic lineage that is distinct from multiple sporadic lineages. Genome Biol Evol. 2014;6(12):3281–3294. Epub 2014/12/07. 10.1093/gbe/evu263 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellais S, Six A, Fouet A, Longo M, Dmytruk N, Glaser P, et al. Capsular switching in group B Streptococcus CC17 hypervirulent clone: a future challenge for polysaccharide vaccine development. J Infect Dis. 2012;206(11):1745–1752. Epub 2012/09/25. 10.1093/infdis/jis605 . [DOI] [PubMed] [Google Scholar]
- 15.Lam TT, Claus H, Frosch M, Vogel U. Analysis of non-typeable Haemophilus influenzae in invasive disease reveals lack of the capsule locus. Clin Microbiol Infect. 2016;22(1):63.e7–8. Epub 2015/10/11. 10.1016/j.cmi.2015.09.027 . [DOI] [PubMed] [Google Scholar]
- 16.Turner CE, Abbott J, Lamagni T, Holden MT, David S, Jones MD, et al. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. MBio. 2015;6(4):e00622 Epub 2015/07/16. 10.1128/mBio.00622-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores AR, Jewell BE, Fittipaldi N, Beres SB, Musser JM. Human disease isolates of serotype m4 and m22 group A streptococcus lack genes required for hyaluronic acid capsule biosynthesis. MBio. 2012;3(6):e00413–12. Epub 2012/11/08. 10.1128/mBio.00413-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottschalk M. Streptococcosis In: Zimmerman J, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of swine: Wiley Publishers; 2012. pp. 841–855 [Google Scholar]
- 19.Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21(6):381–407. Epub 1997/08/01. . [DOI] [PubMed] [Google Scholar]
- 20.Lee GT, Chiu CY, Haller BL, Denn PM, Hall CS, Gerberding JL. Streptococcus suis meningitis, United States. Emerg Infect Dis. 2008;14(1):183–185. Epub 2008/02/09. 10.3201/eid1401.070930 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakkitjaroen N, Takamatsu D, Okura M, Sato M, Osaki M, Sekizaki T. Loss of capsule among Streptococcus suis isolates from porcine endocarditis and its biological significance. J Med Microbiol. 2011;60(Pt 11):1669–1676. Epub 2011/07/23. 10.1099/jmm.0.034686-0 . [DOI] [PubMed] [Google Scholar]
- 22.Weinert LA, Chaudhuri RR, Wang J, Peters SE, Corander J, Jombart T, et al. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat Commun. 2015;6:6740 Epub 2015/04/01. 10.1038/ncomms7740 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott SD, Tai JY. The type-specific polysaccharides of Streptococcus suis. J Exp Med. 1978;148(6):1699–1704. Epub 1978/12/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, et al. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67(4):1750–1756. Epub 1999/03/20. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith HE, de Vries R, van't Slot R, Smits MA. The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb Pathog. 2000;29(2):127–134. Epub 2000/07/25. 10.1006/mpat.2000.0372 . [DOI] [PubMed] [Google Scholar]
- 26.Van Calsteren MR, Gagnon F, Lacouture S, Fittipaldi N, Gottschalk M. Structure determination of Streptococcus suis serotype 2 capsular polysaccharide. Biochem Cell Biol. 2010;88(3):513–525. Epub 2010/06/18. 10.1139/o09-170 . [DOI] [PubMed] [Google Scholar]
- 27.Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl Environ Microbiol. 2013;79(8):2796–2806. Epub 2013/02/19. 10.1128/aem.03742-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fittipaldi N, Segura M, Grenier D, Gottschalk M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012;7(2):259–279. Epub 2012/02/14. 10.2217/fmb.11.149 . [DOI] [PubMed] [Google Scholar]
- 29.Segura M, Gottschalk M, Olivier M. Encapsulated Streptococcus suis inhibits activation of signaling pathways involved in phagocytosis. Infect Immun. 2004;72(9):5322–5330. Epub 2004/08/24. 10.1128/iai.72.9.5322-5330.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benga L, Fulde M, Neis C, Goethe R, Valentin-Weigand P. Polysaccharide capsule and suilysin contribute to extracellular survival of Streptococcus suis co-cultivated with primary porcine phagocytes. Vet Microbiol. 2008;132(1–2):211–9. Epub 2008/06/21. 10.1016/j.vetmic.2008.05.005 . [DOI] [PubMed] [Google Scholar]
- 31.Houde M, Gottschalk M, Gagnon F, Van Calsteren MR, Segura M. Streptococcus suis capsular polysaccharide inhibits phagocytosis through destabilization of lipid microdomains and prevents lactosylceramide-dependent recognition. Infect Immun. 2012;80(2):506–517. Epub 2011/11/30. 10.1128/iai.05734-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benga L, Goethe R, Rohde M, Valentin-Weigand P. Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell Microbiol. 2004;6(9):867–881. Epub 2004/07/27. 10.1111/j.1462-5822.2004.00409.x . [DOI] [PubMed] [Google Scholar]
- 33.Tanabe S, Bonifait L, Fittipaldi N, Grignon L, Gottschalk M, Grenier D. Pleiotropic effects of polysaccharide capsule loss on selected biological properties of Streptococcus suis. Can J Vet Res. 2010;74(1):65–70. Epub 2010/04/02. . [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrando ML, de Greeff A, van Rooijen WJ, Stockhofe-Zurwieden N, Nielsen J, Wichgers Schreur PJ, et al. Host-pathogen interaction at the intestinal mucosa correlates with zoonotic potential of Streptococcus suis. J Infect Dis. 2015;212(1):95–105. Epub 2014/12/20. 10.1093/infdis/jiu813 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tenenbaum T, Papandreou T, Gellrich D, Friedrichs U, Seibt A, Adam R, et al. Polar bacterial invasion and translocation of Streptococcus suis across the blood-cerebrospinal fluid barrier in vitro. Cell Microbiol. 2009;11(2):323–336. Epub 2008/12/03. 10.1111/j.1462-5822.2008.01255.x . [DOI] [PubMed] [Google Scholar]
- 36.Ishida S, Tien le HT, Osawa R, Tohya M, Nomoto R, Kawamura Y, et al. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J Microbiol Methods. 2014;107:66–70. Epub 2014/09/18. 10.1016/j.mimet.2014.09.003 . [DOI] [PubMed] [Google Scholar]
- 37.Okura M, Lachance C, Osaki M, Sekizaki T, Maruyama F, Nozawa T, et al. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J Clin Microbiol. 2014;52(5):1714–1719. Epub 2014/02/28. 10.1128/jcm.03411-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marois C, Bougeard S, Gottschalk M, Kobisch M. Multiplex PCR assay for detection of Streptococcus suis species and serotypes 2 and 1/2 in tonsils of live and dead pigs. J Clin Microbiol. 2004;42(7):3169–3175. Epub 2004/07/10. 10.1128/jcm.42.7.3169-3175.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138(2):179–207. Epub 1980/04/01. . [DOI] [PubMed] [Google Scholar]
- 40.Okura M, Osaki M, Fittipaldi N, Gottschalk M, Sekizaki T, Takamatsu D. The minor pilin subunit Sgp2 is necessary for assembly of the pilus encoded by the srtG cluster of Streptococcus suis. J Bacteriol. 2011;193(4):822–831. Epub 2010/12/15. 10.1128/jb.01555-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mogollon JD, Pijoan C, Murtaugh MP, Kaplan EL, Collins JE, Cleary PP. Characterization of prototype and clinically defined strains of Streptococcus suis by genomic fingerprinting. J Clin Microbiol. 1990;28(11):2462–2466. Epub 1990/11/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, et al. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol. 2002;40(10):3671–3680. Epub 2002/10/02. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tritt A, Eisen JA, Facciotti MT, Darling AE. An integrated pipeline for de novo assembly of microbial genomes. PLoS One. 2012;7(9):e42304 Epub 2012/10/03. 10.1371/journal.pone.0042304 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31(17):2877–2878. Epub 2015/04/29. 10.1093/bioinformatics/btv271 . [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. Epub 2011/05/07. 10.1093/molbev/msr121 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakkitjaroen N, Takamatsu D, Okura M, Sato M, Osaki M, Sekizaki T. Capsule loss or death: the position of mutations among capsule genes sways the destiny of Streptococcus suis. FEMS Microbiol Lett. 2014;354(1):46–54. Epub 2014/03/25. 10.1111/1574-6968.12428 . [DOI] [PubMed] [Google Scholar]
- 47.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. Epub 2014/03/20. 10.1093/bioinformatics/btu153 . [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. PGAP: pan-genomes analysis pipeline. Bioinformatics. 2012;28(3):416–418. Epub 2011/12/02. 10.1093/bioinformatics/btr655 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Athey TB, Auger JP, Teatero S, Dumesnil A, Takamatsu D, Wasserscheid J, et al. Complex population structure and virulence differences among serotype 2 Streptococcus suis strains belonging to sequence type 28. PLoS One. 2015;10(9):e0137760 Epub 2015/09/17. 10.1371/journal.pone.0137760 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–352. Epub 2011/06/16. 10.1093/nar/gkr485 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, et al. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 2015;43:W104–108. Epub 2015/04/29. 10.1093/nar/gkv401 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323(5912):379–382. Epub 2009/01/20. 10.1126/science.1167140 . [DOI] [PubMed] [Google Scholar]
- 53.McLean JS, Lombardo MJ, Ziegler MG, Novotny M, Yee-Greenbaum J, Badger JH, et al. Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. Genome Res. 2013;23(5):867–877. Epub 2013/04/09. 10.1101/gr.150433.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willenborg J, Fulde M, de Greeff A, Rohde M, Smith HE, Valentin-Weigand P, et al. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology. 2011;157(Pt 6):1823–1833. Epub 2011/02/26. 10.1099/mic.0.046417-0 . [DOI] [PubMed] [Google Scholar]
- 55.Charland N, Harel J, Kobisch M, Lacasse S, Gottschalk M. Streptococcus suis serotype 2 mutants deficient in capsular expression. Microbiology. 1998;144 (Pt 2):325–332. Epub 1998/03/11. 10.1099/00221287-144-2-325 . [DOI] [PubMed] [Google Scholar]
- 56.Chabot-Roy G, Willson P, Segura M, Lacouture S, Gottschalk M. Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb Pathog. 2006;41(1):21–32. Epub 2006/05/23. 10.1016/j.micpath.2006.04.001 . [DOI] [PubMed] [Google Scholar]
- 57.Holt ME, Enright MR, Alexander TJ. Immunisation of pigs with live cultures of Streptococcus suis type 2. Res Vet Sci. 1988;45(3):349–352. Epub 1988/11/01. . [PubMed] [Google Scholar]
- 58.Holt ME, Enright MR, Alexander TJ. Studies of the protective effect of different fractions of sera from pigs immune to Streptococcus suis type 2 infection. J Comp Pathol. 1989;100(4):435–442. Epub 1989/05/01. . [DOI] [PubMed] [Google Scholar]
- 59.Blouin C, Higgins R, Gottschalk M, Simard J. Evaluation of the antibody response in pigs vaccinated against Streptococcus suis capsular type 2 using a double-antibody sandwich enzyme-linked immunosorbent assay. Can J Vet Res. 1994;58(1):49–54. Epub 1994/01/01. . [PMC free article] [PubMed] [Google Scholar]
- 60.Onishi H, Sugawara M, Okura M, Osaki M, Takamatsu D. Prevalence of Streptococcus suis genotypes in isolates from porcine endocarditis in East Japan. J Vet Med Sci. 2012;74(12):1681–1684. Epub 2012/08/11. . [DOI] [PubMed] [Google Scholar]
- 61.Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One. 2009;4(7):e6072 Epub 2009/07/16. 10.1371/journal.pone.0006072 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S, Gao M, An T, Liu Y, Jin J, Wang G, et al. Genetic diversity and virulence of novel sequence types of Streptococcus suis from diseased and healthy pigs in China. Front Microbiol. 2015;6:173 Epub 2015/03/19. 10.3389/fmicb.2015.00173 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magistrali C, Dionisi AM, De Curtis P, Cucco L, Vischi O, Scuota S, et al. Contamination of Salmonella spp. in a pig finishing herd, from the arrival of the animals to the slaughterhouse. Res Vet Sci. 2008;85(2):204–207. Epub 2008/01/31. 10.1016/j.rvsc.2007.12.002 . [DOI] [PubMed] [Google Scholar]
- 64.Darch SE, McNally A, Harrison F, Corander J, Barr HL, Paszkiewicz K, et al. Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci Rep. 2015;5:7649 Epub 2015/01/13. 10.1038/srep07649 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 2015;18(3):307–319. Epub 2015/08/25. 10.1016/j.chom.2015.07.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steward KF, Robinson C, Waller AS. Transcriptional changes are involved in phenotype switching in Streptococcus equi subspecies equi. Mol Biosyst. 2016. Epub 2016/02/09. 10.1039/c5mb00780a . [DOI] [PubMed] [Google Scholar]
- 67.Griffiths AJF, Miller JH, Suzuki DT, Lewontin RC, Gelbart WM. An Introduction to genetic analysis. 7th ed New York: W.H. Freeman; 2000. [Google Scholar]
- 68.Harris SR, Robinson C, Steward KF, Webb KS, Paillot R, Parkhill J, et al. Genome specialization and decay of the strangles pathogen, Streptococcus equi, is driven by persistent infection. Genome Res. 2015;25(9):1360–1371. Epub 2015/07/15. 10.1101/gr.189803.115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. Epub 2011/01/29. 10.1126/science.1198545 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies MR, Holden MT, Coupland P, Chen JH, Venturini C, Barnett TC, et al. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet. 2015;47(1):84–87. Epub 2014/11/18. 10.1038/ng.3147 . [DOI] [PubMed] [Google Scholar]
- 71.Palmieri C, Magi G, Mingoia M, Bagnarelli P, Ripa S, Varaldo PE, et al. Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major streptococcal pathogens. Antimicrob Agents Chemother. 2012;56(9):4697–4702. Epub 2012/06/20. 10.1128/aac.00629-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, et al. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci U S A. 2013;110(29):11923–11927. Epub 2013/7/1. 10.1073/pnas.1305559110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All genome sequence data files are available from the DNA Data Bank of Japan Sequence Read Archive under accession number DRA004206.