Abstract
OBJECTIVE
The purpose of this study is to evaluate whether the application of spectral presaturation inversion recovery (SPIR) fat suppression in standard multiecho gradient-echo sequences has a significant effect on hepatic R2* quantitation in patients with iron overload syndromes.
MATERIALS AND METHODS
Eighty patients were scanned with a multiecho gradient-echo sequence without and with the application of SPIR. Six different postprocessing approaches were used to extract R2* values for maximum generality.
RESULTS
SPIR fat suppression lowered R2* values by 3.9–7.0% (p < 0.0001 in all pairwise comparisons), independently of the postprocessing algorithm. Coefficients of variation for R2* ranged from 4.5% to 10.0%. Regardless of the size of the ROI (area of homogeneous tissue or entire liver profile in the slice), pixelwise approaches combined with an exponential-plus-constant fitting model yielded the lowest coefficients of variation (4.5% and 5.1%), whereas truncated exponential fits of the averaged signals produced the highest coefficients of variation (7.8% and 10%). For R2* values exceeding 200 Hz, a Bland-Altman analysis showed a bias that grew linearly for all postprocessing methods.
CONCLUSION
SPIR fat suppression resulted in systematically lower hepatic R2* estimates. Because calibration curves were derived using images without fat suppression, these biases should be corrected when reporting liver iron concentrations estimated from fat-suppressed multiecho T2*-weighted images.
Keywords: fat suppression, iron overload, liver, R2*
Iron overload, resulting from disorders of increased intestinal absorption or from repeated blood transfusions [1], is a problem of increasing clinical significance. The liver is the first organ to accumulate significant amounts of iron [2], and changes in liver iron concentration (LIC) account for 98% of the variation in total iron stores [3]. MRI relaxometry has been increasingly used to quantify liver iron, replacing liver biopsy as the standard of care for monitoring iron chelation therapy [4]. Both spin-echo (R2) and gradient-echo (R2*) estimates correlate highly with liver biopsy [5, 6], but R2* approaches are more widely used because image acquisition is generally faster and easier to perform.
However, the utility of LIC estimation from R2* measurements has been hindered by differences in acquisition and analysis techniques. For example, there is no consensus regarding the effect of fat suppression on R2* LIC estimation. The coexistence of fat and iron in the liver are relatively common in patients with nontransfusional siderosis [7]. The presence of fat introduces additional signal modulations with TE that are manifest as a positive bias in the apparent R2* [8], making the accuracy of R2* assessment dependent on the specific TEs used. The difference in resonance frequency between fat and water allows the TE-dependent signal modulation to be minimized by acquiring images at TEs that are multiples of 2.3 ms (at 1.5 T) where off-resonance precession places water and fat magnetization approximately in phase and in antiphase. However, in heavily iron-overloaded livers, the T2* decay is so rapid that most of the MRI signal will have irreversibly disappeared at the first in-phase TE (4.6 ms), making accurate R2* estimation impossible.
Simultaneous estimation of T2* and fat fractions using multiecho gradient-echo techniques such as IDEAL (iterative decomposition of water and fat with echo asymmetric and least-squares estimation) [9, 10] offers a logical approach to isolating signal from water and fat. Fat has several spectral peaks, and a more accurate multifrequency model of fat can be included in the IDEAL technique, improving T2* estimation in the presence of fat for applications such as quantification of hepatic iron overload [10]. Unfortunately, these techniques are currently not widely available, and implementation remains vendor specific.
Alternatively, conventional fat-suppression techniques, such as frequency-selective fat saturation and inversion recovery fat nulling, can minimize fat signal contributions [11, 12]. Although these approaches have no effect on R2 LIC quantitation in iron-overloaded subjects [13], to our knowledge, there has been no systematic comparison examining the effect of fat suppression on R2* LIC estimation accuracy.
The aim of the current study was to evaluate the effect of spectral presaturation inversion recovery (SPIR) fat suppression on standard multiecho gradient-echo sequences and the derived hepatic R2* and LIC estimates in patients with iron overload syndromes. We examined six R2* postprocessing approaches to ensure generalizability.
Materials and Methods
Study Population
Beginning in January 2013, we acquired paired R2* assessments of liver iron with and without the use of SPIR fat suppression in all patients undergoing clinical iron assessment. Our clinical practice has been to report non-fat-suppressed images unless there was evidence of severe steatosis. With increasing reports of fat suppression use with LIC quantification in the literature, we decided to review our existing data to determine whether fat suppression altered image quality or quantification. We obtained a final cohort of 80 patients (28 with thalassemia major, 33 with sickle cell disease, and 19 with other transfusional and nontransfusional iron overload diseases). The mean (± SD) age was 18.9 ± 10.4 years (age range, 2–53 years), and 25 patients were male.
The protocol for the conduct of this study was approved by The Children’s Hospital of Los Angeles’ Committee for the Protection of Human Subjects (CCI#12–00087). Waiver of informed consent was granted for the review of existing clinical data.
MRI Acquisition
All patients were scanned using a five-element torso coil on a single 1.5-T scanner (Achieva, Philips Healthcare). A multiecho gradient-echo sequence was used to collect the images for R2* analysis in a single end-expiratory breath-hold. A single midhepatic slice was acquired at 16 bipolar gradient-echoes evenly spaced from 1.0 to 13.4 ms. Image parameters were as follows: TR, 50 ms; slice thickness, 10 mm; matrix, 128 × 128 pixels; receiver bandwidth, 2273 Hz/pixel; and flip angle, 20°. The same sequence with identical parameters was obtained with SPIR fat suppression. This preparation sequence applies a frequency-specific inversion pulse, ideally inverting only the fat magnetization while leaving water resonances undisturbed. SPIR inversion uses a nonselective asymmetric sync pulse, with a flip angle of 120°, bandwidth of 1 kHz, and an off-resonance frequency of −638 Hz. The gradient-echo excitation occurs as the fat magnetization passes through its null point at an inversion time of 180 ms [14].
From here onward, we will refer to the R2* values obtained from the standard sequence (reference) as non-fat-suppressed R2* values and to those obtained from the sequence with the application of fat suppression as fat-suppressed R2* values.
MRI Analysis
All R2* images were processed centrally by the same operator (with 6 years of experience) using custom-written software developed in Matlab (release 7.5, MathWorks).
Different analysis approaches were used to improve the generalizability of the conclusions. Each analysis approach was defined by three different parameters, described here along with the possible alternatives.
The first parameter is area of liver analyzed. Alternatives are global analysis including the full extent of the liver within the slice, but excluding obvious major hilar vessels [5, 15], and ROI defined in an area of homogeneous liver tissue, avoiding blood vessels and other sources of artifacts [16–18].
The second parameter is type of fitting. Alternatives are averaged-signal or region-based analysis, where pixels within the ROI are averaged together and fit is performed for this averaged decay curve [17, 18], and pixelwise analysis, where fitting is performed for each pixel within the ROI and the median of the distribution of R2* values is calculated [5, 15].
The third parameter is curve-fitting models. One alternative to the curve-fitting model is a three-parameter model, which includes an exponential with a variable offset [5, 16, 18], calculated as follows:
| (1) |
where S is signal intensity, S0 is signal intensity at TE of 0, TE is TE, and C is constant offset that corrects for contributions for blood, bile, motion artifact, and noise bias. The other alternative to the curve-fitting model is a two-parameter model, with a single exponential model (Equation 1 without the constant C) [17]. For moderate and high R2* values, fitting is performed after the manual exclusion of later TEs with high iron-mediated signal loss.
The two-parameter model cannot be combined with pixelwise fitting because there are no validated automatic truncation algorithms for liver signal decay, resulting in a total of six different algorithms (23 - 2), including those most often used clinically by different centers worldwide: PW_GA(3P) (i.e., pixelwise global analysis with three parameters) and Avg_ ROI(2P) (i.e., average ROI with two parameters). The algorithms are summarized in Table 1.
TABLE 1.
Description of Different Approaches Used to Calculate R2* Values and Nomenclature for the Respective R2* Algorithms
| Algorithm Name | Type of ROI | Type of Fitting | Curve-Fitting Model | Reference |
|---|---|---|---|---|
|
| ||||
| R2*_PW_GA(3P) | Entire liver profile in the slice | Pixelwise approach | Offset model | [5] |
| R2*_Avg_GA(3P) | Entire liver profile in the slice | Averaged-signal approach | Offset model | [15] |
| R2*_Avg_GA(2P) | Entire liver profile in the slice | Averaged-signal approach | Truncation model | Not applicable |
| R2*_PW_ROI(3P) | Area of homogeneous liver tissue | Pixelwise approach | Offset model | [16] |
| R2*_Avg_ROI(3P) | Area of homogeneous liver tissue | Averaged-signal approach | Offset model | [18] |
| R2*_Avg_ROI(2P) | Area of homogeneous liver tissue | Averaged-signal approach | Truncation model | [17] |
Note—Each approach is defined by three different aspects and two approaches differ for at least one of these aspects. PW = pixelwise, GA = global analysis, 3P = three parameter, Avg = average, 2P = two parameter.
Statistical Analysis
All data were analyzed using SPSS (version 16.0, SPSS) and MedCalc (version 7.2.1.0 for Windows, MedCalc Software) statistical packages. Continuous variables were described as mean ± SD.
For each approach, the following statistical comparison was made between fat-suppressed and non-fat-suppressed R2* estimates: the Wilcoxon signed-rank test was applied to detect statistically significant differences between two datasets. Summary data were displayed using scatterplots, and linear regression was performed. Linear regression models provided slope and intercept estimates and the coefficient of determination (R2), which measures the goodness of the linear fit. The coefficient of variation was obtained as the ratio of the SD of the half mean square of the differences between the repeated values, to the general mean. A coefficient of variation less than 10% was considered good. The agreement between the variables was determined by Bland-Altman technique plotting the difference versus the average of the two measurements. Bias (systematic difference between methods) was the mean of the difference between the two R2* measurements and agreement was the mean ± 1.96 SD. In each Bland-Altman plot, the regression line of difference was shown. All tests were performed with a significance level of α = 0.05.
Results
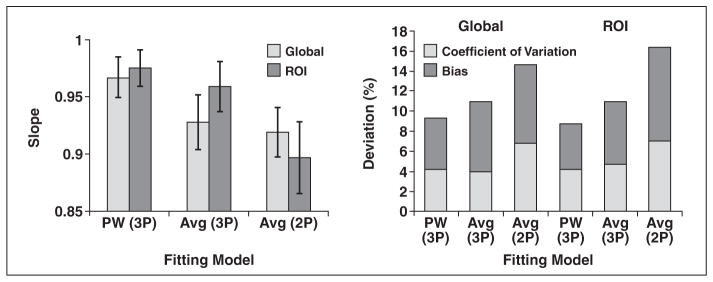
The R2* values obtained in this study completely spanned the clinically relevant range, varying from 34 to 1777 Hz, using our standard approach (pixelwise three-parameter fit). Table 2 and Figure 1 summarize the effect of fat suppression on R2* estimation. Fat suppression lowered R2* values by 3.9–7.0% (p< 0.0001 in all the pairwise comparisons), independently of the postprocessing algorithm.
TABLE 2.
Comparison Between Hepatic R2* Values and Hepatic R2* Values With Application of Fat Suppression for Each Approach Adopted for R2* Calculation
| Algorithm | Paired-Sample Wilcoxon Signed-Rank Test | Regression Analysis | Difference (%) | Coefficient of Variation (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Fat-Suppressed vs Non-Fat-Suppressed (Hz) | Difference (Hz) | p | Slope, Coefficient (SE) | p for Slope | Intercept (Hz), Coefficient (SE) | R2 | |||
| R2*_PW_GA(3P) | 677.5 ± 472.3 vs 706.0 ± 486.7 | −28.5 ± 41.5 | < 0.0001 | 0.967 (0.009) | 0.00029 | −5.388 (7.620) | 0.993 | −4.2 ± 5.1 | 5.1 |
| R2*_Avg_GA(3P) | 605.9 ± 388.5 vs 638.6 ± 416.1 | −32.7 ± 52.2 | < 0.0001 | 0.928 (0.012) | < 0.0001 | 13.283 (8.861) | 0.988 | −3.9 ± 6.7 | 7.0 |
| R2*_Avg_GA(2P) | 468.3 ± 276.2 vs 506.6.6 ± 298.9 | −38.4 ± 37.7 | < 0.0001 | 0.919 (0.011) | < 0.0001 | 2.684 (6.437) | 0.989 | −6.8 ± 7.7 | 7.8 |
| R2*_PW_ROI(3P) | 674.3 ± 466.5 vs 698.6 ± 477.2 | −24.3 ± 36.6 | < 0.0001 | 0.975 (0.008) | 0.0025 | −6.692 (6.936) | 0.994 | −4.2 ± 6.6 | 4.5 |
| R2*_Avg_ROI(3P) | 665.5 ± 458.6 vs 695.5 ± 475.6 | −30.2 ± 51.6 | < 0.0001 | 0.959 (0.011) | 0.0004 | −1.593 (9.568) | 0.989 | −4.7 ± 7.4 | 6.2 |
| R2*_Avg_ROI(2P) | 557.2 ± 361.9 vs 599.7 ± 398.4 | −42.5 ± 70.3 | < 0.0001 | 0.897 (0.016) | < 0.0001 | 19.229 (11.669) | 0.975 | −7.0 ± 9.4 | 10.0 |
Note—Except where noted otherwise, data are mean ± SD. SE = standard error, PW = pixelwise, GA = global analysis, 3P = three parameter, Avg = average, 2P = two parameter.
Fig. 1.
Effect of fat suppression on R2* estimation. Regression slope (left) is shown as function of fitting model. Error bars denote 95% CIs. Bias and coefficient of variation are shown for images collected with and without saturation (right). Bias is displayed as absolute value to facilitate plotting (fat saturation consistently lowered R2* estimates). 2P = two parameter, 3P = three parameter, Avg = average, PW = pixelwise.
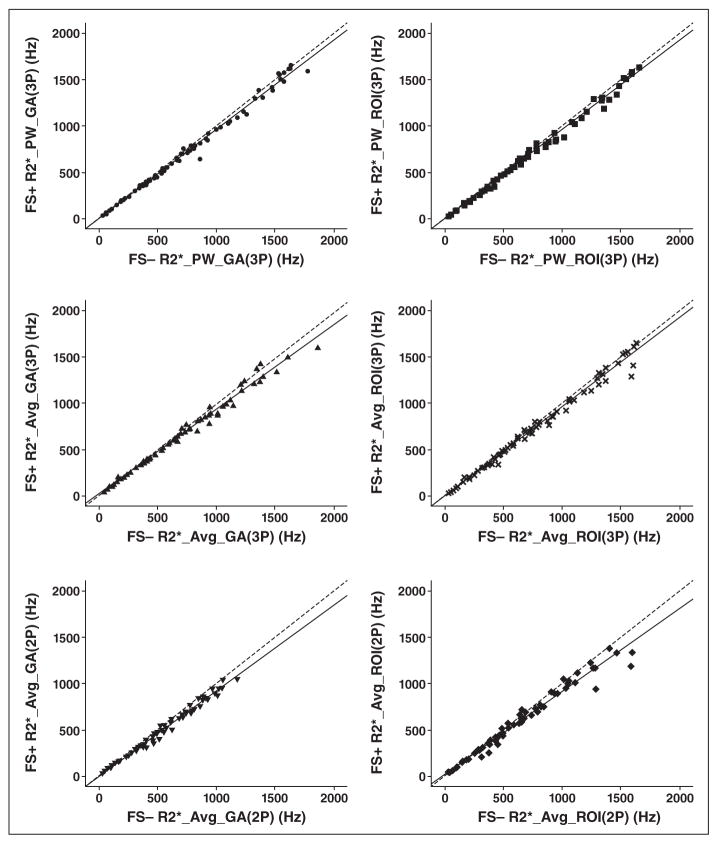
Figure 2 shows the fat-suppressed R2* values as a function of non-fat-suppressed R2* values for each postprocessing algorithm along with its relative regression line. The results of each regression analysis are indicated in Table 2. All R2 values for the fit were near unity (lowest value = 0.975; highest value = 0.994). However, all the slopes were significantly less than 1 (range, 0.897–0.975).
Fig. 2.
Scatterplot comparison between fat-suppressed (FS+) R2* and non-fat-suppressed (FS−) R2* values. Regression lines (solid lines) are statistically lower than unity (dashed lines) in all cases. Different symbols represent R2* values obtained with different algorithms. 2P = two parameter, 3P = three parameter, Avg = average, GA = global analysis.
Coefficients of variation ranged from 4.5% to 10.0% (Table 2). Pixelwise approaches yielded the lowest coefficient of variation (4.5% and 5.1%); truncated exponential fits produced higher coefficients of variation (7.8% and 10%).
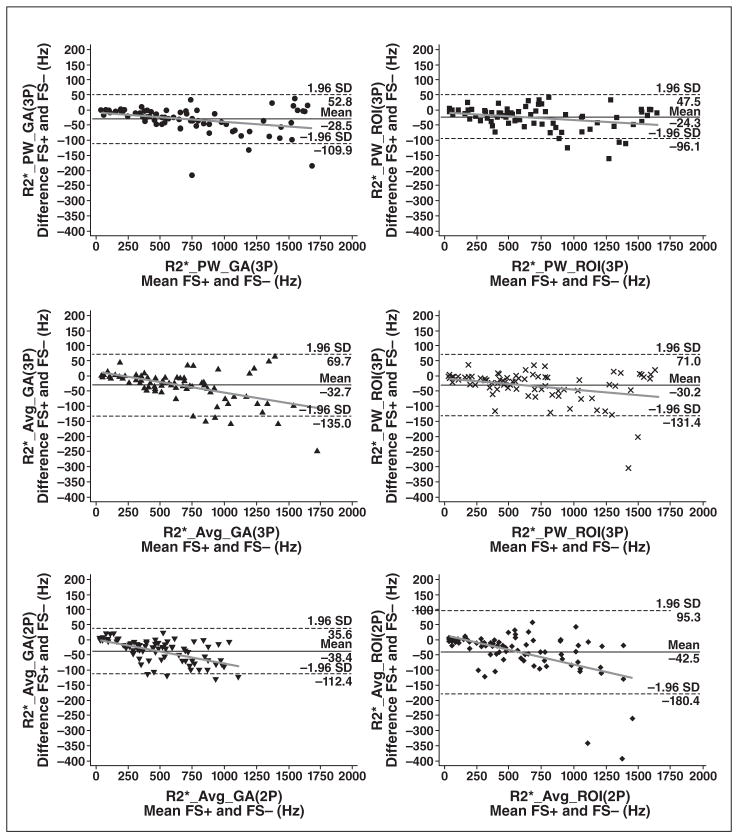
Figure 3 shows the Bland-Altman plots. No bias was apparent until R2* exceeded 200 Hz, but it grew linearly thereafter for all six postprocessing methods.
Fig. 3.
Bland–Altman plots of absolute differences between fat-suppressed (FS+) R2* and non-fat-suppressed (FS−) R2* values for all algorithms used for R2* assessment. In each Bland-Altman plot dashed lines indicate limits of agreement, and gray line is regression line of difference. Different symbols represent R2* values obtained with different algorithms. 2P= two parameter, 3P = three parameter, Avg = average, GA = global analysis, PW = pixelwise.
Discussion
To our knowledge, this is the first study to compare the effect of SPIR fat suppression on R2* quantitation in transfusional siderosis. We have shown that SPIR fat suppression systematically lowered R2* estimates by 3.9–7.0% and introduced comparable random uncertainty between the two measurements. Because calibration curves have been derived using non-fat-saturated images [5, 17, 19], these biases should be corrected when reporting LIC values from fat-saturated images.
There are two explanations for the difference in fat-suppressed and non-fat-suppressed R2* estimates. First, fat and water cannot be separated by radiofrequency excitations in iron-overloaded subjects. Iron causes overlap of the water and fat line width’s for LIC concentrations greater than approximately 5 mg/g (R2* > 220 Hz) at 1.5 T. As a result, the SPIR preparation suppresses significant amounts of the water signal from heavily iron-loaded tissues, lowers the signal-to-noise ratio in an iron-dependent manner, and biases R2* estimates toward lower values. Figure 3 shows that bias and variability increase dramatically once liver R2* exceeds 200 Hz. Second, the fat-water signal modulation with TE influences whether weak echoes are retained or truncated. Because SPIR preparation reduces both the fat and water signals, this truncation process becomes less reliable. This is quite evident in Figure 1, where the combined deviation (bias plus coefficients of variation) is much higher (≈15%) for truncation (two-parameter model) methods. The three-parameter model generally is more robust to fat suppression because it fits the entire echo train, where in-phase and out-of-phase oscillations have contributed three full cycles, contributing approximately equal positive and negative fitting errors to the dominant exponential.
This study was limited by the use of a single type of fat suppression approach (SPIR), which is implemented by the vendor. SPIR does not account for the multispectral nature of fat resonances. Although the fundamental interactions between iron, fat, and fitting models are a physical reality, the effect size observed in this study may vary across suppression methods and platforms. For example, fat-suppression techniques exploiting T1 differences between fat and liver will suffer from incomplete separation as liver T1 shortens with iron overload, but the magnitude of the bias may be different than observed in our study. Similar caveats must be made for other fitting models, such as those that attempt to correct for noise bias through statistical methods. This study would also have been strengthened by formal signal-to-noise ratio assessment, but the use of a proprietary surface correction and background noise suppression (CLEAR [Constant LEvel AppeaRance], Philips Healthcare) create spatially varying noise characteristics.
Fat suppression has not been routine in R2* LIC estimation, but some groups have recently been advocating its use, particularly with the ROI_Avg(2P) (average ROI with two parameters) model [12]. Our data suggest that SPIR fat suppression is inappropriate for patients with transfusional siderosis because it reduces signal to noise, biases the R2* estimate, and adds unnecessary measurement variability into the R2* estimate. Errors introduced by fat saturation (worst case 95% CIs, −25.8% to 11.8%) are not large enough to completely alter patient management, but sufficiently large to warrant controlling. However, SPIR suppression could be beneficial in patients with high hepatic lipid content and low liver iron burden, such as patients with metabolic syndrome [20]; however, reasonable R2* estimation can be performed in these subjects by restricting TEs to in-phase echoes only.
The optimal way to correct for fat-water signal modulations is to perform simultaneous modeling of the T2* decay and fat interactions, using models that account for the spectral off-resonance characteristics of mobile lipids [9, 10]. We think that these methods will gradually replace simple multiecho gradient-echo R2* techniques over time, but limited availability and platform specificity prohibit their widespread use presently. Fortunately, patients undergoing long-term transfusion rarely have significant hepatic steatosis because chronic anemia states are generally hypermetabolic [21], explaining why R2* LIC approaches are so robust in these patients. If fat suppression is used at individual centers, paired analysis detailing the effect of fat saturation on the calibration curve should be performed.
Acknowledgments
This work was supported in part by the National Center for Advancing Translational Sciences, National Institutes of Health (grants 1RC HL099412-01 and UL1TR000130 to Children’s Hospital Los Angeles).
References
- 1.McLaren GD, Muir WA, Kellermeyer RW. Iron overload disorders: natural history, pathogenesis, diagnosis, and therapy. Crit Rev Clin Lab Sci. 1983;19:205–266. doi: 10.3109/10408368309165764. [DOI] [PubMed] [Google Scholar]
- 2.Noetzli LJ, Carson SM, Nord AS, Coates TD, Wood JC. Longitudinal analysis of heart and liver iron in thalassemia major. Blood. 2008;112:2973–2978. doi: 10.1182/blood-2008-04-148767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelucci E, Brittenham GM, McLaren CE, et al. Hepatic iron concentration and total body iron stores in thalassemia major. N Engl J Med. 2000;343:327–331. doi: 10.1056/NEJM200008033430503. [DOI] [PubMed] [Google Scholar]
- 4.Wood JC. Impact of iron assessment by MRI. Hematology Am Soc Hematol Educ Program. 2011;2011:443–450. doi: 10.1182/asheducation-2011.1.443. [DOI] [PubMed] [Google Scholar]
- 5.Wood JC, Enriquez C, Ghugre N, et al. MRI R2 and R2* mapping accurately estimates hepatic iron concentration in transfusion-dependent thalassemia and sickle cell disease patients. Blood. 2005;106:1460–1465. doi: 10.1182/blood-2004-10-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St Pierre TG, Clark PR, Chuaanusorn W, et al. Noninvasive measurement and imaging of liver iron concentrations using proton magnetic resonance. Blood. 2005;105:855–861. doi: 10.1182/blood-2004-01-0177. [DOI] [PubMed] [Google Scholar]
- 7.Sirlin CB, Reeder SB. Magnetic resonance imaging quantification of liver iron. Magn Reson Imaging Clin N Am. 2010;18:359–381. ix. doi: 10.1016/j.mric.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeder SB, Sirlin CB. Quantification of liver fat with magnetic resonance imaging. Magn Reson Imaging Clin N Am. 2010;18:337–357. ix. doi: 10.1016/j.mric.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeder SB, Pineda AR, Wen Z, et al. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn Reson Med. 2005;54:636–644. doi: 10.1002/mrm.20624. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multiecho water-fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med. 2008;60:1122–1134. doi: 10.1002/mrm.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwenzer NF, Machann J, Haap MM, et al. T2* relaxometry in liver, pancreas, and spleen in a healthy cohort of one hundred twenty-nine subjects-correlation with age, gender, and serum ferritin. Invest Radiol. 2008;43:854–860. doi: 10.1097/RLI.0b013e3181862413. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, He T, Gatehouse PD, et al. Improved MRI R2* relaxometry of iron-loaded liver with noise correction. Magn Reson Med. 2013;70:1765–1774. doi: 10.1002/mrm.24607. [DOI] [PubMed] [Google Scholar]
- 13.Papakonstantinou O, Foufa K, Benekos O, et al. Use of fat suppression in R2 relaxometry with MRI for the quantification of tissue iron overload in beta-thalassemic patients. Magn Reson Imaging. 2012;30:926–933. doi: 10.1016/j.mri.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Jackson A, Sheppard S, Laitt RD, Kassner A, Moriarty D. Optic neuritis: MR imaging with combined fat- and water-suppression techniques. Radiology. 1998;206:57–63. doi: 10.1148/radiology.206.1.9423652. [DOI] [PubMed] [Google Scholar]
- 15.Positano V, Salani B, Pepe A, et al. Improved T2* assessment in liver iron overload by magnetic resonance imaging. Magn Reson Imaging. 2009;27:188–197. doi: 10.1016/j.mri.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Maris TG, Papakonstantinou O, Chatzimanoli V, et al. Myocardial and liver iron status using a fast T2* quantitative MRI (T2*qMRI) technique. Magn Reson Med. 2007;57:742–753. doi: 10.1002/mrm.21204. [DOI] [PubMed] [Google Scholar]
- 17.Anderson LJ, Holden S, Davis B, et al. Cardiovascular T2-star (T2*) magnetic resonance for the early diagnosis of myocardial iron overload. Eur Heart J. 2001;22:2171–2179. doi: 10.1053/euhj.2001.2822. [DOI] [PubMed] [Google Scholar]
- 18.Meloni A, Luciani A, Positano V, et al. Single region of interest versus multislice T2* MRI approach for the quantification of hepatic iron overload. J Magn Reson Imaging. 2011;33:348–355. doi: 10.1002/jmri.22417. [DOI] [PubMed] [Google Scholar]
- 19.Hankins JS, McCarville MB, Loeffler RB, et al. R2* magnetic resonance imaging of the liver in patients with iron overload. Blood. 2009;113:4853–4855. doi: 10.1182/blood-2008-12-191643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchesini G, Brizi M, Bianchi G, et al. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 2001;50:1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- 21.Hibbert JM, Creary MS, Gee BE, Buchanan ID, Quarshie A, Hsu LL. Erythropoiesis and myocardial energy requirements contribute to the hyper-metabolism of childhood sickle cell anemia. J Pediatr Gastroenterol Nutr. 2006;43:680–687. doi: 10.1097/01.mpg.0000228120.44606.d6. [DOI] [PMC free article] [PubMed] [Google Scholar]