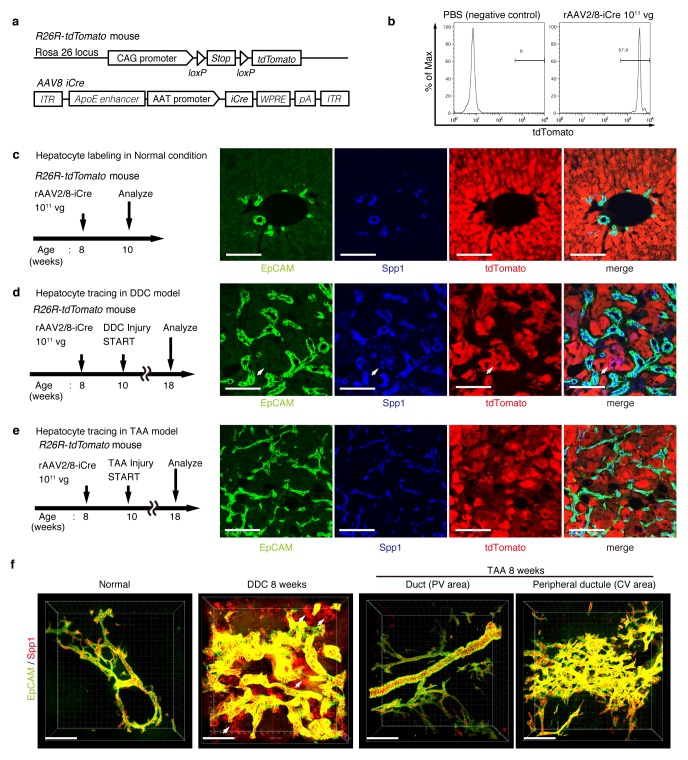
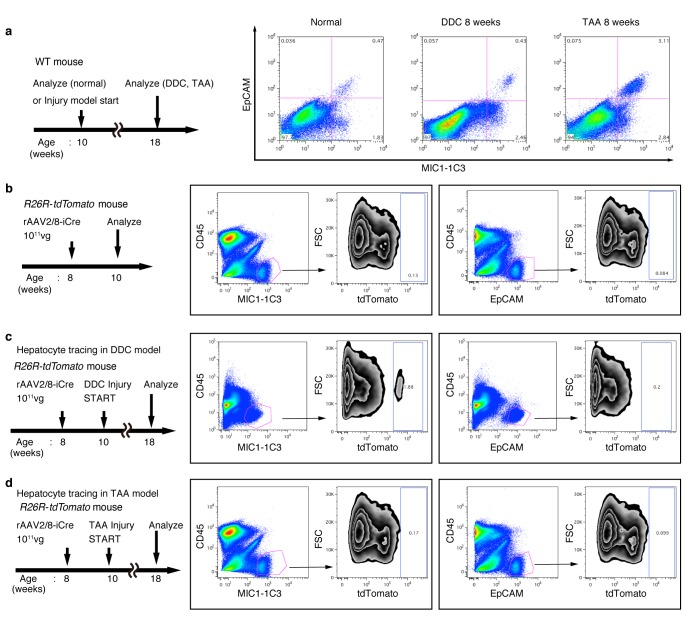
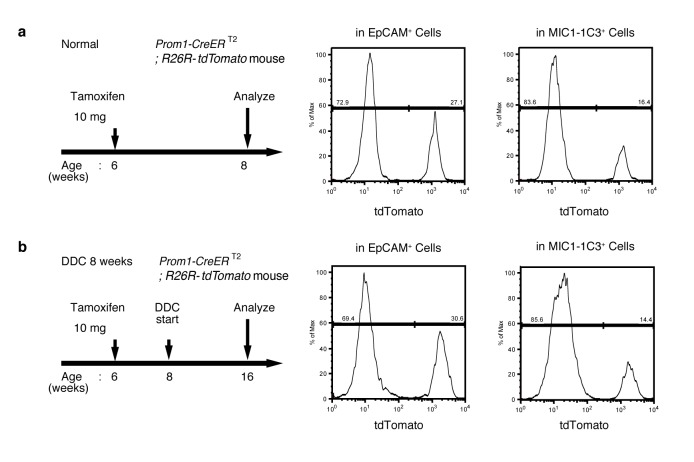
Figure 3. Lineage tracing of hepatocytes.
(a) R26R-tdTomato mice were used in combination with rAAV2/8-iCre for the labeling of hepatocytes. rAAV2/8-iCre is designed to transduce only hepatocytes. (b) Representative image of FACS analysis of hepatocytes labeled by rAAV2/8-iCre. These histogram images show the result of serial purification gates (FSC/SSC, pulse width, DAPI-). (c) Adult R26R-tdTomato mice were injected with rAAV2/8-iCre (1x1011 vg /mouse). 2 weeks after injection, the mice were sacrificed and the livers were stained with anti-EpCAM and anti-Spp1 antibody (scale bar, 100 um). (d and e) Mice were injected with rAAV2/8-iCre (1011 vg /mouse) and then subjected to a 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) or TAA injury model. tdTomato+ Spp1+ EpCAM- cells were only observed in DDC-fed mouse liver sections (white arrows). Analysis was done with 5 mice per each injury model. More than 6 sections were made per mouse. (f) 3D imaging was performed with WT mice (normal state, DDC for 8 weeks, TAA for 8 weeks). Acquired z-stack data is displayed as maximum-intensity projection after contrast adjustment with IMARIS software. In the DDC liver, Spp1+ EpCAM- cells were observed (white arrow) around main biliary tubular structures that were composed of EpCAM+ cells.