Abstract
Tuberculosis is a major neglected disease for which the quest to find new treatments continues. There is an abundance of data from large phenotypic screens in the public domain against Mycobacterium tuberculosis (Mtb). Since machine learning methods can learn from past data, we were interested in addressing whether more data builds better models. We now describe using Bayesian machine learning to assess whether we can improve our models by combining the large quantities of single-point data with the much smaller (higher quality) dual-event datasets, which use both dose-response data for both whole-cell antitubercular activity and Vero cell cytotoxicity. We have evaluated 12 models ranging from different single-point, dual-event dose response, single-point and dual-event dose response as well as combined datasets for three distinct datasets from the same laboratory. We used a fourth dataset of active and inactive compounds from the same group as well as a smaller set of 177 active compounds from GlaxoSmithKline as test sets. Our data suggest combining single-point with dual-event dose response data does not diminish the internal or external predictive ability of the models based on the receiver operator curve (ROC) for these models (internal ROC range 0.83-0.91, external ROC range 0.62-0.83) compared to the orders of magnitude smaller dual event models (internal ROC range 0.6-0.83 and external ROC 0.54-0.83). In conclusion, models developed with 1200-5000 compounds appear to be as predictive as those generated with 25,000 to 350,000 molecules. Our results have implications for justifying further HTS versus focused testing based on model predictions.
Keywords: Bayesian models, Bigger data, Collaborative Drug Discovery Tuberculosis database, Dual-event models, Function class fingerprints, Mycobacterium tuberculosis, Tuberculosis
INTRODUCTION
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis (TB). This bacterium has infected approximately one-third of the world’s population, and kills 1.3 million people annually.1 Additional therapeutic agents are needed that are active against Mtb to overcome resistance, shorten treatment and avoid toxicity that may occur in patients co-infected with HIV. 2-4 Over the last decade there has been considerable investment in TB drug discovery and development, such that at least $500 million was spent in 2013 according to one estimate. 5 While the sequencing of the Mtb genome has provided metabolic insights and potential targets,6 genomic data have not led directly to any drugs. 7, 8,9 Target-based design of antibacterial agents has been declared a failure,8 and whole-cell phenotypic high-throughput screens (HTS) of libraries of thousands to hundreds of thousands of molecules is now in vogue.3, 10-12 Whole-cell phenotypic HTS against Mtb has gained much support, having led to the clinical-stage candidate SQ109 13 and the drug bedaquiline.14 On the other hand, the general process is characterized by very low hit rates,15 and the approach does not usually provide information on the potential target/s leading to complications in lead optimization and final drug approval. These HTS typically employ a single point, or concentration, primary screen to identify hits that are then evaluated in a dose-response format in concert with parallel testing to assess cytotoxicity in a model mammalian cell line (e.g, Vero, HepG2 or other cells). 10-12 This phenotypic screening format produces a wealth of data that can be used for computational machine learning. 16
Building on an initial report leveraging HTS data through Bayesian models,17 we have focused on the development and utilization of machine-learning models in the discovery of novel chemical probes and drug discovery hits and leads. 18-25 We have made extensive use of the public datasets coming out of the MLSMR (derived from Molecular Libraries Screening Center Network and also called the MLSCN/MLPCN library elsewhere), TAACF-CB2, and TAACF-kinase screens conducted by Southern Research Institute under contract from the National Institute of Allergy and Infectious Diseases (NIAID).10-12 The outcome has been single- (antitubercular efficacy) and dual-event (antitubercular efficacy and lack of relative Vero cell cytotoxicity) models with both single-point and dose-response data to uncover promising antituberculars (with hit rates in excess of 20%) of the pyrazolopyrimidine, triazine, benzothiazole, sulfonamide, and aminoquinoline classes 25. Additional follow--up studies have provided similar hit rates. 19, 24
Parallel efforts in our laboratories have in part focused on the optimization of our use of machine-learning model performance. Critical metrics are the model’s ability to predict the data set from which it was trained (measured with leave out groups and receiver operator characteristic statistics (ROC)) and to correctly identify both actives and inactives from a compound library distinct from its training set (whether using retrospective or prospective analysis). Recently we explored the impact of the type of machine-learning algorithm. 22 We reported the examination of Support Vector Machine (SVM) and Recursive Partitioning (RP) single tree and forest models to compare with dual-event Bayesian models of in vitro antitubercular efficacy and acceptable Vero cell cytotoxicity (selectivity index SI = (MIC or IC90)/CC50 ≥ 10; where MIC = minimum compound concentration to inhibit growth of organism usually by 90 or 99%, IC90 = compound concentration necessary to inhibit 90% of the organism’s growth, CC50 = compound concentration that inhibits growth of the cells by 50%).22 We did not find a dramatic difference between the Bayesian and other models for the same individual datasets when performing 5-fold cross validation. The ability of a model to predict hits amongst the GlaxoSmithKline (GSK) set of 177 antituberculars 26, in fact, appeared to depend more on the identity of the training set than on the method used. Therefore, we probed the effect of combining datasets and realized that larger datasets (as judged solely by number of compounds) do not necessarily afford more predictive models 20. This effort clearly involved not just increasing the size of the data set, but also altering the ratio of actives to inactives and perhaps their respective distributions in chemical property space. We hypothesized that a better trained model could arise by fusion of single-point screening inactives with dual-event dose-response actives and inactives. Studies evaluating this novel hypothesis in drug discovery machine-learning strategy are described herein.
Experimental
CDD database and SRI datasets
The Tuberculosis Antimicrobial Acquisition and Coordinating Facility (TAACF), Molecular Libraries Small Molecule Repository (MLSMR) screening datasets and TB: ARRA 10-12 library were collected and uploaded in the CDD TB database (Collaborative Drug Discovery Inc. Burlingame, CA)18 from sdf files and mapped to custom protocols.27 All Mtb datasets used in model building are available for free public read-only access and mining upon registration in the CDD database 23, 27-29 as well as in PubChem.30
Building and validating dual-event machine learning models with novel bioactivity and cytotoxicity data
In our previous publications we described the generation and validation of the Laplacian-corrected Bayesian classifier models developed with cytotoxicity data to create dual-event models 19, 24, 25 using Discovery Studio 3.5 (San Diego, CA).17, 31-34 These individual models were developed based on several unique datasets: a. MLSMR dose-response and cytotoxicity; b. TAACF-CB2 dose-response and cytotoxicity; and c. TAACF-kinase dose-response and cytotoxicity, where cytotoxicity was determined for Vero cells for each set. The models were all generated using the following molecular descriptors: molecular function class fingerprints of maximum diameter 6 (FCFP_6) 35, AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area which were all calculated from input sdf files.
We have now expanded the range of models by using the previously described single-point screening datasets 18, 23 (Figure 1) and removing any compounds classed as active. The corresponding dual-event dataset 24, 25 was then combined with it to provide the actives as well as additional inactives. The resulting single- and dual-event datasets were used to generate new, larger models that were also validated using leave-one-out cross-validation, 5 fold validation and by leaving out 50% of the data and rebuilding the model 100 times using a custom protocol to generate the receiver operator curve area under the curve (ROC AUC), concordance, specificity and selectivity as described previously.19, 24, 25 In the current study, as well as using the datasets individually, we also combined the three larger datasets, which combined single-point and dual-event data (MLSMR, TAACF-CB2, TAACF-kinase).
Figure 1.
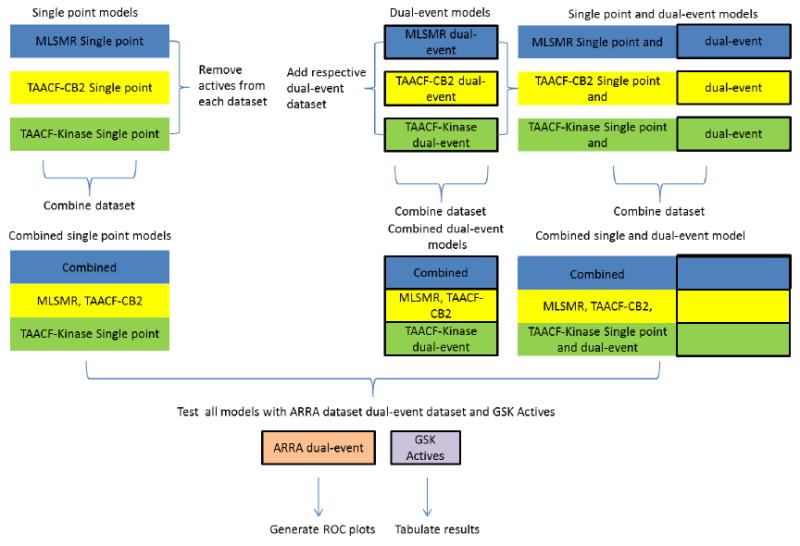
Schema to show models built and evaluated (bold outlined = dose-response data)
Testing Bayesian models trained with external datasets
The models were further evaluated by predicting a set of 1924 analogs described previously in the ARRA dataset. 21 Additionally, a set of 177 antitubercular leads (actives) disclosed by GSK 26 was scored with all of the models generated in this study to determine how many hits could be predicted. The mean closest distance for each model’s training set to the ARRA or GSK datasets was calculated to provide a measure of training set similarity to the test set. In Discovery Studio this was set to the default to use the Euclidian distance function with mean-center and scale and scale by number of dimensions turned on. The proximity of two molecules (and of the training sets) scales inversely with the calculated distance.
Assessing Mtb HTS chemistry property space
The GSK and ARRA datasets were compared to the 345,011-member dataset, used to train the combined dose-response and cytotoxicity plus single-point inactives model used in this study, as to their relative placement in chemistry property space. A Principal Component Analysis (PCA) using Discovery Studio was generated with the interpretable descriptors chosen previously (AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area). These libraries were also compared through the “compare libraries” protocol in Discovery studio via the use of assemblies (Murcko Assemblies) 36.
Statistical Analysis
The mean descriptor values for in vitro active and inactive antitubercular compounds were compared using two tailed t-test with JMP v. 8.0.1 (SAS Institute, Cary, NC).
RESULTS
Combining single-point and dose-response data
Novel machine learning datasets were created for the MLSMR, TAACF-CB2, TAACF-kinase and combined libraries by merging the respective dose-response dual-event and single-point antitubercular efficacy (single-event) inactives datasets. The percent of actives in a dataset ranges from 0.07% to 56.42% (Table 1). Bayesian models were constructed for each novel dataset and they exhibited Bayesian 5 fold (leave out 20%) ROC AUC values (Table 1 and Supplemental data) and leave out 50% x 100 ROC AUC values (Table 1 and Supplemental Table 1) greater than 0.8 (range 0.83-0.91). These metrics of a model’s ability to predict its training set are improved or equivalent to those for models constructed with dose-response dual-event data (ROC AUC range 0.6-0.83) and single-point antitubercular efficacy data alone (ROC AUC range 0.84-0.88).
Table 1.
Individual Bayesian models, leave out testing and external prediction with the ARRA dataset (N = 1924 molecules). Mean-closest distance scales inversely with the similarity to training set (highlighting number of compounds that overlap between the training set and test set).
| Mtb models (training set N) [number actives (percent)] | Bayesian (5 fold ROC) | Bayesian (leave out 50% × 100 ROC) | predicting ‘ARRA dose response and cytotoxicity’ data set (N = 1924) ROC | Mean closest distance of training set to test set |
|---|---|---|---|---|
| MLSMR single-point data (220463) [4096 actives (1.86)] | 0.87 | 0.86 | 0.58 | 0.36 (2 in set) |
| TAACF-CB2 single-point data (102633) [1783 actives (1.74)] | 0.85 | 0.84 | 0.75 | 0.32 (281 in set) |
| TAACF-kinase single-point (23797) [1308 actives (5.50)] | 0.88 | 0.88 | 0.55 | 0.43 (123 in set) |
| Combined single-point (346893) [7187 actives (2.07)] | 0.87 | 0.85 | 0.61 | 0.23 (401 in set) |
| MLSMR dose-response and cytotoxicity (2273)a,b [165 actives (7.26)] | 0.83 | 0.82 | 0.82 | 0.51(1 in training set) |
| TAACF-CB2 dose-response and cytotoxicity (1783) a, b [1006 actives (56.42)] | 0.60 | 0.64 | 0.54 | 0.50 (66 in training set) |
| TAACF-kinase dose-response and cytotoxicity (1248) a, b [182 actives (14.58)] | 0.76 | 0.74 | 0.74 | 0.56 (52 in training set) |
| Combined dose-response and cytotoxicity (5304) a, b [1352 actives (25.49)] | 0.75 | 0.74 | 0.83 | 0.40 (81 in training set) |
| MLSMR dose-response and cytotoxicity and single-point (218640) [165 actives (0.07)] | 0.86 | 0.84 | 0.83 | 0.37 (2 in set) |
| TAACF-CB2 dose-response and cytotoxicity and single-point (102634) [1006 actives (0.98)] | 0.85 | 0.83 | 0.74 | 0.32 (281 in set) |
| TAACF-kinase dose-response and cytotoxicity and single-point (23737) [182 actives (0.77)] | 0.91 | 0.90 | 0.62 | 0.43 (118 in set) |
| Combined dose-response and cytotoxicity and single-point (345011) [1353 actives (0.39)] | 0.88 | 0.87 | 0.79 | 0.23 (396 in set) |
Where: IC90 < 10 μg/ml (TAACF-CB2 only) or 10 μM (other datasets) and a selectivity index (SI) greater than ten where the SI is calculated from SI = CC50/IC90.
Previously published 22
The features important for separate single- or dual-event models have been previously described.18, 20, 23-25 For the new Bayesian models developed in this study we now briefly describe these molecular features found in actives or inactives. For the MLSMR model, we can identify those FCFP_6 substructure descriptors consistent with both activity and lack of cytotoxicity including alkyl 2-thioacetate, 1,3,4-oxadiazole 2-thioether, alkyl 2-alkoxyacetate, 4-oxo-1,4-dihydropyridine-3-carboxylic acid, and pyridine 2-thioacetate (Figure S1). Features of inactives include sulfonamide, hydrazine/hydrazone, piperidine, and 3-aminotetrahydrothiophene 1,1-dioxide (Figure S2). For the TAACF-CB2 model, substructure descriptors consistent with both activity and lack of cytotoxicity include alkyl 2-thioacetate, N-alkylimidazole, 5-substituted-2-nitrofuran, 8-acetoxyquinoline, 4-aminoketone, and 2-ketosubstituted thiophene (Figure S3). Inactives features are 1,2,4-triazole 3-thioether, sulfonate ester, 4-substituted morpholine, 2-substituted tetrahydrofuran, sulfonamide, N-cyclopropylacetamide, and 1-(pyrrolidin-1-yl)ethanone (Figure S4). For the TAACF-kinase model, substructure descriptors consistent with both activity and lack of cytotoxicity include N-(1,3,4-oxadiazol-2-yl)thiophene-2-carboxamide, N-(thiazol-2-yl)furan-2-carboxamide, 3-(1H-pyrrol-1-yl)propan-1-amine, and 2-amino-5-aryl-1,3,4-oxadiazole (Figure S5). Features of inactives contained pyridone, N-alkyl-2-thioacetamide, 2,3-disubstituted benzothiophene, pyrrolidin-2-one, and 3-amino-2-substituted benzofuran (Figure S6). For the combined model, substructure descriptors consistent with both activity and lack of cytotoxicity include alkyl 2-alkoxyacetate, 5-nitrofuran 2-carboxamide, 8-acetoxyquinoline, N-butylimidazole, N-propylaminoimidazole, 2-amino-5-phenyl-1,3,4-oxadiazole, thiazole 2-amide (Figure S7). Inactives features are 1,2,4-triazole 3-thioacetamide, trisubstituted isoxazole, N-cyclopropylacetamide, thiazole 2-imine, pyrimidin-2-one, sulfonate ester, 1-(piperidin-1-yl)ethanone, 2-hydroxypyridine, sulfonamide, 3,4-dihydropyrrolo[2,3-d]pyrimidin-2-one, pyrimidin-2,4-dione, and 1,3,4-triazole 2-sulfide (Figure S8). For comparison, the combined single-point model substructure descriptors consistent with both activity and lack of cytotoxicity are 2-aryloxazole, thiazole 2-amide, 3-aminopropylpyrrole, 5-nitrofuran 2-amide, 5-nitrofuran 2-imine, 2-amino-5-thienyl 1,3,4-oxadiazole, 6-fluoro-8-alkoxyquinolin-4-one, and pyridine 4-carboxamide (Figure S9). Inactive features are sulfonamide, 1,2,4-triazole 2-sulfide, benzothiadiazole, 2-aminobenzamide, 3-hydroxy-1-pyrrol-2-one, benzoic acid, piperidine 1-amide, N-alkyl-2-(alkylamino)acetamide, 1,2,4-triazin-5-one, 1,2,4-triazole, and piperidine 4-carboxamide (Figure S10).
Testing Models with the ARRA dataset
With the demonstrated slightly enhanced or at least equivalent statistical robustness of the novel MLSMR, TAACF-CB2, TAACF-Kinase and combined models due to addition of the single-point inactives, we turned to assessing their predictive value with antitubercular datasets. The ARRA dataset consists of a set of 1924 whole-cell actives chosen as commercially available analogs of hits from the cumulative screening of >300,000 compounds 24. The ability of each new dual-event model to predict the activity (or lack of activity) of the ARRA compounds was quantified through an ROC AUC value. These were calculated to range from 0.62-0.83. These values are indicative of general improvements over the dose-response dual-event models (ROC AUC range 0.54-0.83) and the single-point single-event models (ROC AUC range 0.55-0.75) (Table 1). It is noteworthy that some compounds (up to 21%) were present in the model training set and the ARRA set and that this varied between models tested. These molecules were retained as otherwise the number in this set would vary from model to model.
Testing models with the GSK dataset
In 2013, GSK disclosed a set of 177 small molecule antitubercular lead compounds 26. Very few of these (≤ 10) compounds were present in any of the model training sets. Each model was then used to predict hits in the known GSK set: single-point models capture 38.4–58.7%, the dual-event models capture 18.6-48%, and the models incorporating dual-event dose-response and single-point data return 20.3-61% (Table 2). The GSK test set represents a useful test of the models, but since it only contains actives, an ROC AUC cannot be calculated. The best performing model was the TAACF-CB2 dual-event dose-response with single-point data. This model was not close to the test set as measured by the mean closest distance of training set to the GSK dataset. The second best model (combined single-point model) was ranked first based on this parameter, which is a measure of similarity for the test and training sets.
Table 2.
The number of molecules predicted as active out of 177 GSK 26 lead compounds (%) and mean closest distance (smaller is more similar) to training set. Out of the 177 GSK compounds, only a small number were in the models and were included in the table for ease of comparison.
| Mtb model (training set N) | Number of molecules predicted as active (Percent) | Mean closest distance of training set to test set | Rank by number predicted correctly | Rank by mean closest distance of training set to test set |
|---|---|---|---|---|
| MLSMR single point data (220463) | 100 (56.5) | 0.38 | 4 | 3 |
| TAACF-CB2 single point data (100100) | 102 (57.6) 2 in set | 0.46 | 3 | 5 |
| TAACF-kinase single point (23797) | 68 (38.4) 3 in set | 0.51 | 7 | 7 |
| Combined single point (344360) | 104 (58.7) 5 in set | 0.36 | 2 | 1 |
| MLSMR dose response and cytotoxicity (2273) a | 66 (37.3)* 5 in set | 0.50 | 8 | 6 |
| TAACF-CB2 dose response and cytotoxicity (1783) a | 85 (48)* 2 in set | 0.58 | 6 | 8 |
| TAACF-kinase dose response and cytotoxicity (1248) a | 33 (18.6)* 3 in set | 0.62 | 11 | 9 |
| Combined dose response and cytotoxicity (5304) a | 65 (36.7)* 10 in set | 0.46 | 9 | 5 |
| MLSMR dose response and cytotoxicity and negatives (218640) | 65 (36.7) 5 in set | 0.39 | 9 | 4 |
| TAACF-CB2 dose response and cytotoxicity and negatives (102634) | 108 (61.0) 2 in set | 0.46 | 1 | 5 |
| TAACF-kinase dose response and cytotoxicity and negatives (23737) | 36 (20.3) 3 in set | 0.51 | 10 | 7 |
| Combined dose response and cytotoxicity and negatives (345011) | 95 (53.7) 10 in test set | 0.37 | 5 | 2 |
Where: IC90 < 10 μg/ml (TAACF-CB2) or 10 μM and a selectivity index (SI) greater than ten were the SI is calculated from SI = CC50/IC90.
Assessing Mtb HTS Chemistry Property Space
The analysis of the test set compounds in this study using PCA mirrored our previous analysis of the much smaller dual-event datasets.21 In this case, the ARRA dataset of 1924 molecules is enclosed in the main cluster of the plot with the 345,011 compounds (Figure 2A). 74% of the variance was explained by first 3 principal components. The 177 GSK compounds were also predominantly enclosed within the main cluster, although a couple of molecules are outside of this cluster (Figure 2B); previously this dataset was shown to be well distributed amongst the combined dual-event dataset. 22 The ARRA data set was compared with the other 345,011 compounds using Murcko Assemblies (a published approach that can be used for library comparison 36), resulting in a Tanimoto similarity score of 0.47 (Table S2) suggesting that the datasets are dissimilar (a value close to 1 would be identical, and for our purposes a value less than 0.6 represents dissimilar). The GSK compounds were also compared with the 345,011-member dataset using Murcko Assemblies; the Tanimoto similarity score was again low at 0.13 (Table S3), indicating a greater dissimilarity to the training set than the ARRA dataset.
Figure 2.
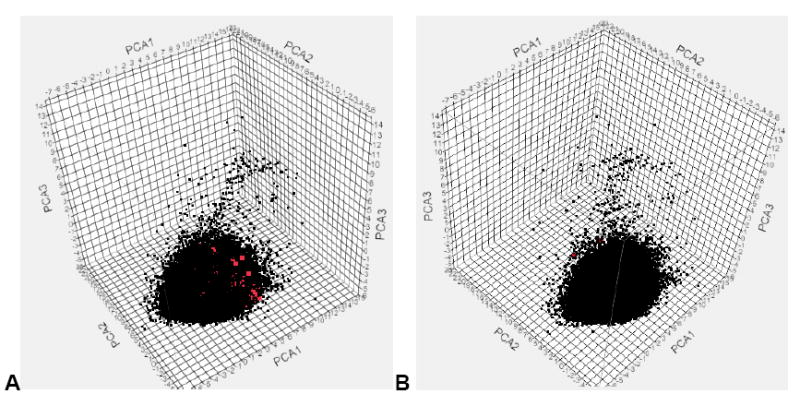
PCA. A. ARRA (red) and combined dose-response and cytotoxicity and single-point inactives (black), 74% of variance explained by first 3 PCs, B. 177 GSK (red) and combined dose-response and cytotoxicity and negatives (black), 74% of variance explained by the first 3 PCs.
Comparing actives and inactives using simple molecular descriptors
The mean value for each molecular descriptor used in the Bayesian model for the combined dose-response and cytotoxicity and single-point inactives dataset, was used to compare actives and inactives (Table 3). The molecular descriptors appeared to be normally distributed (Figure S11). AlogP, the number of rings, and the number of aromatic rings were all statistically higher (using the two tailed t-test) in the active compounds, while the number of hydrogen bond donors, the number of hydrogen bond acceptors and the fractional polar surface area were all statistically significantly lower in the actives.
Table 3.
Mean (Standard Deviation) of molecular descriptors for the combined dose-response and cytotoxicity and single-point dataset, comparing actives and inactives.
| MW | AlogP | HBD | HBA | Num Rings | Num Arom Rings | FPSA | RBN | |
|---|---|---|---|---|---|---|---|---|
| Active (1353) | 350.43 ± 67.73 | 3.84 ± 1.11 ** | 0.97 ± 0.84 ** | 3.93 ± 1.63 ** | 3.05 ± 0.96 * | 2.47 ± 0.84 ** | 0.24 ± 0.09 ** | 5.12 ± 2.18 |
|
| ||||||||
| Inactive (343658) | 353.12 ± 75.60 | 3.07 ± 1.33 | 1.14 ± 0.87 | 4.27 ± 1.62 | 2.96 ± 1.01 | 2.19 ± 0.93 | 0.25 ± 0.09 | 5.09 ± 2.24 |
MWT = molecular weight, HBD = hydrogen bond donor, HBA = Hydrogen bond acceptor, Num Rings = Number of Rings, Num Arom Rings = Number of Aromatic Rings, FPSA = Fractional polar surface area, RBN = rotatable bond number.
p < 0.05,
p < 0.0001
Fractional polar surface area = Total partially positively charged molecular surface area divided by the total molecular surface.
DISCUSSION
When generating computational machine learning models18, 20-25, 37, 38 or quantitative structure-activity relationship (QSAR) models,39 the assumption is that higher quality and well balanced datasets will usually yield the best models. Therefore, if given the choice, one would opt for using multipoint dose-response data over single-point screening data. In addition, one would generally expect that computational models containing the greatest number of molecules would likely be the most predictive for an external library of compounds, because they likely cover more chemical property space and they are likely more diverse. There are other factors to consider that involve assay details ranging from the culture medium used 40 to the mode of compound dispensing 41.41 In the domain of phenotypic screening,42 for each organism we face sizeable challenges when considering “ideal” in vitro assay conditions as well as the optimal computational model.
Traditionally, with QSAR and machine learning applied to tuberculosis, scientists have focused on relatively small datasets (<100 to several thousand compounds).16, 43 As more data has become available in the public domain,26, 44 we are faced with many questions around how we handle and use the accumulating comparably and relatively ‘massive’ (by comparison) datasets. While these datasets are really not ‘big data’ by today’s definition,45 they are far bigger than usually used for drug discovery computational modeling efforts.16, 43 Their size presents challenges for some of the algorithms used in terms of speed, processing requirements, and assessing data quality46-50. When is the training set for a model big enough? Is the model good enough? Is the model universal and predictive for all prospective compounds, or do limitations exist as to the relevant chemical or molecular property space covered? In essence, when are the models robust enough that further HTS will not add value considering the low hit rates and excessive cost? Trade-offs between data quantity, model predictivity and possibly cost are likely. Outside of this discussion is the separate opinion that in tuberculosis research we may possess sufficient random HTS identified hits to occupy many person-years for hit-to-lead optimization 10-12, 26. This concern is particularly important considering the downstream effort and expense to identify targets, carry out medicinal chemistry optimization 51 and bring novel and interesting leads through the full drug discovery pipeline 4.
We have already noted the lack of correlation between the ROC for a computational Mtb model, the mean closest similarity of test set molecules to the training set and its predictive capability.21, 22 We have confirmed this finding with novel bigger models trained with datasets combining dual-event (antitubercular efficacy and Vero cell cytotoxicity) dose-response actives and inactives with single-point screening inactives, all arising from the same screening workflow and the same laboratory. The predictive value of our novel models with respect to actives and inactives in the ARRA dataset 21 (as judged by ROC) and the GSK actives26 (as determined by the number of active hits correctly predicted) failed to correlate with measures of the similarity of the model training set with the test set. There does not appear to be any clear relationship between internal or external ROC with the number of molecules (Figure S12) or percent of actives (Figure S13) in the training set. Although we observe three Bayesian models that show a decrease in internal testing ROC values with increasing percent actives.
We have demonstrated both here and previously 22 via PCA that these external test sets overlap with the combined 345,011-member or the much smaller combined dual-event dose response training sets. This would suggest coverage of similar chemical property space. At the same time we may be able to extend beyond the property space of the much smaller individual model training sets based on the activity predictions for the GSK set and the relatively low mean closest distance metrics (and using Murcko assemblies). Thus, the machine learning models are able to correctly identify novel active antituberculars outside the chemical property space of our current HTS data. The limit of this ability to extend beyond the training set is currently being probed. However, there is still a need for a more in-depth understanding of the training set and model parameters that influence their predictive value with external datasets.
We have now explored the fusion of single-point screening data and dual-event dose-response data to assess whether addition of orders of magnitude more negative data can impact the predictive value of the Bayesian models. The dual-event dose-response model already significantly refines the concept of an active: a molecule with sufficient antitubercular efficacy as judged via an MIC or IC90 value in addition to its comparison to Vero cell cytotoxicity such that the SI is greater than or equal to 10. However, it may be asserted that a dual-event model based on solely dose-response data has a limited knowledge of inactives. For example, the SRI dose response datasets represent limited subsets (~1,200-5,000 compounds) derived from the actives in an initial single-point screen (~23,000-340,000 compounds) for antitubercular efficacy. Thus, addition of the single-point inactives to the dose response inactives should significantly enhance a model’s knowledge of antitubercular “inactivity.” The largest combined model we can create from these datasets has 345,011 molecules. These new models are enhanced with regard to their number of inactives (Table 1) and their coverage of chemical property space as assessed using PCA plots generated with all training data and the ARRA compounds or GSK actives (Figure 2), compared with a similar plot generated earlier with all dose-response compounds 22.
Our analyses in this study suggest that the models combining single-point and dual-event data are at least as good as the dual-event dose-response models based on internal testing (higher ROC range) and predicting outcomes with the ARRA dataset (narrower ROC range). We could not see a clear relationship between the internal or external ROC and the number of molecules or percent of actives in the model training set. Again, we suggest dataset dependencies. For example, the 177 GSK compounds have minimal overlap with the data used for modeling and the TAACF-CB2 models appear to perform consistently better than models trained with other datasets. The latter also has the highest percentage of actives in the dual-event training set.
We are not aware of any antitubercular screening models larger than our 345,011 compound-trained models that have been evaluated for tuberculosis or other neglected diseases in general. However, we can estimate that to date over 5 million compounds have been screened between the NIH funded efforts, GSK, Novartis, and other Bill and Melinda Gates Foundation supported projects. Unfortunately to date, only a small fraction of the data is publically available. We are not aware of any analysis of the total chemistry property space of compounds tested against Mtb to date. Our analysis of the largest model (Table 3) suggests that several simple molecular descriptors show statistically significant differences between actives and inactives, such as AlogP 52, 53. We have however shown previously that reliance on individual descriptors may not be adequate to predict antitubercular activity. 18, 23
Our data suggest the biggest models created are statistically comparable (based on ROC values) to the orders of magnitude smaller dual-event dose-response models. Possibly this result suggests that existing Bayesian models have maximally learned about molecular features that are inconsistent with sufficient whole-cell activity (and also relative Vero cell cytotoxicity) from the smaller datasets. This point should however be considered limited to the Bayesian approach and fingerprints used in this study, as we have not compared this approach with other machine learning algorithms or descriptors. It is likely that our results may not be extrapolated to other diseases or targets. Therefore, it may be useful to repeat this type of evaluation with data from malaria (Plasmodium spp.)44, 54-58 or other diseases for which there is now also plentiful phenotypic screening data. In addition, further assessment of the chemistry property space using more recent methods such as graph indices 59 may be of value for comparison to the simple PCA visualization. Of course these approaches could be repeated with different machine learning algorithms although we have shown little effect across models for this data previously. 22
In conclusion, the utilization of dose-response dual-event Bayesian models to select compounds from available libraries for prospective testing 19, 24, 25 is not diminished by increasing the model size to include available single-point screening data for inactive compounds. We propose this strategy for model development facilitates a greater understanding of the chemical features and physiochemical properties of inactives via single-point and dose-response data, while more tightly defining those for active compounds through solely dual-event data. While further training set optimization of its size and/or diversity may be of questionable value, the existing models can be used to reliably identify additional actives in unscreened libraries with success rates at least an order of magnitude better than current empirical methods. Future efforts will continue to explore the application of machine learning models to identifying novel antitubercular chemical probes, drug discovery hits and leads and use them to prioritize the thousands of hits already identified for in vivo testing 60. In addition we will endeavor to make these models accessible to the scientific community. 61 Our results may have further implications for not justifying further random HTS for Mtb as we have shown that we probably already have enough data that can be used to find new active molecules from other libraries using a focused testing strategy based on model predictions 24, 25, 62.
Supplementary Material
Acknowledgments
S.E. acknowledges colleagues at CDD. Accelrys are kindly acknowledged for providing Discovery Studio. The Bayesian models created in Discovery Studio are available from the authors upon written request.
The CDD TB has been developed thanks to funding from the Bill and Melinda Gates Foundation (Grant#49852 “Collaborative drug discovery for TB through a novel database of SAR data optimized to promote data archiving and sharing”).
S.E. and J.S.F acknowledges that the Bayesian models described were developed with support from Award Number R43 LM011152-01 and R44 TR000942-02 “Biocomputation across distributed private datasets to enhance drug discovery” from the National Library of Medicine.
J.S.F. acknowledges funding from Rutgers University – NJMS.
Abbreviations Used:
- AUC
area under the curve
- FCFP_6
molecular function class fingerprints of maximum diameter 6
- GSK
GlaxoSmithKline
- HTS
high-throughput screens
- MLSMR
Molecular Libraries Small Molecule Repository
- Mtb
Mycobacterium tuberculosis
- NIAID
National Institute of Allergy and Infectious Diseases
- PCA
Principal components analysis
- QSAR
Quantitative Structure Activity Relationship
- RP
Recursive partitioning
- SI
selectivity index
- SVM
Support Vector Machine
- TB
Tuberculosis
- ROC
receiver operator curve
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information Available: Supplemental information includes thirteen figures, three tables and supplemental data. This material is available free of charge via the Internet at http://pubs.acs.org.
Data and materials availability: All computational models are available from the authors upon request. All molecules used in the models are available in CDD.
Conflicts of Interest
SE is a consultant for Collaborative Drug Discovery, Inc.
References
- 1.Anon Global tuberculosis report 2013. http://www.who.int/tb/publications/global_report/en/
- 2.Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol. 2005;45:529–64. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 3.Ballell L, Field RA, Duncan K, Young RJ. New small-molecule synthetic antimycobacterials. Antimicrob Agents Chemother. 2005;49:2153–2163. doi: 10.1128/AAC.49.6.2153-2163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zumla AI, Gillespie SH, Hoelscher M, Philips PP, Cole ST, Abubakar I, McHugh TD, Schito M, Maeurer M, Nunn AJ. New antituberculosis drugs, regimens, and adjunct therapies: needs, advances, and future prospects. Lancet Infect Dis. 2014;14:327–340. doi: 10.1016/S1473-3099(13)70328-1. [DOI] [PubMed] [Google Scholar]
- 5.Ponder EL, Freundlich JS, Sarker M, Ekins S. Computational Models for Neglected Diseases: Gaps and Opportunities. Pharm Res. 2014;31:271–7. doi: 10.1007/s11095-013-1170-9. [DOI] [PubMed] [Google Scholar]
- 6.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–90. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 8.Payne DA, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Disc. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 9.Wei JR, Krishnamoorthy V, Murphy K, Kim JH, Schnappinger D, Alber T, Sassetti CM, Rhee KY, Rubin EJ. Depletion of antibiotic targets has widely varying effects on growth. Proc Natl Acad Sci U S A. 2011;108:4176–81. doi: 10.1073/pnas.1018301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maddry JA, Ananthan S, Goldman RC, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Reynolds RC, Secrist JA, 3, Sosa MI, White EL, Zhang W. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinb) 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ananthan S, Faaleolea ER, Goldman RC, Hobrath JV, Kwong CD, Laughon BE, Maddry JA, Mehta A, Rasmussen L, Reynolds RC, Secrist JA, 3, Shindo N, Showe DN, Sosa MI, Suling WJ, White EL. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds RC, Ananthan S, Faaleolea E, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Sosa MI, Thammasuvimol E, White EL, Zhang W, Secrist JA., 3rd High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2012;92:72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee RE, Protopopova M, Crooks E, Slayden RA, Terrot M, Barry CE., 3rd Combinatorial lead optimization of [1,2]-diamines based on ethambutol as potential antituberculosis preclinical candidates. J Comb Chem. 2003;5:172–87. doi: 10.1021/cc020071p. [DOI] [PubMed] [Google Scholar]
- 14.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–7. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 15.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DV, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011;10:188–95. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 16.Ekins S, Freundlich JS, Choi I, Sarker M, Talcott C. Computational Databases, Pathway and Cheminformatics Tools for Tuberculosis Drug Discovery. Trends in Microbiology. 2011;19:65–74. doi: 10.1016/j.tim.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prathipati P, Ma NL, Keller TH. Global Bayesian models for the prioritization of antitubercular agents. J Chem Inf Model. 2008;48:2362–70. doi: 10.1021/ci800143n. [DOI] [PubMed] [Google Scholar]
- 18.Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin B. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- 19.Ekins S, Casey AC, Roberts D, Parish T, Bunin BA. Bayesian Models for Screening and TB Mobile for Target Inference with Mycobacterium tuberculosis. Tuberculosis (Edinb) 2014;94:162–169. doi: 10.1016/j.tube.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekins S, Freundlich JS. Validating new tuberculosis computational models with public whole cell screening aerobic activity datasets. Pharm Res. 2011;28:1859–69. doi: 10.1007/s11095-011-0413-x. [DOI] [PubMed] [Google Scholar]
- 21.Ekins S, Freundlich JS, Hobrath JV, White EL, Reynolds RC. Combining Computational Methods for Hit to Lead Optimization in Mycobacterium tuberculosis Drug Discovery. Pharm Res. 2014;31:414–435. doi: 10.1007/s11095-013-1172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekins S, Freundlich JS, Reynolds RC. Fusing dual-event datasets for Mycobacterium Tuberculosis machine learning models and their evaluation. J Chem Inf Model. 2013;53:3054–63. doi: 10.1021/ci400480s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekins S, Kaneko T, Lipinksi CA, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Ernst S, Yang J, Goncharoff N, Hohman M, Bunin B. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis. Mol BioSyst. 2010;6:2316–2324. doi: 10.1039/c0mb00104j. [DOI] [PubMed] [Google Scholar]
- 24.Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models. PLOSONE. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekins S, Reynolds RC, Kim H, Koo MS, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian models leveraging bioactivity and cytotoxicity information for drug discovery. Chem Biol. 2013;20:370–8. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, Gonzalez R, Lozano S, Huss S, Santos-Villarejo A, Martin-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuinan-Blanco M, Lavandera JL, Perez-Herran E, Gamo-Benito FJ, Garcia-Bustos JF, Barros D, Castro JP, Cammack N. Fueling Open-Source Drug Discovery: 177 Small-Molecule Leads against Tuberculosis. ChemMedChem. 2013;8:313–21. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anon Collaborative Drug Discovery, Inc. http://www.collaborativedrug.com/register.
- 28.Ekins S, Gupta RR, Gifford E, Bunin BA, Waller CL. Chemical space: missing pieces in cheminformatics. Pharm Res. 2010;27:2035–9. doi: 10.1007/s11095-010-0229-0. [DOI] [PubMed] [Google Scholar]
- 29.Hohman M, Gregory K, Chibale K, Smith PJ, Ekins S, Bunin B. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Disc Today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Anon The PubChem Database. http://pubchem.ncbi.nlm.nih.gov/
- 31.Bender A, Scheiber J, Glick M, Davies JW, Azzaoui K, Hamon J, Urban L, Whitebread S, Jenkins JL. Analysis of Pharmacology Data and the Prediction of Adverse Drug Reactions and Off-Target Effects from Chemical Structure. ChemMedChem. 2007;2:861–873. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- 32.Klon AE, Lowrie JF, Diller DJ. Improved naive Bayesian modeling of numerical data for absorption, distribution, metabolism and excretion (ADME) property prediction. J Chem Inf Model. 2006;46:1945–56. doi: 10.1021/ci0601315. [DOI] [PubMed] [Google Scholar]
- 33.Hassan M, Brown RD, Varma-O’brien S, Rogers D. Cheminformatics analysis and learning in a data pipelining environment. Mol Divers 2006. 10:283–99. doi: 10.1007/s11030-006-9041-5. [DOI] [PubMed] [Google Scholar]
- 34.Rogers D, Brown RD, Hahn M. Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J Biomol Screen. 2005;10:682–6. doi: 10.1177/1087057105281365. [DOI] [PubMed] [Google Scholar]
- 35.Jones DR, Ekins S, Li L, Hall SD. Computational approaches that predict metabolic intermediate complex formation with CYP3A4 (+b5) Drug Metab Dispos. 2007;35:1466–75. doi: 10.1124/dmd.106.014613. [DOI] [PubMed] [Google Scholar]
- 36.Bemis GW, Murcko MA. The properties of known drugs 1. molcular frameworks. J Med Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 37.Periwal V, Rajappan JK, Jaleel AU, Scaria V. Predictive models for anti-tubercular molecules using machine learning on high-throughput biological screening datasets. BMC Res Notes. 2011;4:504. doi: 10.1186/1756-0500-4-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Periwal V, Kishtapuram S, Consortium OS, Scaria V. Computational models for in-vitro anti-tubercular activity of molecules based on high-throughput chemical biology screening datasets. BMC Pharmacol. 2012;12:1. doi: 10.1186/1471-2210-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura C, Latino DA, Martins F. Comparison of Multiple Linear Regressions and Neural Networks based QSAR models for the design of new antitubercular compounds. Eur J Med Chem. 2013;70:831–45. doi: 10.1016/j.ejmech.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 40.Franzblau SG, DeGroote MA, Cho SH, Andries K, Nuermberger E, Orme IM, Mdluli K, Angulo-Barturen I, Dick T, Dartois V, Lenaerts AJ. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis (Edinb) 2012;92:453–88. doi: 10.1016/j.tube.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Ekins S, Olechno J, Williams AJ. Dispensing processes impact apparent biological activity as determined by computational and statistical analyses. PLoS One. 2013;8:e62325. doi: 10.1371/journal.pone.0062325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng W, Thorne N, McKew JC. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today. 2013;18:1067–73. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekins S, Freundlich JS. Computational models for tuberculosis drug discovery. Methods Mol Biol. 2013;993:245–62. doi: 10.1007/978-1-62703-342-8_16. [DOI] [PubMed] [Google Scholar]
- 44.Gamo F-J, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera J-L, Vanderwall DE, Green DVS, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 45.Anon Big data. http://en.wikipedia.org/wiki/Big_data.
- 46.Southan C, Williams AJ, Ekins S. Challenges and Recommendations for Obtaining Chemical Structures of Industry-Provided Repurposing Candidates. Drug Disc Today. 2013;18:58–70. doi: 10.1016/j.drudis.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Williams AJ, Ekins S, Tkachenko V. Towards a Gold Standard: Regarding Quality in Public Domain Chemistry Databases and Approaches to Improving the Situation. Drug Disc Today. 2012;17:685–701. doi: 10.1016/j.drudis.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Williams AJ, Ekins S. A quality alert and call for improved curation of public chemistry databases. Drug Disc Today. 201116:747–750. doi: 10.1016/j.drudis.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Ekins S, Williams AJ. Meta-analysis of molecular property patterns and filtering of public datasets of antimalarial “hits” and drugs. MedChemComm. 2010;1:325–330. [Google Scholar]
- 50.Ekins S, Williams AJ. When Pharmaceutical Companies Publish Large Datasets: An Abundance Of Riches Or Fool’s Gold? Drug Disc Today. 2010;15:812–815. doi: 10.1016/j.drudis.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Dartois V, Barry CE., 3rd A medicinal chemists’ guide to the unique difficulties of lead optimization for tuberculosis. Bioorg Med Chem Lett. 2013;23:4741–50. doi: 10.1016/j.bmcl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldman RC. Why are membrane targets discovered by phenotypic screens and genome sequencing in Mycobacterium tuberculosis? Tuberculosis (Edinb) 2013;93:569–88. doi: 10.1016/j.tube.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Barry CE, 3, Slayden RA, Sampson AE, Lee RE. Use of genomics and combinatorial chemistry in the development of new antimycobacterial drugs. Biochem Pharmacol. 2000;59:221–31. doi: 10.1016/s0006-2952(99)00253-1. [DOI] [PubMed] [Google Scholar]
- 54.Derbyshire ER, Prudencio M, Mota MM, Clardy J. Liver-stage malaria parasites vulnerable to diverse chemical scaffolds. Proc Natl Acad Sci U S A. 2012;109:8511–6. doi: 10.1073/pnas.1118370109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ekland EH, Schneider J, Fidock DA. Identifying apicoplast-targeting antimalarials using high-throughput compatible approaches. FASEB J. 2011;25:3583–93. doi: 10.1096/fj.11-187401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plouffe D, Brinker A, McNamara C, Henson K, Kato N, Kuhen K, Nagle A, Adrian F, Matzen JT, Anderson P, Nam TG, Gray NS, Chatterjee A, Janes J, Yan SF, Trager R, Caldwell JS, Schultz PG, Zhou Y, Winzeler EA. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci U S A. 2008;105:9059–64. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Fourches D, Sedykh A, Zhu H, Golbraikh A, Ekins S, Clark J, Connelly MC, Sigal M, Hodges D, Guiguemde A, Guy RK, Tropsha A. Discovery of Novel Antimalarial Compounds Enabled by QSAR-Based Virtual Screening. J Chem Inf Model. 2013;53:475–92. doi: 10.1021/ci300421n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, Jimenez-Diaz MB, Martinez MS, Wilson EB, Tripathi AK, Gut J, Sharlow ER, Bathurst I, El Mazouni F, Fowble JW, Forquer I, McGinley PL, Castro S, Angulo-Barturen I, Ferrer S, Rosenthal PJ, Derisi JL, Sullivan DJ, Lazo JS, Roos DS, Riscoe MK, Phillips MA, Rathod PK, Van Voorhis WC, Avery VM, Guy RK. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–5. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fourches D, Tropsha A. Using graph indices for the analysis and comparison of chemical datasets. Mol Inf. 2013;32:2–17. doi: 10.1002/minf.201300076. [DOI] [PubMed] [Google Scholar]
- 60.Ekins S, Pottorf R, Reynolds RC, Williams AJ, Clark AM, Freundlich JS. Looking Back To The Future: Predicting In vivo Efficacy of Small Molecules Versus Mycobacterium tuberculosis. J Chem Inf Model. 2014;54:1070–82. doi: 10.1021/ci500077v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta RR, Gifford EM, Liston T, Waller CL, Bunin B, Ekins S. Using open source computational tools for predicting human metabolic stability and additional ADME/TOX properties. Drug Metab Dispos. 2010;38:2083–2090. doi: 10.1124/dmd.110.034918. [DOI] [PubMed] [Google Scholar]
- 62.Ekins S, Casey AC, Roberts D, Parish T, Bunin BA. Bayesian Models for Screening and TB Mobile for Target Inference with Mycobacterium tuberculosis. Tuberculosis (Edinb) 2014;94:162–9. doi: 10.1016/j.tube.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


