Abstract
Intracellular accumulation of wild type tau is a hallmark of sporadic Alzheimer's disease (AD). However, the molecular mechanisms underlying tau toxicity is not fully understood. Here, we detected mitophagy deficits evidenced by the increased levels of mitophagy markers, including COX IV, TOMM20, and the ratio of mtDNA to genomic DNA indexed as mt-Atp6/Rpl13, in the AD brains and in the human wild type full-length tau (htau) transgenic mice. More interestingly, the mitophagy deficit was only shown in the AD patients who had an increased total tau level. Further studies demonstrated that overexpression of htau induced mitophagy deficits in HEK293 cells, the primary hippocampal neurons and in the brains of C57 mice. Upon overexpression of htau, the mitochondrial membrane potential was increased and the levels of PTEN-induced kinase 1 (PINK1) and Parkin decreased in the mitochondrial fraction, while upregulation of Parkin attenuated the htau-induced mitophagy deficits. Finally, we detected a dose-dependent allocation of tau proteins into the mitochondrial outer membrane fraction along with its cytoplasmic accumulation. These data suggest that intracellular accumulation of htau induces mitophagy deficits by direct inserting into the mitochondrial membrane and thus increasing the membrane potential, which impairs the mitochondrial residence of PINK1/Parkin. Our findings reveal a novel mechanism underlying the htau-induced neuronal toxicities in AD and other tauopathies.
Keywords: tau, mitophagy, PINK1, Parkin, Alzheimer's disease, Gerotarget
INTRODUCTION
Mitochondria play crucial roles in energy production, the synthesis of key metabolites, apoptosis regulation, calcium buffering, and generation of endogenous reactive oxygen species [1, 2]. Since the neuronal activities, such as synaptic transmission, axonal transport, dendritic transport, ion channels and ion pump activity, are extremely energy-dependent [3], neurons are particularly sensitive to the changes of mitochondrial functions. Therefore, mitochondrial damage has been reported to be associated with many neurodegenerative diseases, including Alzheimer's disease (AD), Parkinson disease, and Huntington's disease [4-6].
The control of mitochondrial mass is through mitochondrial biogenesis and degradation. PINK1/parkin-dependent mitophagy contributes to mitochondrial quality control by selectively eliminating the dysfunctional mitochondria and maintain mitochondrial integrity and function [7]. PINK1 and the E3 ubiquitin-protein ligase parkin are two important protein mediators in the process of mitophagy. When the membrane potential of a mitochondrial segment depolarized, PINK1 accumulates on the outer mitochondrial membrane, then its kinase activity recruits parkin to the mitochondria [8, 9]. After mitochondrial residing, the parkin-mediated ubiquitination of mitochondrial substrates, such as voltage-dependent anion-selective channel protein (VDAC1) and mitofusins (MFN1/2), will lead to the recruitment of p62/SQSTRM and LC3 [10-12]. Finally, the mitochondrion whose membrane potential dissipated is engulfed into the autophagosome and eliminated through mitophagy.
Abnormal accumulation of wild type tau proteins is a hallmark of sporadic AD [13]. Expression of full-length human tau alone causes intracellular tau pathologies and behavioral deficits in mice [14, 15], while turning off htau expression attenuates the pathologies [16]. Reduction of endogenous tau also ameliorates memory deficits caused by β-amyloid (Aβ) [17]. However, how intracellular accumulation of the wild type tau impairs cells' function and eventually leads to neurodegeneration is currently not fully understood.
Mitochondrial dysfunction is an early pathological event of AD [18, 19], and abnormal mitochondrial morphology and distribution are detected in the postmortem AD brains and their fibroblasts [4, 20]. Recent studies suggest that there is an intrinsic link between tau and mitochondria. For instance, an N-terminal truncated tau (20-22kDa) is largely enriched in mitochondria of the AD brains and its amount in nerve terminal fields correlates with the pathological synaptic changes [21]. Mitochondrial dysfunction is also detected in P301L tau transgenic mice [22]. Deregulation of mitochondrial complex I with aging is tau dependent [23]. Tau phosphorylation can antagonize cell apoptosis with the mechanisms involving Bcl-2 and caspase-3 in mitochondria [24-26], while expression of fusion proteins attenuates apoptosis [27]. These observations suggest that intracellular accumulation of tau may cause neurodegeneration through disrupting mitochondrial functions, but the direct evidence for the role of wild type full-length tau in the mitophagy is still lacking.
In the present study, we show that intracellular accumulation of human wild type full-length tau, as seen in the sporadic AD brains, induces mitophagy deficits, with the mechanisms involving a direct insertion of htau into the outer membrane fractions of mitochondria, an increased mitochondrial membrane potential and an impaired mitochondrial residence of PINK1/Parkin.
RESULTS
Mitophagy deficit is associated with abnormal tau accumulation in the brains of AD patients and the AD-like htau transgenic mice
To explore whether mitophagy deficit is involved in AD pathogenesis, we analyzed the levels of mitophagy markers, including COX IV, TOMM20, and the ratio of mtDNA to genomic DNA, indexed as mt-Atp6/Rpl13 by Western blotting or real-time PCR. We found that levels of COX IV, TOMM20 and the ratio of mt-Atp6/Rpl13 were all significantly increased in the homogenates of AD brains (Figure 1A-1C), indicating increases of mitochondrial number. Interestingly, we observed that the mitophagy deficit seemed correlated with abnormal tau accumulation, i.e., the increase of COX IV and TOMM20 was only shown in those AD patients who had high level of tau proteins (AD group), not AD patients (AD1 group) whose total tau had no significant change compared with the nondemented control (Figure 1A-1C). The mitophagy deficits were also detected in the hippocampus of 6m-old htau transgenic mice (STOCK Mapttm1(EGFP)Klt Tg(MAPT)8cPdav/J) when compared with the age-matched littermates (Figure 1E, 1F), with significant tau accumulation (Figure 1D). The perinuclear mitochondrial accumulations were also shown by electron microscopy (unpublished data). These data suggest an intrinsic association of intracellular tau accumulation with mitophagy deficits.
Figure 1. Tau-correlated mitophagy deficits were found in the brains of AD patients and the htau transgenic mice.
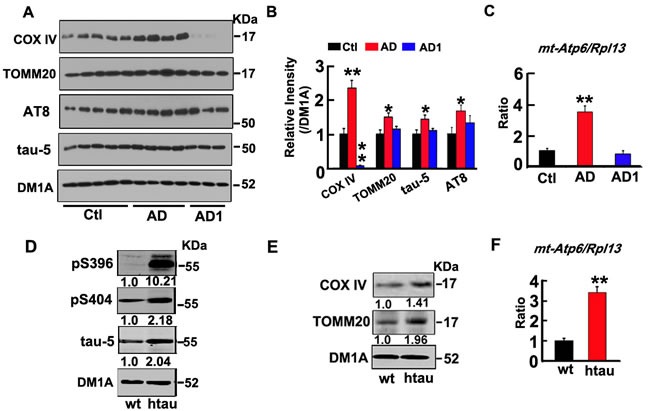
A., B. Compared with the age-matched non-AD controls, the levels of mitochondrial marker proteins (COX IV and TOMM20), total tau proteins (tau-5) and phosphorylated tau (AT8) were increased in the homogenates of AD brains measured by Western blotting. DM1A against tubulin was used as a loading control. C. The increased ratio of mt-Atp6/Rpl13 (mitochondria-encoded DNA/nucleus-encoded DNA) measured by real-time PCR. D. The increased total tau (tau-5), and phosphorylated tau (pS396, pS404), and E. mitochondrial marker proteins (COX IV and TOMM20) in htau transgenic mice (htau) compared with the age-matched wild type littermates (wt) measured by Western blotting. F. The increased ratio of mt-Atp6/Rpl13 in htau mice measured by real-time PCR. Data were expressed as mean±SD. *, p < 0.05, **, p < 0.01 vs Ctl (the control) or wt (age-matched wild type littermates).
Overexpression of htau induces mitophagy deficits both in vitro and in vivo
To verify the role of tau in mitophagy deficits, we overexpressed htau plasmid in HEK293 cells for 48 h (termed T293tau, Figure 2A, 2B), or the eGFP-labeled AAV2/8-htau in primary hippocampal neurons (7 div) for 48 h (Figure 2C, 2D), or in the hippocampal CA3 region of 3m-old c57 mice for 1 month by stereotaxic injection (Figure 2E, 2F), and the image of AAV2/8-eGFP transfected hippocampal CA3 region was shown as Supplementary Figure 1. Then, the levels of mitophagy markers together with tau proteins were measured by Western blotting. Compared with the vector transfected controls, the levels of COX IV, TOMM20 (Figure 2A-2F) and the ratio of mt-Atp6/Rpl13 (Figure 2G, 2H) were increased by overexpression of htau both in vitro and in vivo. We also found that the autophagy proteins Lc-3 II and p62 significantly increased (Supplementary Figure 2). Furthermore, a time-dependent increase of mt-Atp6/Rpl13 (~3.4-fold and 4.5-fold respectively at 48 h and 72 h) was detected in T293tau cells compared to the vector-transfected controls (T293vec) (Figure 2H). To confirm whether the increase of mitochondria is due to the mitophagy deficit or mitochondrial biogenesis, we detected mRNA levels of peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and mitochondrial transcription factor A (TFAM), two key transcription factors involved in the mitochondrial biogenesis [28, 29]. The results showed that overexpression of htau did not change the mRNA levels of PGC-1a and TFAM (Figure 2I). These are direct evidence showing that the htau accumulation blocks mitophagy.
Figure 2. Overexpression of htau induces mitophagy deficits in vitro and in vivo.
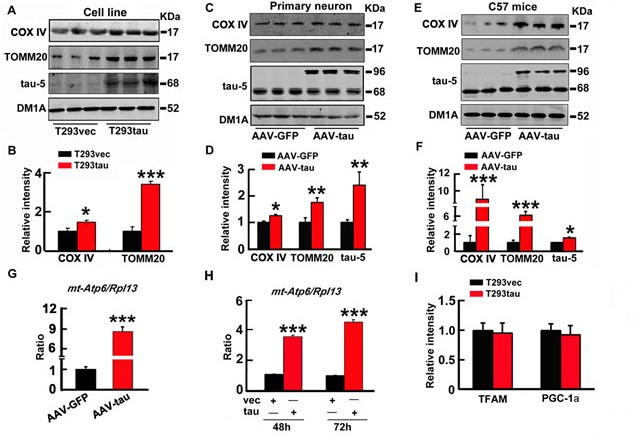
A., B. The human wild type full-length tau (htau) plasmid was transiently expressed in HEK293 cells for 48 h (T293tau), C., D. EGFP-labeled AAV-htau was overexpressed in primary hippocampus neurons (7 div) for 48 h, or E., F. The eGFP-labeled AAV-htau was stereotaxically infused into the hippocampal CA3 of 3m-old C57 mice for 1 month, and then, levels of mitochondrial marker proteins COX IV and TOMM20, and total tau (tau-5) in the cell or brain lysates were detected by Western blotting. G., H. The ratio of mt-Atp6/Rpl13 in the hippocampus of c57 mice (G) or HEK293 cells at 48 h or 72 h after serum deprivation (H) measured by real-time PCR. I. Human wild type full-length tau (T293tau) or the vector (T293vec) plasmid was transiently expressed in HEK293 cells for 48 h, then the mRNA levels of PGC-1α (peroxisome proliferator-activated receptor γ coactivator 1α) and TFAM (mitochondrial transcription factor A) were measured by real-time PCR. Each experiment was repeated for at least 3 times, and data were expressed as mean±SD. *, p < 0.05; **, p < 0.01; ***, p < 0.001 vs T293vec or AAV-GFP.
Overexpression of htau increases mitochondrial membrane potential with mitophagy deficits and a decreased mitochondrial residence of PINK1/Parkin
The membrane potential is critical for the mitochondrial residence of PINK1/Parkin and the subsequent ubiquitination-associated mitophagy [7-12]. Therefore, we measured the mitochondrial membrane potential. Unexpectedly, we observed a significant increase of mitochondrial membrane potential in the cells with overexpression of htau (Figure 3A). To confirm the influence of htau on mitochondrial membrane potential, we used carbonyl cyanide m-chlorophenylhydrazone (CCCP), a mitochondrial membrane depolarizer that can decrease mitochondrial membrane potential and thus causes proteolysis of the mitochondrial proteins by mitophagy [30]. We observed that CCCP treatment remarkably decreased the membrane potential in control cells, but only a slight reduction was shown in the htau-expressing cells measured by JC-1 or tetramethyl rhodamine methyl ester (TMRM) (Figure 3A, 3B). We also found that levels of COX IV and TOMM20 were significantly decreased, while LC-3 II significantly increased after CCCP treatment in the control cells, whereas only a slight reduction of COX IV and TOMM20 was seen in the htau-expressing cells (Figure 3C, 3D, and Supplementary Figure 3). Similarly, CCCP treatment reduced mtDNA in both control and htau transfected cells, but the reduction in the tau-expressing cells was much less than the control cells (Figure 3E). The mitochondrial membrane potential affects the residence of PINK1 and Parkin in mitochondria [7-12]. With an increased mitochondrial membrane potential by htau accumulation, we observed a diminished PINK1 and a significantly decreased level of Parkin in the mitochondrial fraction (Figure 3F). These data together suggest that htau accumulation may induce mitophagy deficits by increasing mitochondrial membrane potential and disrupting the mitochondrial residence of PINK1/Parkin.
Figure 3. Tau increases mitochondrial membrane potential and induces mitofusin accumulation.
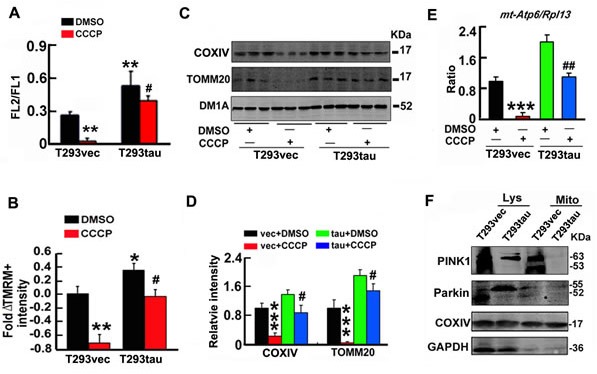
A. T293tau or T293vec cells were treated with CCCP (20 μM) or DMSO (vehicle control) for 30 min, then the membrane potential (FL2/FL1) was measured by JC-1 staining (n = 10). B.T293tau or T293vec cells were incubated with TMRM (20 nM) for 30 min, and then the fold ΔTMRM+ intensity (ΔΨm) was analyzed. C.-E. Levels of mitochondrial marker proteins (COX IV and TOMM20) and ratio of mt-Atp6/Rpl13 were detected by Western blotting (C, D) and real-time PCR (E), respectively, in T293tau and T293vec cells after treatment with CCCP (20 μM for 30 min). (F) Levels of PINK1 and Parkin in cell lysates (Lys) and mitochondria (Mito) fractions measured by Western blotting. Data were expressed as mean±SD. *, p < 0.05; **, p < 0.01, ***, p < 0.001 vs T293vec+DMSO; #, p < 0.05, ##, p < 0.01 vs T293tau+DMSO.
Upregulating parkin rescues the htau-induced mitophagy deficits
To validate the role of the reduced Parkin in htau-induced mitophagy deficits, we overexpressed wild-type parkin (PARK2) in T293tau cells. The results showed that simultaneous upregulation of Parkin attenuated the htau-induced mitophagy impairment (Figure 4A-4C), which confirm the role of Parkin reduction in the htau-induced mitophagic deficits. We also found that total tau and phosphorylated tau levels had no significant change while parkin overexpression (Supplementary Figure 4).
Figure 4. Up-regulating parkin rescues the htau-induced mitophagy deficits.
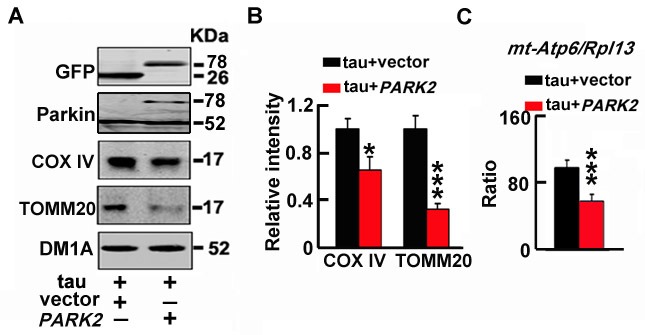
A., B. T293tau cells were transfected with wild type parkin (PARK2) plasmid for 48 h, and then the mitochondrial marker proteins (COX IV and TOMM20) were measured by Western blotting. C. The ratio of mt-Atp6 and Rpl13 was assessed by real time PCR. Each experiment was repeated at least 3 times. The data are expressed as mean±SD. *, p < 0.05, ***, p < 0.001 vs vec.
Overexpression of htau increases tau levels in mitochondrial outer membrane fraction
To further explore whether tau directly interacts with mitochondria, we first measured the level of tau in the mitochondrial fraction after transient expression of tau in HEK293 cells. Interestingly, the total tau (tau-5), unphosphorylated tau at tau-1 epitope and the phosphorylated tau at Ser262 (located at the microtubule binding domain of tau) were detected in the mitochondrial fraction, while the C-terminal Ser396- and Ser404-phosphorylated tau were not detected (Figure 5A). Immunofluorescence staining data show co-localization of tau-1 epitope tau with MitoTracker Red in the elongated mitochondria (Figure 5B). Furthermore, N-terminal tau-1 epitope tau was co-immunoprecipitated with mitochondrial protein OPA1 and Mfn1 (Figure 5C). We confirmed the above results in htau transgenic mice by showing tau-1- and tau-5-postive tau proteins in the mitochondrial fraction (Figure 5D), and tau-1 co-immunoprecipitation with OPA1 and Mfn1 (Figure 5E). These data provide strong evidence for a direct interaction of tau with mitochondria.
Figure 5. A direct interaction of tau with mitochondrial marker proteins.
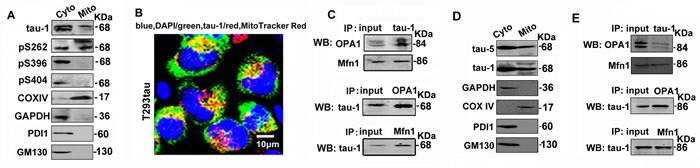
A. The cytoplasmic (Cyto) and mitochondrial (Mito) fractions were prepared from T293tau cells, and tau was analyzed by using a panel of phosphorylation site-specific antibodies, including tau-1 (reacts with the unphosphorylated tau at Ser198/199/202), pS262, pS396 and pS404. B. The representative image shows co-staining of tau-1 (green) with MitoTracker Red in T293tau cells. C. Co-immunoprecipitation data show association of tau with mitochondrial marker proteins OPA1 and Mfn1. D. The cytoplasmic (Cyto) and mitochondrial (Mito) fractions were prepared from the hippocampus of htau transgenic mice, and tau was analyzed by using anti-tau-1 antibody. E. Co-immunoprecipitation data show association of tau with mitochondrial marker proteins OPA1 and Mfn1 in hippocampal extracts of htau transgenic mice. GAPDH is a marker of cytoplasmic proteins, while TOMM40 and COXIV are respectively markers of mitochondrial outer membrane, inner membrane and intermembrane space proteins. PDI or GM130 is the marker of ER or Golgi apparatus used to prove the purity of the mitochondrial fraction.
To identify the relationship of mitochondrial location of tau with its cytoplasmic accumulation in neural cells, we first transiently expressed htau in a selected Neuro 2a (N2a) cell line, which expresses negligible endogenous tau protein (Figure 6A). With the cytoplasmic tau accumulation, apparent tau signals were detected in the mitochondrial fraction (Figure 6A). To better mimic the AD-like intracellular tau accumulation, we transfected two different dosages of htau plasmid into another N2a cell line that expresses endogenous tau proteins [24]. Along with a dose-dependent cytosolic accumulation of tau, exogenous overexpression of htau increased the mitochondrial levels of total tau (tau-5), phosphorylated tau at pT205, pS214, and pS262, and the unphosphorylated tau at tau-1 epitope with major molecular mass of ~90 KDa and ~55 KDa (Figure 6B). These data confirm that intracellular tau accumulation can be dose-dependently allocated into the mitochondria.
Figure 6. A dose-dependent allocation of tau into the mitochondria accompany with the intracellular tau accumulation.
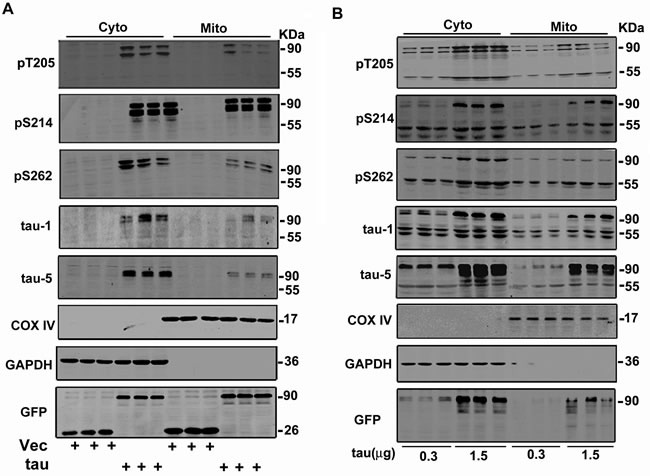
A. Cytoplasmic expression of htau in a N2a cell line that only expresses negligible endogenous tau protein causes mitochondrial location of tau proteins. B. Cytoplasmic overexpression of different concentrations of htau in a N2a cell line that expresses low level of endogenous tau protein causes a dose-dependent elevation of mitochondrial tau protein.
To validate the topology of tau in mitochondria, we treated the purified mitochondria with a high-pH wash (Na2CO3). Like cytochrome c (cyt c), a protein marker of mitochondrial intermembrane space, the majority of tau-1 reactive tau was dissolved into the supernatant fraction after Na2CO3 treatment, suggesting the mitochondrial intermembrane space location of the majority unphosphorylated tau (Figure 7A). Trypsin can digest mitochondrial outer membrane proteins, such as TOMM40, while trypsin plus Triton-X100 can remove the mitochondrial inner membrane proteins, such as COX IV [31]. We found that treating the purified mitochondria with trypsin alone completely removed tau proteins, as seen in TOMM40 (Figure 7B). These data strongly suggest that tau may span on the mitochondrial outer membrane with some domains exposing to the inter-membrane spaces.
Figure 7. Tau may span on the mitochondrial outer membrane with some domains exposing to the inter-membrane spaces.
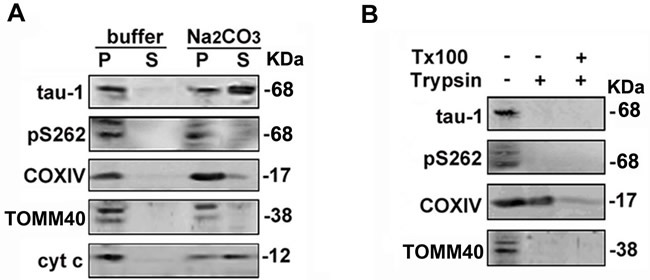
A. The mitochondrial fractions prepared from T293tau cells were re-suspended in buffers with or without Na2CO3 (0.1 mol/L, pH 11.5) followed by centrifugation, and the supernatants (S) or membrane pellets (P) were detected with the indicated antibodies by Western blotting. B.Mitochondrial fractions were digested with trypsin in the absence or presence of 1% Triton X-100 (Tx-100) for 30 min or with mock control, and the protein level of tau was analyzed. TOMM40, COXIV and cyt c are respectively markers of mitochondrial outer membrane, inner membrane and intermembrane space proteins.
DISCUSSION
Intracellular accumulation of wild type tau is the major cause of neurodegeneration in sporadic AD; however, the mechanism is not fully elucidated. Tau aggregation is associated with mitochondrial damage, oxidative stress as well as structural and functional alterations of neurons in AD [32-34]. As efficient mitochondria clearance by mitophagy is a key mechanism for mitochondrial quality control, here, we investigated the effects of tau on mitophagy and the molecular mechanisms. We find that intracellular accumulation of human wild type full-length tau, as seen in sporadic AD brains, results in mitophagy deficits. Although mitochondrial abnormality has been observed in AD patients and in transgenic mice expressing P301L mutated tau (the latter is only seen in patients of FTDP-17 but not AD), and the N-terminal truncated tau proteins were detected in mitochondria [18-23], our current study provides the first direct evidence showing that intracellular accumulation of wild type full-length tau inhibits mitophagy, which reveal a novel mechanism underlying tau-induced neurodegeneration in sporadic AD patients.
The mitophagy deficits can be evaluated by accumulation of mitochondrial marker proteins and the increase of mtDNA [35]. In neurons, autophagosomes actively form along neurites and in synapses but efficient clearance of the autophagic vesicles requires their retrograde transport towards the neuronal cell body, where the lysosomes are enriched. Microtubules serve as tracks for the intracellular transportations and tau accumulation blocks the transport. We speculate that the increased htau may block transport of the autophagosomes, thus lead to a massive accumulation of the impaired mitochondria into somatodendritic compartment of the cells, which is consistent with the reports that damaged mitochondria with altered morphology per neuronal perikarya were significantly increased in AD brains [36]. Mitochondrial fusion and fission also play a crucial role in mitophagy, in which, fission has been considered as a prerequisite for mitophagy [37, 38], because autophagy cargos with a large size cannot be efficiently engulfed by phagophores [7, 39-41]. A previous study in drosophila and fibroblast showed that overexpression of human R406W mutant tau could elongate mitochondria with the mechanisms involving a reduced fission [42]. The effect of wild type full-length tau accumulation on mitochondrial fusion or fission deserves further investigation.
The PINK1/Parkin signaling is critical for degradation of the mitochondrial fusion proteins and a selective mitophagy processing [7-12]. Parkin is responsible for ubiquitinating substrates on the mitochondrial outer membrane and subsequent recruitment of autophagy machinery [8-10]. We found that reduced PINK1 and Parkin in the mitochondrial fraction of the htau-transfected cells. Although the mechanisms underlying PINK1 recruiting Parkin to the mitochondria are not fully understood, Mfn2 phosphorylation by PINK1 was considered as a key step [43]. The important role of Parkin deficits in htau-induced mitochondrial impairments was further demonstrated by up-regulating Parkin, which rescued the htau-induced mitophagy deficits. Previous studies reported that overexpression of Parkin decreased intracellular Aβ levels and extracellular plaque deposition, attenuated caspase activity, prevented mitochondrial dysfunction and oxidative stress and restored neurotransmitter synthesis [36, 44]. Here, we show that overexpression of Parkin attenuates the htau-induced mitophagy deficits, suggesting that parkin may serve as a potential target for AD therapy.
A previous study showed that overproduction of β-amyloid reduced membrane potential [45]. It was also reported that the triple transgenic AD mice or the aged P301L tau transgenic mice exhibited mitochondrial dysfunction but with no changes on mitochondria membrane potential [23, 24]. In the current study, we observed unexpectedly a significantly increased mitochondrial membrane potential upon overexpression of htau. Furthermore, treatment with CCCP significantly reduced the membrane potential in control cells but not to the same levels in htau expressing cells. Low membrane potential is required for the mitochondrial residency of PINK1/Parkin [30], therefore, the increased membrane potential may underlie the htau-induced reduction of PINK1/Parkin in mitochondria. Since the mitochondrial tau level was significantly increased accompanying with a dose-dependent overexpression of htau, we speculate that tau may increase the membrane potential by direct insertion into the outer membrane fraction. Interestingly, we only detected N-terminal tau but not C-terminal phospho-tau in the mitochondria. A most recent study detects N-terminal tau in the mitochondrial membrane fraction [46], which supports our findings of the increased membrane potential by the mitochondrial tau insertion. Mitochondrial abnormality is undoubtedly associated with the pathogenesis of AD. There are some contradictory studies about the levels of mitochondrial proteins and mtDNA in the AD brains. It was shown that COX activity in post mortem AD brains was decreased [47, 48]. Moreover, the reduced cellular expression of COX subunit II and IV more pronounced in AD [49]. Hirai et al also found that some neurons showing increased oxidative damage have a striking and significant decrease in mtDNA and cytochrome oxidase in the brain of AD patients. However, they also found that pyramidal neurons show increased mtDNA and cytochrome oxidase in the hippocampus of AD [50]. In the present study, we found that tau accumulation induced mtDNA and mitochondrial proteins increased. Why the mitochondrial study in the AD had these contradictory reports, we speculated that, though Aβ and tau aggregation are associated with mitochondrial damage, oxidative stress as well as structural and functional alterations of neurons in AD [32-34], it seems that Aβ and tau proteins induces mitochondrial dysfunction and mitochondrial number with different mechanisms, which waiting for further study.
In non-apoptotic cells, JC-1 shows green color in cytoplasm (maximal excitation 510 nm, maximal emission 527 nm) and it turns red in mitochondria (maximal excitation 585 nm, maximal emission 590 nm). Treatment with 10 μM CCCP for 20 min can significantly reduce the red/green ratio, suggesting decrease of membrane potential [51]. However, we did not see significant change of JC-1 staining in htau-expressing cells by using 10 μM CCCP for 20 min, thus we used 20 μM CCCP for 30 min. Mitochondrial fusion occurred in daughter-mitochondria that show high membrane potential [52]. We speculate that expression of htau may increase mitochondrial fusion, which may explain why we do not see significant reduction of the membrane potential in htau-expressing cells when low concentration of CCCP was used.
Taken together, we find in the present study that intracellular accumulation of human wild type full-length tau, as seen in sporadic AD brains, induces mitophagy deficits with the mechanisms involving in causal-correspondently an increased mitochondrial membrane potential and an impeded mitochondrial residency of PINK1/Parkin.
MATERIALS AND METHODS
Plasmids, antibodies and reagents
Human pIRES-eGFP-Tau40 plasmid was a gift of Dr. Khalid Iqbal (New York State Institute for Basic Research in Developmental Disabilities, Staten Island, NY). The PCI-neo-Tau40 was constructed in our lab. For GFP-parkin (PK2, the full-length mouse parkin cDNA) was cloned into pEFGP-C1 plasmid (Clontech). EGFP-labeled AAV2/8-htau and its control virus were purchased from OBio Biologic Technology Co., Ltd.
Mouse monoclonal antibody (mAb) anti-OPA1 and anti-GM130 were from BD Bioscience; mAb anti-Mfn1 was from Santa Cruz; mAb anti-COXIV, anti-GAPDH, anti-Cytc, anti-GFP, anti-PDI1 and pAb anti-ubiquitin, anti-PINK1, anti-Parkin, anti-Mfn2, anti-SQSTM/p62, anti-LC and anti-TOMM40 were from Abcam; mAb anti-α-tubulin was from Sigma; two kinds of mAb anti-tau-5 were from NeoMarkers or Millopore, mAb tau-1 was from Chemicon, pAbs against phosphorylated tau (pT205, pS214, pS262, pS396 or pS404) were from BioSource; mAb AT8 (tau phosphorylated at Ser202 and Thr205) was from Thermo Fisher Scientific, pAb anti-TOMM20 was from Anbo Biotechnology, CCCP/JC-1 was from Sigma; Lipofectamine2000 and TMRM was from Invitrogen.
Cell culture
The human embryonic kidney293 (HEK293) were grown in Dulbecco's Modified Eagle's medium (DMEM medium) (12491-015, Gibco), supplemented with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin, in a humid 5% CO2 incubator at 37°C. After growing 24 h in plates or flasks, the cells were transfected with the indicated plasmid(s) using Lipofectamine2000 according to the manufacturer's instructions.
For primary neuron cultures, 18 days embryonic (E18) rat hippocampus were seeded at 30,000-40,000 cells per well on 6-well plates coated with Poly-D-Lysine/Laminin (Bioscience) in neurobasal medium (Invitrogen) supplemented with 2% B27/0.5 mM glutamine/25 mM glutamate. Half the culture medium was changed every 3 days with neurobasal medium supplemented with 2% B27 and 0.5 mM glutamine. All cultures were kept at 37°C in a humidified 5% CO2 containing atmosphere. More than 90% of the cells were neurons after they were cultured for 7 to 17 div; this was verified by positive staining for the neuronal specific markers microtubule-associated protein-2 (MAP2, dendritic marker, Millipore). At 7 to 10 div, neurons were transfected with tau plasmids and mito-DsRed2 2:1 using NeuroFECTTM according to the manufacturer's protocol.
Human tissue samples
Post-mortem brain samples were dissected from frozen brains of 7 AD cases (age 72.3±11.7 years, means ±s.d.) and 5 nondemented controls (ages 79.3±11.0 years) from the Emory Alzheimer's Disease Research Center. The study was approved by the Biospecimen Committee. AD was diagnosed according to the criteria of the Consortium to Establish a Registry for AD and the National Institute on Aging. Diagnoses were confirmed by the presence of amyloid plaques and neurofibrillary tangles in formalin-fixed tissue. Informed consent was obtained from the subjects.
The mitochondrial membrane potential assay
The mitochondrial membrane potential (Δψm) was assayed by JC-1 following the previous procedure [53]. Briefly, the cells were grown on a 96-well plate to ~70% density, and then incubated for 20 min with 5 mg/ml JC-1 in culture mediums. The red and green fluorescence were detected with the BioTek multifunctional microplater (Ascent Fluoroscan, Tecan, Durham, NC). The ratio of red (excitation 550 nm, emission 600 nm) to green fluorescence (excitation 485 nm, emission 535 nm) (FL2/FL1) was calculated in triplicates.
HEK293 cells were incubated with low concentrations (20 nM) of TMRM+ in experimental medium for 30 min. At equilibrium, the fluorescence produced by this low concentration of TMRM+ (excitation 543/emission red photomultiplier channel) is a direct function of Δψm, and complications attributable to self-quenching of the dye are eliminated [54, 55]. Dye intensities in images were quantified by measuring the raw pixel intensities in cells somas with the region tool of MetaMorph software. The dye intensity measured in each cell was normalized to the average dye intensity of control cells receiving the same concentration of dye for the same period as the experimental cells. Normalized data are shown as fold change from the average intensity of the dye measured in control cultures at the same time.
Preparation of mitochondria
Mitochondria were isolated as described [56]. Briefly, HEK293 cells were washed two times with Phosphate-Buffered Saline (PBS), and resuspended in ice cold permeabilization buffer containing 200 mM mannitol, 70 mM sucrose, 1 mM EGTA, 10 mM Hepes, 1 mM PMSF (Sigma) and protease inhibitor cocktail (Sigma). Cells were homogenized on ice with a 1 ml Insulin syringe 27G1/2, drawing through the needle 20 times. Following centrifugation at 600 g at 4°C for 10 min, the supernatant was collected and centrifuged at 8,000 g at 4°C for 15 min. The supernatant containing cytoplasmic proteins was collected in 12.5% trichloroacetic acid (Sigma) and stored at −20°C. The pellet (mitochondrial fraction) was washed two times with permeabilization buffer and the mitochondria were lysed with 1×cell lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100), plus 1 mM PMSF and protease inhibitor cocktail.
The mitochondrial topology assay of tau
To examine the topology of tau in mitochondria, the isolated mitochondria were resuspended in sodium carbonate (pH 11.5) or the lysis buffer and incubated on ice for 30 min, and then centrifuged for 5 min at 20,000×g, the supernatant were collected in 12.5% trichloroacetic acid. For trypsin digestion, the isolated mitochondria (60 μg) were incubated with trypsin (60 μg/ml) for 30 min on ice in a final volume of 200 μl in the presence or absence of 1% triton X-100. The reaction was stopped by addition of 10 μl of trypsin inhibitor (10 mg/ml stock), or separation of the organelles from the trypsin solution by centrifugation. The resulting pellets were then resuspended in lysis buffer, briefly sonicated prior to determining the protein concentration using the BCA assay (Pierce) with bovine serum albumin as standard. Equal amount of protein (20 μg) from each reaction was analyzed by Western blotting. The antibodies against, cytochrome c oxidase (COXIV) and cyt c were used as markers of the outer mitochondrial membrane, inner membrane proteins and intermembrane space protein markers, respectively [31].
Western blotting
Equal amounts of protein were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. The membranes were blocked in 5% non-fat milk for 1 h at room temperature and then incubated with primary antibody at 4°C overnight. Then the blots were incubated with IRDye 800CW-conjugated affinity-purified anti-mouse IgG (Rockland) and IRDye 800CW anti-rabbit IgG secondary antibody (Rockland) for 1 h at room temperature. Immunoreactive bands were visualized using the Odyssey Infrared Imaging System (Licor Biosciences, Lincoln, NE, USA).
Reverse transcription and real-time quantitative PCR
Reverse transcription and real-time quantitative PCR were carried out according to manufacturer's instruction (TaKaRa, Dalian, China). The PCR system contains 3 mM MgCl2, 0.5 μM forward and reverse primers, 2 μl SYBR Green PCR master mixes and 2 μl cDNA, and the standards for each gene were prepared using appropriate primers by a conventional PCR. The samples were assayed on a Rotor Gene 300 Real-time Cycler (Corbett Research, Sydney, Australia). The expression level of the interest gene was normalized by the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was not changed by the treatments. PCR primers employed in the present study are as follow:
Atp6 forward and reverse primers, 5′-TTTCCCCCTCTATTGATCCC-3′ and 5′-GTGGCCTTGGTATGTGCTTT-3′; Rpl13 forward and reverse primers, 5′-CCGGCATTCACAAGAAGGTG-3′ and 5′-CGAGCTTTCTCCTTCTTATAGACGT-3′; GAPDH forward primer 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse primer 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Ethics statement
Investigation has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki and according to national and international guidelines and has been approved by the authors' institutional review board.
Statistical analysis
All data were collected and analyzed in a blinded manner. Data were expressed as means±SD and analyzed using SPSS 10.0 statistical software (SPSS Inc. Chicago, IL, USA). P-values were calculated with unpaired Student's t-test (two-tailed, type 2) for one-way comparisons and with ANOVA followed by post hoc test (Student's t-test, two-tailed, type 2) for multiple comparisons.
SUPPLEMENTARY MATERIAL FIGURES
Acknowledgments
This work was supported in parts by National Natural Science Foundation of China (81171195, 91132305, 81271402, 81261120570 and 81528007).
Abbreviations
- AD
Alzheimer's disease
- htau
wild type human full-length tau protein
- Aβ
β-amyloid
- Mfn1
mitofusin 1
- Mfn2
mitofusin 2
- OPA1
optic atrophy type 1
- DLP1
dynamin like protein 1
- PINK1
PTEN-induced kinase 1
- HEK293
human embryonic kidney 293
- DMEM
Dulbecco's Modified Eagle's medium
- IP
immunoprecipitation
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- TMRM
etramethyl rhodamine methyl ester
- PGC-1α
peroxisome proliferator-activated receptor coactivator 1α
- TFAM
mitochondrial transcription factor A
Footnotes
CONFLICTS OF INTEREST
All authors disclose: (a) no actual or potential conflicts of interest including any financial, personal or other relationships with other people or organizations within three years of beginning the work submitted that could inappropriately influence (bias) their work. (b) When applicable, provide statements verifying that appropriate approval and procedures were used concerning animals.
Author contributions
J.Z.W., and G.P.L. designed research; Y.H., X.C.L., Z.H.W., Y.L., X.P.L., Q.F., X.Z. and Q.W. performed experiments; X.C.L., Y.H., Z.C., Z.Y., K.Y., G.P.L., and J.Z.W. analyzed data. J.Z.W., G.P.L., and K.Y., wrote the paper.
REFERENCES
- 1.Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, Rossignol R. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 2.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 3.Kann O, Kovacs R. Mitochondria and neuronal activity. Am J Physiol Cell Physiol. 2007;292:C641–C657. doi: 10.1152/ajpcell.00222.2006. [DOI] [PubMed] [Google Scholar]
- 4.Zhu X, Perry G, Moreira PI, Aliev G, Cash AD, Hirai K, Smith MA. Mitochondrial abnormalities and oxidative imbalance in Alzheimer disease. J Alzheimer's Dis. 2006;9:147–153. doi: 10.3233/jad-2006-9207. [DOI] [PubMed] [Google Scholar]
- 5.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 6.Reddy PH, Mao P, Manczak M. Mitochondrial structural and functional dynamics in Huntington's disease. Brain Res Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andorfer C, Kress Y, Espinoza M, de Silva R, Tucker KL, Barde YA, Duff K, Davies P. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J Neurochem. 2003;86:582–590. doi: 10.1046/j.1471-4159.2003.01879.x. [DOI] [PubMed] [Google Scholar]
- 15.Van der Jeugd A, Ahmed T, Burnouf S, Belarbi K, Hamdame M, Grosjean ME, Humez S, Balschun D, Blum D, Buée L, D'Hooge R. Hippocampal tauopathy in tau transgenic mice coincides with impaired hippocampus-dependent learning and memory, and attenuated late-phase long-term depression of synaptic transmission. Neurobiol Learn Mem. 2011;95:296–304. doi: 10.1016/j.nlm.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Boutajangout A, Quartermain D, Sigurdsson EM. Immunotherapy targeting pathological tau prevents cognitive decline in a new tangle mouse model. J Neurosci. 2010;30:16559–16566. doi: 10.1523/JNEUROSCI.4363-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vossel KA, Zhang K, Brodbeck J, Daub AC, Sharma P, Finkbeiner S, Cui B, Mucke L. Tau reduction prevents Abeta-induced defects in axonal transport. Science. 2010;330:198. doi: 10.1126/science.1194653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Su B, Fujioka H, Zhu X. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer's disease patients. Am J Pathol. 2008;173:470–482. doi: 10.2353/ajpath.2008.071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baloyannis SJ. Mitochondrial alterations in Alzheimer's disease. J Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- 21.Amadoro G, Corsetti V, Stringaro A, Colone M, D'Aguanno S, Meli G, Ciotti M, Sancesario G, Cattaneo A, Bussani R, Mercanti D, Calissano P. A NH2 tau fragment targets neuronal mitochondria at AD synapses: possible implications for neurodegeneration. J Alzheimers Dis. 2010;21:445–470. doi: 10.3233/JAD-2010-100120. [DOI] [PubMed] [Google Scholar]
- 22.David DC, Hauptmann S, Scherping I, Schuessel K, Keil U, Rizzu P, Ravid R, Dröse S, Brandt U, Müller WE, Eckert A, Götz J. Proteomic and functional analyses reveal a mitochondrial dysfunction in P301L tau transgenic mice. J Biol Chem. 2005;280:23802–23814. doi: 10.1074/jbc.M500356200. [DOI] [PubMed] [Google Scholar]
- 23.Rhein V, Song X, Wiesner A, Ittner LM, Baysang G, Meier F, Ozmen L, Bluethmann H, Dröse S, Brandt U, Savaskan E, Czech C, Götz J, et al. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc Natl Acad Sci USA. 2009;106:20057–20062. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li HL, Wang HH, Liu SJ, Deng YQ, Zhang YJ, Tian Q, Wang XC, Chen XQ, Yang Y, Zhang YJ, Wang Q, Xu H, Liao FF, et al. Phosphorylation of tau antagonizes apoptosis by stabilizing beta-catenin, a mechanism involved in Alzheimer's neurodegeneration. Proc Natl Acad Sci USA. 2007;104:3591–3596. doi: 10.1073/pnas.0609303104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan DX, Chai GS, Ni ZF, Hu Y, Luo Y, Cheng XS, Chen NN, Wang JZ, Liu GP. Phosphorylation of tau by death-associated protein kinase 1 antagonizes the kinase-induced cell apoptosis. J Alzheimers Dis. 2013;37:795–808. doi: 10.3233/JAD-130377. [DOI] [PubMed] [Google Scholar]
- 26.Luo DJ, Feng Q, Wang ZH, Sun DS, Wang Q, Wang JZ, Liu GP. Knockdown of phosphotyrosyl phosphatase activator induces apoptosis via mitochondrial pathway and the attenuation by simultaneous tau hyperphosphorylation. J Neurochem. 2014;130:816–25. doi: 10.1111/jnc.12761. [DOI] [PubMed] [Google Scholar]
- 27.Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 28.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 29.Austin S, St-Pierre J. PGC1α and mitochondrial metabolism—emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 30.Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb. Perspect Biol. 2012;pii(4):a011338. doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, Lich JD, Heise MT, Chen Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 32.Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 33.McGeer PL, Rogers J, McGeer EG. Inflammation, anti-inflammatory agents and Alzheimer disease: the last 12 years. J Alzheimers Dis. 2006;9:271–276. doi: 10.3233/jad-2006-9s330. [DOI] [PubMed] [Google Scholar]
- 34.Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12:1005–15. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Yan H, Yuan Y, Gao J, Shen Z, Cheng Y, Shen Y, Wang RR, Wang X, Hu WW, Wang G, Chen Z. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013;9:1321–1333. doi: 10.4161/auto.25132. [DOI] [PubMed] [Google Scholar]
- 36.Ye X, Sun X, Starovoytov V, Cai Q. Parkin-mediated mitophagy in mutant hAPP neurons and Alzheimer's disease patient brains. Hum Mol Genet. 2015;24:2938–2951. doi: 10.1093/hmg/ddv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao K, Klionsky DJ. Mitochondrial fission facilitates mitophagy in Saccharomyces cerevisiae. Autophagy. 2013;9:1900–1901. doi: 10.4161/auto.25804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, Dawson VL, Dawson TM. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci USA. 2004;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 41.Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, Gajendiran M, Roth BL, Chesselet MF, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 42.DuBoff B, Götz J, Feany MB. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75:618–632. doi: 10.1016/j.neuron.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khandelwal PJ, Herman AM, Hoe HS, Rebeck GW, Moussa CE. Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Abeta in AD models. Hum Mol Genet. 2011;20:2091–2102. doi: 10.1093/hmg/ddr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci USA. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pooler AM, Usardi A, Evans CJ, Philpott KL, Noble W, Hanger DP. Dynamic association of tau with neuronal membranes is regulated by phosphorylation. Neurobiol Aging. 2012;33:431. doi: 10.1016/j.neurobiolaging.2011.01.005. e27-38. [DOI] [PubMed] [Google Scholar]
- 47.Cooper JM, Wischik C, Schapira AH. Mitochondrial function in Alzheimer's disease. Lancet. 1993;341:969–970. doi: 10.1016/0140-6736(93)91269-r. [DOI] [PubMed] [Google Scholar]
- 48.Parker WD, Jr, Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer's disease brain. Neurology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 49.Ojaimi J, Masters CL, McLean C, Opeskin K, McKelvie P, Byrne E. Irregular distribution of cytochrome c oxidase protein subunits in aging and Alzheimer's disease. Ann Neurol. 1999;46:656–660. [PubMed] [Google Scholar]
- 50.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nuydens R, Novalbos J, Dispersyn G, Weber C, Borgers M, Geerts H. A rapid method for the evaluation of compounds with mitochondria-protective properties. J Neurosci Methods. 1999;92:153–159. doi: 10.1016/s0165-0270(99)00107-7. [DOI] [PubMed] [Google Scholar]
- 52.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu BY, Turner N, Li YY, Gu M, Huang MW, Wu F, Pang T, Nan FJ, Ye JM, Li JY. High-throughput assay for modulators of mitochondrial membrane potential identifies a novel compound with beneficial effects on db/db mice. Diabetes. 2010;59:256–265. doi: 10.2337/db09-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McManus MJ, Murphy MP, Franklin JL. The mitochondria-targeted antioxidant MitoQ prevents loss of spatial memory retention and early neuropathology in a transgenic mouse model of Alzheimer's disease. J Neurosci. 2011;31:15703–15715. doi: 10.1523/JNEUROSCI.0552-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi DC, Bakowska JC. Determination of mitochondrial membrane potential and reactive oxygen species in live rat cortical neurons. J Vis Exp. 2011;51 doi: 10.3791/2704. pii: 2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


