Abstract
Triple-negative breast cancer (TNBC) is a subtype of breast cancer with poor prognosis and high heterogeneity. The aim of this study was to screen patients for single-nucleotide polymorphisms (SNPs) associated with the prognosis of TNBC. Database-derived SNPs (NextBio, Ensembl, NCBI and MirSNP) located in the 3′-untranslated regions (3′-UTRs) of genes that are differentially expressed in breast cancer were selected. The possible associations between 111 SNPs and progression risk among 323 TNBC patients were investigated using a two-step case-control study with a discovery cohort (n=162) and a validation cohort (n=161). We identified the rs1054135 SNP in the adipocyte fatty acid binding protein 4 (FABP4) gene as a predictor of TNBC recurrence. The G allele of rs1054135 was associated with a reduced risk of disease progression as well as a prolonged disease-free survival time (DFS), with a hazard ratio (HR) for recurrence in the combined sample of 0.269 [95%CI: 0.098−0.735;P=0.001]. Notably, for individuals having the rs1054135 SNP with the AA/AG genotype, the magnitude of increased tumour recurrence risk for overweight patients (BMI≥25kg/m2) was significantly elevated (HR2.53; 95%CI: 1.06–6.03). Immunohistochemical staining of adipocytes adjacent to TNBC tissues showed that the expression level of FABP4 was statistically significantly lower in patients with the rs1054135-GG genotype and those in the disease-free group (P=0.0004 and P=0.0091, respectively). These results suggested that the expression of a lipid metabolism-related gene and an important SNP in the 3′-UTR of FABP4 are associated with TNBC prognosis, which may aid in the screening of high-risk patients with TNBC recurrence and the development of novel chemotherapeutic agents.
Keywords: triple-negative breast cancer, FABP4, genetic variant, 3′-untranslated regions, prognosis
INTRODUCTION
Triple-negative breast cancers (TNBCs) are a diverse and heterogeneous group of tumours that, by definition, lack estrogen and progesterone receptors and amplification of the HER2 (human epidermal growth factor receptor-2) gene [1–2]. The majority of the tumours classified as TNBCs are highly malignant, characterized by their aggressive behavior, young age of onset, and early relapse [3–4]. Transcriptional profiling studies suggest that there is further heterogeneity within triple-negative breast cancers, and these tumours can be categorized into six or more groups using genomic analysis [5–6]. However, the high expense of these detection methods and instability of their prognostic efficacy makes this profiling applicable only within laboratories. Therefore, more effective and sensitive prognostic markers are urgently needed to further subdivide TNBCs and to guide clinical practice more accurately.
A significant fraction of cancer patients have occult disseminated tumours at the time of primary diagnosis, which usually progress to clinically relevant lesions [7]. Since the majority of cancer mortality is associated with metastatic disease, biomarkers with the ability to predict metastatic risk in tumours would be of great value. Recent advances have led to the recognition that microRNAs (miRNAs) can act as key genetic regulators of a wide variety of biological processes, including tumour growth, proliferation, and survival [8–9]. Indeed, a number of miRNAs have been identified as potent oncogenes and tumour suppressors, playing crucial roles in the metastatic process of breast cancer [10–11]. More importantly, a series of studies has revealed strong correlations between altered miRNA expression and distant disease-free survival (DDFS), as well as overall survival (OS) of TNBC, suggesting their prognostic value for TNBC [12–13].
miRNAs are small, noncoding RNAs that regulate gene expression by degrading and/or suppressing the translation of target messenger RNAs (mRNA) by base pairing with sequences within the 3′-untranslated region (UTR) of mRNA [14]. On the one hand, miRNAs regulate gene expression in a post-transcriptional manner [15]. On the other hand, a reciprocal feedback loop between miRNAs and their target genes is often observed. Emerging evidence reveals that miRNA expression maybe regulated by single-nucleotide polymorphisms (SNPs) in the 3′-UTRs of their target genes [16–17]. In other words, SNPs located in the 3′-UTRs of target genes may influence not only the expression of the targeted genes but that of miRNAs as well. Therefore, these findings raise the possibility that some SNPs located in the complementary miRNA-binding sites of the 3′-UTRs of target genes may influence the biological properties of tumour cells through their impact on the expression of both targeted genes and miRNAs. Together, these may eventually determine individual susceptibility to tumour metastasis.
Although increasing evidence suggests that polymorphisms in such areas could act as strong predictors of cancer risk and prognosis, including that of breast cancer [18–20], none of the previously identified miRNA-altering polymorphisms have been specifically associated with the outcome of triple-negative breast cancer. Thus, in this case-control study, we identified the main SNPs located in the 3′-UTRs of differentially expressed genes in breast cancer in an attempt to discover the genetic variants in the 3′-UTR that potentially influence interactions with miRNAs and are associated with TNBC recurrence in a Chinese Han population.
RESULTS
Subject characteristics
The detection rates in 12 samples were less than 90% and were excluded from the final analysis of the discovery cohort. The overall median follow-up times of the discovery and validation cohorts were 89.8 and 47.3 months, respectively. Table 1 describes the characteristics of the study population. Surprisingly, the distributions of some clinical characteristics differed between the two cohorts. However, there was a consistently higher incidence of lymph node metastasis in the relapse group.
Table 1. Pretreatment characteristics of the discovery and validation cohorts.
| Discovery cohort | Validation cohort | |||||
|---|---|---|---|---|---|---|
| Disease –free n=113(%) |
Relapse n=36(%) |
Pa | Disease –free n=114(%) |
Relapse n=48(%) |
Pa | |
| Age(y) | 0.012 | 0.403 | ||||
| ≦40 | 17(15.0) | 12(33.3) | 23(20.2) | 7(14.6) | ||
| >40 | 96(85.0) | 24(66.7) | 91(79.8) | 41(85.4) | ||
| BMI(Body Mass Index) | 0.995 | 0.456 | ||||
| 25.02+-0.32 | 25.01+-0.62 | 24.04+-0.32 | 24.65+-0.75 | |||
| Breast cancer/Ovarian cancer history | 1.000 | 0.750 | ||||
| yes | 6(5.3) | 2(5.6) | 8(7.0) | 4(8.3) | ||
| no | 107(94.7) | 34(94.4) | 106(93.0) | 44(91.7) | ||
| Menopausal status at diagnosis | 0.027 | 0.395 | ||||
| premenopausal | 72(63.7) | 30(83.3) | 63(55.3) | 30(62.5) | ||
| postmenopausal | 41(36.3) | 6(16.7) | 51(44.7) | 18(37.5) | ||
| Operation method | 0.887 | 0.421 | ||||
| modified radical mastectomy | 93(82.3) | 30(83.3) | 86(75.4) | 39(81.3) | ||
| breast conserving surgery | 20(17.7) | 6(16.7) | 28(24.6) | 9(18.7) | ||
| Histological type | 0.404 | 0.438 | ||||
| infiltrative nonspecific cancer | 88(85.4) | 33(91.7) | 107(93.9) | 47(97.9) | ||
| others | 15(14.6) | 3(8.3) | 7(6.1) | 1(2.1) | ||
| Histological grade | 0.299 | 0.342 | ||||
| I-II | 46(49.5) | 12(38.7) | 46(47.4) | 15(38.5) | ||
| III | 47(50.5) | 19(61.3) | 51(52.6) | 24(61.5) | ||
| unknown | 19 | 5 | 17 | 9 | ||
| Lymphatic vessel invasion | 0.190 | 0.082 | ||||
| yes | 12(10.6) | 1(2.8) | 5(4.4) | 6(12.8) | ||
| no | 101(89.4) | 35(97.2) | 109(95.6) | 41(87.2) | ||
| Unknown | 0 | 0 | 0 | 1 | ||
| Tumour size | 0.894 | 0.036 | ||||
| ≦2cm | 43(38.4) | 13(37.1) | 54(47.8) | 14(29.8) | ||
| >2cm | 69(61.6) | 22(62.9) | 59(52.2) | 33(70.2) | ||
| Unknown | 1 | 1 | 1 | 1 | ||
| Lymph-node involvement | 0.043 | 0.039 | ||||
| no | 74(66.1) | 17(47.2) | 76(67.3) | 24(50.0) | ||
| yes | 38(33.9) | 19(52.8) | 37(32.7) | 24(50.0) | ||
| Unknown | 1 | 0 | 1 | 0 | ||
| Taxane/anthracycline-based chemotherapy | 0.303 | 0.454 | ||||
| no | 12(10.7) | 1(2.9) | 5(4.5) | 4(8.3) | ||
| yes | 100(89.3) | 34(97.1) | 107(95.5) | 44(91.7) | ||
| unknown | 1 | 1 | 2 | 0 | ||
| Radiotherapy | 0.097 | 0.671 | ||||
| no | 68(60.2) | 16(44.4) | 42(36.8) | 16(33.3) | ||
| yes | 45(39.8) | 20(55.6) | 72(63.2) | 32(66.7) | ||
Two-sided χ2 test
Genotyping and association analysis of SNPs and TNBC prognosis
To identify SNPs with potential prognostic value, 149 samples in the discovery cohort were tested initially. Sixteen SNPs were not in Hardy–Weinberg equilibrium (data not shown). Results from association analyses for 111 SNPs and the risk of disease progression are presented in Table 2. Fourteen SNPs were associated with the risk of TNBC recurrence and metastasis (P<0.05).
Table 2. Association between SNPs in differentially expressed genes and the risk of disease progression.
| Gene | SNP | Alleles (major/minor) | MAF | HRa (95% CI) | P | Genetic model | |
|---|---|---|---|---|---|---|---|
| Disease-free | Relapse | ||||||
| CCND1 | rs678653 | G/C | 0.1 | 0.07 | 18.09 (0.66-495.58) | 0.1 | REC |
| ESR1 | rs3798577 | T/C | 0.40 | 0.46 | 1.35 (0.50-3.60) | 0.55 | DOM |
| ADAMTS1 | rs2738 | C/A | 0.17 | 0.08 | 0.46 (0.17-1.29) | 0.12 | ADD |
| ADAMTS1 | rs9636786 | T/C | 0.17 | 0.21 | 1.21 (0.48-3.06) | 0.69 | DOM |
| ADH1B | rs1042026 | G/A | 0.27 | 0.29 | 0.44 (0.07-2.67) | 0.35 | REC |
| ADH1B | rs17033 | A/G | 0.14 | 0.11 | 0.77 (0.27-2.20) | 0.63 | DOM |
| ATM | rs227092 | G/T | 0.44 | 0.41 | 1.58 (0.44-5.68) | 0.49 | REC |
| BACH1 | rs15092 | A/G | 0.08 | 0.06 | 0.00 (0.00-NA) | 0.20 | REC |
| BRCA1 | rs12516 | C/T | 0.35 | 0.36 | 1.46 (0.42-5.05) | 0.56 | REC |
| BRIP1 | rs7213430 | A/G | 0.31 | 0.39 | 2.14 (1.09-4.22) | 0.026 | ADD |
| C8orf4 | rs10199 | A/G | 0.42 | 0.47 | 1.75 (0.91-3.34) | 0.086 | ADD |
| CASP8 | rs1045494 | T/C | 0.18 | 0.24 | 1.44 (0.59-3.53) | 0.42 | DOM |
| CCDC170 | rs3734806 | G/A | 0.39 | 0.42 | 1.43 (0.35-5.87) | 0.62 | REC |
| CCDC170 | rs3757322 | T/G | 0.40 | 0.42 | 0.76 (0.29-2.01) | 0.58 | DOM |
| CCDC170 | rs9383935 | C/T | 0.38 | 0.43 | 1.52 (0.39-5.90) | 0.55 | REC |
| CCDC170 | rs6932260 | T/C | 0.50 | 0.42 | 0.53 (0.27-1.02) | 0.052 | ADD |
| CCDC170 | rs9383589 | A/G | 0.36 | 0.32 | 0.44 (0.17-1.11) | 0.078 | DOM |
| CCND1 | rs7177 | A/C | 0.12 | 0.06 | 19.20 (0.71-520.31) | 0.093 | REC |
| CDH1 | rs13689 | T/C | 0.17 | 0.15 | 1.24 (0.27-5.82) | 0.78 | REC |
| CDS1 | rs6827228 | C/T | 0.11 | 0.06 | 0.39 (0.10-1.50) | 0.14 | ADD |
| CLDN5 | rs12628900 | C/T | 0.09 | 0.14 | 1.94 (0.69-5.43) | 0.21 | REC |
| COL10A1 | rs1059277 | G/A | 0.02 | 0.03 | 1.66 (0.23-11.82) | 0.62 | REC |
| COL11A1 | rs9659030 | T/C | 0.20 | 0.21 | 1.89 (0.77-4.65) | 0.16 | DOM |
| COL1A1 | rs1061947 | C/T | 0.04 | 0.03 | 1.21 (0.16-8.93) | 0.85 | REC |
| COL1A1 | rs1061237 | T/C | 0.48 | 0.46 | 0.43 (0.12-1.49) | 0.16 | REC |
| COL4A2 | rs1049977 | T/C | 0.19 | 0.12 | 0.41 (0.15-1.16) | 0.08 | DOM |
| CSMD1 | rs583087 | C/T | 0.06 | 0.07 | 1.36 (0.41-4.58) | 062 | REC |
| CYYR1 | rs17002176 | A/G | 0.04 | 0.07 | 1.79 (0.44-7.32) | 0.43 | REC |
| CYYR1 | rs17002187 | G/A | 0.04 | 0.07 | 2.97 (0.78-11.36) | 0.12 | REC |
| CYYR1 | rs219643 | C/T | 0.00 | 0.00 | 0.00 (0.00-NA) | 0.59 | REC |
| CYYR1 | rs2830239 | A/G | 0.29 | 0.32 | 0.39 (0.04-3.79) | 0.38 | REC |
| ERBB4 | rs1595064 | C/G | 0.43 | 0.44 | 0.69 (0.20-2.44) | 0.56 | REC |
| ERBB4 | rs1595065 | T/C | 0.28 | 0.29 | 1.25 (0.50-3.12) | 0.64 | DOM |
| ERBB4 | rs10932374 | G/A | 0.30 | 0.36 | 1.76 (0.50-6.22) | 0.39 | REC |
| ERBB4 | rs1836724 | T/C | 0.25 | 0.24 | 0.61 (0.05-6.90) | 0.68 | REC |
| ERBB4 | rs12467225 | C/T | 0.33 | 0.26 | 0.54 (0.23-1.28) | 0.16 | DOM |
| ERBB4 | rs1972820 | T/C | 0.27 | 0.21 | 0.66 (0.31-1.44) | 0.29 | ADD |
| ERBB4 | rs11895168 | C/A | 0.28 | 0.25 | 0.76 (0.31-1.85) | 0.54 | DOM |
| ERBB4 | rs1595066 | G/A | 0.33 | 0.33 | 0.81 (0.34-1.95) | 0.64 | DOM |
| ERBB4 | rs3748960 | T/C | 0.07 | 0.14 | 3.51(1.07-11.46) | 0.039 | DOM |
| ERBB4 | rs4672612 | G/A | 0.24 | 0.24 | 0.00 (0.00-NA) | 0.19 | REC |
| ERBB4 | rs16845990 | T/C | 0.33 | 0.27 | 0.63 (0.23-1.73) | 0.37 | DOM |
| ESR1 | rs3798758 | G/T | 0.30 | 0.26 | 2.81 (0.37-21.24) | 0.33 | REC |
| ETV6 | rs1062298 | G/T | 0.42 | 0.44 | 1.38 (0.46-4.12) | 0.56 | REC |
| ETV6 | rs1573613 | T/C | 0.46 | 0.53 | 1.35 (0.45-4.08) | 0.59 | DOM |
| ETV6 | rs2156932 | A/G | 0.03 | 0.07 | 2.30 (0.55-9.53) | 0.25 | REC |
| ETV6 | rs1573612 | T/C | 0.46 | 0.50 | 1.73 (0.54-5.54) | 0.36 | REC |
| FABP4 | rs1054135 | A/G | 0.47 | 0.33 | 0.35 (0.15-0.80) | 0.0084 | ADD |
| FBN1 | rs11070641 | T/C | 0.13 | 0.15 | 1.16 (0.47-2.84) | 0.74 | ADD |
| FBN1 | rs12050562 | C/T | 0.22 | 0.39 | 2.54 (1.26-5.13) | 0.0077 | ADD |
| FGFR2 | rs1047057 | C/T | 0.43 | 0.37 | 0.45 (0.11-1.81) | 0.24 | REC |
| GNAI1 | rs17153599 | C/T | 0.11 | 0.15 | 1.27 (0.52-3.09) | 0.6 | ADD |
| KRAS | rs9266 | C/T | 0.26 | 0.25 | 0.68 (0.32-1.49) | 0.33 | ADD |
| KRAS | rs1137282 | T/C | 0.11 | 0.07 | 0.42 (0.13-1.39) | 0.13 | ADD |
| KRAS | rs12587 | C/A | 0.28 | 0.24 | 0.00 (0.00-NA) | 0.091 | REC |
| KRAS | rs13096 | G/A | 0.23 | 0.25 | 0.38 (0.04-3.62) | 0.35 | REC |
| KRAS | rs7973450 | A/G | 0.08 | 0.07 | 0.65 (0.20-2.16) | 0.47 | ADD |
| KRAS | rs712 | G/T | 0.25 | 0.18 | 0.00 (0.00-NA) | 0.014 | REC |
| KRAS | rs7960917 | T/C | 0.12 | 0.08 | 0.55 (0.19-1.56) | 0.24 | ADD |
| KCNMB3 | rs3976507 | G/A | 0.07 | 0.09 | 1.00 (0.28-3.59) | 1.00 | DOM |
| KHDRBS3 | rs3184618 | A/G | 0 | 0.01 | 2.52 (0.05-121.99) | 0.64 | REC |
| MSRB3 | rs7711 | A/G | 0.16 | 0.13 | 0.93 (0.35-2.48) | 0.89 | REC |
| MSRB3 | rs7316024 | T/A | 0.49 | 0.47 | 1.41 (0.50-3.99) | 0.51 | DOM |
| NTRK2 | rs11140793 | A/C | 0.05 | 0.06 | 0.74 (0.15-3.78) | 0.72 | REC |
| NTRK2 | rs3654 | A/G | 0.18 | 0.14 | 0.00 (0.00-NA) | 0.046 | REC |
| NTRK2 | rs2013566 | A/G | 0.17 | 0.13 | 0.00 (0.00-NA) | 0.058 | REC |
| NTRK2 | rs3739570 | T/C | 0.50 | 0.49 | 0.49 (0.18-1.32) | 0.16 | DOM |
| NTRK2 | rs1047896 | T/C | 0.06 | 0.08 | 0.00 (0.00-NA) | 0.49 | REC |
| NTRK2 | rs1624327 | C/T | 0.11 | 0.17 | 23.08 (0.46-NA) | 0.11 | REC |
| NTRK2 | rs1221 | G/A | 0.05 | 0.04 | 0.18 (0.02-1.71) | 0.18 | REC |
| NTRK2 | rs1627784 | A/G | 0.33 | 0.38 | 1.78 (0.52-6.06) | 0.36 | REC |
| NTRK2 | rs7020204 | C/T | 0.16 | 0.15 | 0.78 (0.35-1.76) | 0.54 | ADD |
| NTRK2 | rs3780634 | A/G | 0.07 | 0.04 | 0.15 (0.02-1.35) | 0.04 | REC |
| NTRK2 | rs10780691 | C/T | 0.3 | 0.2 | 0.00(0.00-NA) | 0.04 | REC |
| NTRK2 | rs7816 | T/A | 0.09 | 0.21 | 3.67 (1.41-9.60) | 0.0064 | ADD |
| PHB | rs1049620 | A/G | 0.43 | 0.56 | 1.81 (0.85-3.83) | 0.12 | ADD |
| PID1 | rs3771286 | C/T | 0.52 | 0.35 | 0.34 (0.16-0.76) | 0.0051 | ADD |
| RHOU | rs1062060 | C/T | 0.05 | 0.07 | 0.66 (0.15-2.96) | 0.58 | DOM |
| RHOU | rs13349 | A/G | 0.24 | 0.26 | 0.30 (0.03-3.00) | 0.26 | REC |
| RHOU | rs11578216 | T/A | 0.06 | 0.04 | 1.05 (0.23-4.66) | 0.95 | DOM |
| RHOU | rs11580020 | G/A | 0.07 | 0.04 | 0.00 (0.00-NA) | 0.69 | ADD |
| RHOU | rs2058703 | T/C | 0.01 | 0.03 | 2.30 (0.31-17.13) | 0.43 | DOM |
| RND3 | rs10185950 | A/C | 0.07 | 0.03 | 0.23 (0.03-2.04) | 0.13 | DOM |
| SASH1 | rs8641 | A/G | 0.33 | 0.31 | 1.84 (0.35-9.68) | 0.48 | REC |
| SFRP1 | rs3242 | C/T | 0.05 | 0.04 | 1.39 (0.31-6.23) | 0.67 | REC |
| SLC24A2 | rs3739481 | G/C | 0.49 | 0.54 | 0.66 (0.24-1.78) | 0.41 | DOM |
| SLC24A2 | rs4977544 | C/T | 0.09 | 0.04 | 0.26 (0.06-1.10) | 0.045 | REC |
| SLC24A2 | rs4977545 | G/T | 0.22 | 0.13 | 0.50 (0.20-1.22) | 0.11 | ADD |
| SLC24A2 | rs7864646 | A/G | 0.07 | 0.04 | 0.66 (0.15-2.98) | 0.58 | REC |
| SLC24A2 | rs7872265 | T/C | 0.46 | 0.36 | 0.56 (0.23-1.37) | 0.20 | DOM |
| SLC24A2 | rs7867513 | C/T | 0.35 | 0.34 | 1.72 (0.43-6.90) | 0.46 | REC |
| SLC24A2 | rs7022987 | C/T | 0.33 | 0.4 | 0.68 (0.26-1.75) | 0.42 | DOM |
| SLC24A2 | rs7854673 | A/T | 0.07 | 0.00 | 0.00 (0.00-NA) | 8e-04 | REC |
| SLC24A2 | rs1556000 | G/T | 0.08 | 0.05 | 0.67 (0.15-2.88) | 0.58 | REC |
| SORCS1 | rs12359404 | C/T | 0.13 | 0.13 | 13.15 (0.43-406.54) | 0.16 | REC |
| SORCS1 | rs10491050 | T/C | 0.22 | 0.25 | 1.36 (0.55-3.33) | 0.51 | DOM |
| SORCS1 | rs11192963 | T/C | 0.26 | 0.33 | 2.06 (0.95-4.49) | 0.065 | ADD |
| TAB2 | rs2744434 | G/A | 0.41 | 0.53 | 1.52 (0.78-2.95) | 0.22 | ADD |
| TAB2 | rs7896 | C/G | 0.08 | 0.09 | NA (0.00-NA) | 0.011 | REC |
| TACSTD2 | rs7333 | G/A | 0.15 | 0.12 | 0.54(0.19-1.53) | 0.23 | ADD |
| THSD4 | rs12594531 | C/A | 0.37 | 0.43 | 2.21 (0.80-6.11) | 0.11 | DOM |
| THSD4 | rs3087532 | C/T | 0.18 | 0.15 | 0.00 (0.00-NA) | 0.088 | REC |
| THSD4 | rs7402189 | A/G | 0.24 | 0.21 | 1.46 (0.59-3.60) | 0.41 | DOM |
| THSD4 | rs10468050 | G/C | 0.13 | 0.11 | 0.84 (0.28-2.53) | 0.76 | DOM |
| THSD4 | rs1054260 | C/T | 0.25 | 0.19 | 0.51 (0.23-1.10) | 0.075 | ADD |
| THSD4 | rs4776575 | G/A | 0.20 | 0.20 | 1.81 (0.16-19.79) | 0.63 | REC |
| TPM1 | rs6738 | A/G | 0.05 | 0.03 | 0.47 (0.09-2.50) | 0.35 | REC |
| TPM1 | rs7178040 | G/T | 0.02 | 0.00 | 0.00 (0.00-NA) | 0.13 | REC |
| ZNF365 | rs11819488 | A/G | 0.19 | 0.31 | 2.74 (1.28-5.86) | 0.0075 | ADD |
| ZNF365 | rs729739 | G/A | 0.09 | 0.06 | 0.00 (0.00-NA) | 0.41 | REC |
| ZNF365 | rs729738 | C/A | 0.08 | 0.07 | 1.18 (0.34-4.09) | 0.79 | REC |
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SNPs and recurrence & metastasis risk was adjusted for age, tumour size (≤2cm and >2 cm), lymph-node involvement (no and yes), histological type, menopausal status (no and yes), vascular invasion (no and yes), breast or ovarian cancer history (no and yes), taxane/anthracycline-based chemotherapy (yes or no) and radiotherapy (no and yes)
Independent validation was conducted in the second cohort, which included 162 TNBCs with 114 disease-free cases and 48 relapsed cases. Three SNPs were significantly associated with TNBC recurrence (P<0.05), including FABP4 rs1054135, KRAS rs712 and NTRK2 rs7816 (Table 3). The statistical significance was retained after multiple comparisons only for FABP4rs1054135. The G allele of rs1054135 was associated with a reduced risk of disease progression with an adjusted hazard ratio of 0.14(0.03-0.66) in a recessive model (Table 4).
Table 3. Association of SNPs rs1054135, rs712, rs7816, rs12050562, rs3748960, rs3654, rs11819488, rs10780691, rs7213430, rs3771286, rs4977544, rs7896, rs7854673, rs3780634, and the risk of disease progression.
| Gene | SNP | Alleles (major/minor) |
MAF | HRa (95% CI) | P | Genetic model | FDR | |
|---|---|---|---|---|---|---|---|---|
| Disease-free | Relapse | |||||||
| FABP4 | rs1054135 | A/G | 0.48 | 0.32 | 0.36 (0.19-0.69) | 0.0012 | ADD | 0.017 |
| KRAS | rs712 | G/T | 0.21 | 0.33 | 2.11 (1.12-3.95) | 0.019 | ADD | 0.247 |
| NTRK2 | rs7816 | T/A | 0.18 | 0.14 | 0.00 (0.00-NA) | 0.031 | REC | 0.372 |
| FBN1 | rs12050562 | C/T | 0.24 | 0.27 | 2.00 (0.92-4.36) | 0.077 | DOM | 0.847 |
| ERBB4 | rs3748960 | T/C | 0.08 | 0.02 | 0.36 (0.08-1.61) | 0.12 | ADD | 1.20 |
| NTRK2 | rs3654 | A/G | 0.17 | 0.22 | 1.55 (0.77-3.12) | 0.22 | ADD | 1.98 |
| ZNF365 | rs11819488 | A/G | 0.19 | 0.21 | 2.56 (0.31-21.06) | 0.39 | REC | 3.12 |
| NTRK2 | rs10780691 | C/T | 0.22 | 0.24 | 0.50 (0.09-2.90) | 0.42 | REC | 2.94 |
| BRIP1 | rs7213430 | A/G | 0.28 | 0.26 | 0.83 (0.47-1.47) | 0.52 | ADD | 3.12 |
| PID1 | rs3771286 | C/T | 0.5 | 0.45 | 0.85 (0.49-1.45) | 0.54 | ADD | 2.70 |
| SLC24A2 | rs4977544 | C/T | 0.05 | 0.03 | 0.69 (0.17-2.82) | 0.59 | REC | 2.26 |
| TAB2 | rs7896 | C/G | 0.06 | 0.05 | 0.64 (0.06-6.86) | 0.71 | REC | 2.13 |
| SLC24A2 | rs7854673 | A/T | 0.06 | 0.06 | 1.25 (0.37-4.20) | 0.72 | REC | 1.44 |
| NTRK2 | rs3780634 | A/G | 0.05 | 0.04 | 1.13 (0.30-4.31) | 0.85 | REC | 0.85 |
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SNPs and recurrence & metastasis risk was adjusted for age, tumour size (≤2cm and >2 cm), lymph-node involvement (no and yes), histological type, menopausal status (no and yes), vascular invasion (no and yes), breast or ovarian cancer history (no and yes), taxane/anthracycline-based chemotherapy (yes or no) and radiotherapy (no and yes)
Table 4. Association between the rs1054135 genotype and the risk of TNBC relapse (validation cohort).
| SNP | genotype | Disease-free (%) | Relapse (%) | HRa (95% CI) | P value |
|---|---|---|---|---|---|
| rs1054135 | AA+AG | 88(80.0) | 42 (95.5) | 1.00(Reference) | |
| GG | 22 (20.0) | 2(4.5) | 0.14(0.03-0.66) | 0.0026 |
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SNPs and recurrence & metastasis risk was adjusted for age, tumour size (≤2cm and >2 cm), lymph-node involvement (no and yes), histological type, menopausal status (no and yes), vascular invasion (no and yes), breast or ovarian cancer history (no and yes), taxane/anthracycline-based chemotherapy (yes or no) and radiotherapy (no and yes).
The data also showed a covariate effect of BMI and FABP4 on TNBC reccurence. For patients with the AA/AG genotype, the magnitude of increased tumour recurrence risk for overweight patients (BMI≥25kg/m2) was significantly elevated (HR, 2.53; 95%CI, 1.06–6.03).
Positive results were obtained for the associations between the rs1054135 genotype and DFS using the Kaplan–Meier method (Figure 1). The discovery and validation sets were subsequently combined for analysis, and showed that individuals with the rs1054135-GG genotype were associated with a prolonged DFS, with a HR of 0.269 (95%CI = 0.098−0.735; P= 0.001).
Figure 1. Relationship between the FABP4 SNP rs1054135 and DFS in TNBC patients.
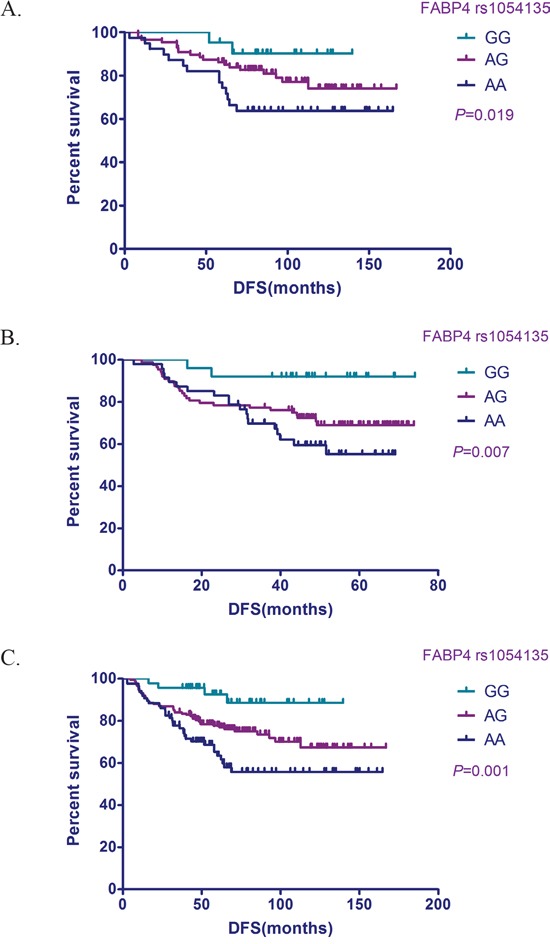
Kaplan–Meier survival probability plots stratified by FABP4rs1054135 genotype. A. Discovery cohort. B. Validation cohort. C. Combined sample.
Correlations between FABP4 expression, the rs1054135 genotype and TNBC prognosis
In order to test the hypothesis that the rs1054135 genotype facilitates tumour metastasis by regulating FABP4 expression, its protein expression was analyzed by immunohistochemistry in 52 TNBC tissues (disease-free group, n=34; relapsed group, n=18) with the associated genotype data.
Remarkably, adipocytes adjacent to breast tissue exhibited higher FABP4 protein levels, when compared with those located more distantly from breast tissues (Figure 2). Quantitative determination with IOD also showed that FABP4 protein levels were significantly higher in adipocytes with the rs1054135-AA/AG genotype (P<0.01). The median IOD of the rs1054135-AA/AG group was 0.149, while more than half of patients with the rs1054135-GG genotype did not express FABP4 (IOD=0). A significant difference in FABP4 protein levels was also observed between the disease-free and relapse groups (with a median IOD of 0.1240 and 0.1498, respectively), which is consistent with our hypothesis (Figure 3).
Figure 2. Example of immunohistochemical staining with FABP4.
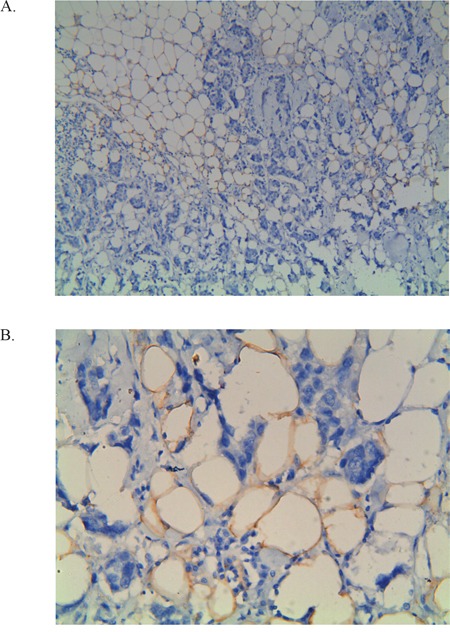
A. 100x magnification and the same area at 400x magnification B. The IOD counts were performed by the computer using 400x magnification of these images.
Figure 3. Scatterplot of FABP4 expression.
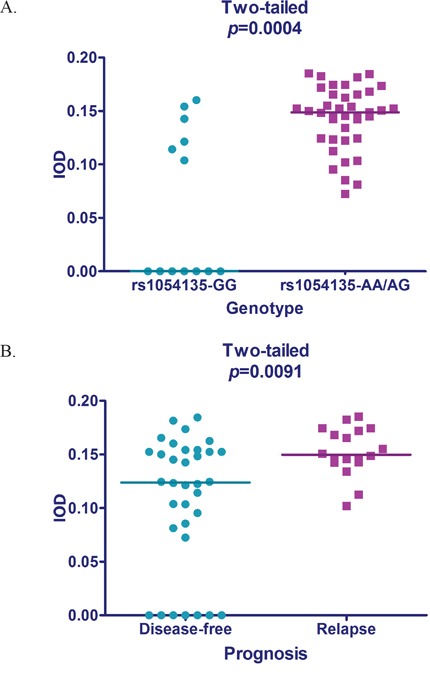
A. Intergroup difference of FABP4 expression in patients with different prognosis tested by the Mann-Whitney U test. B. Intergroup difference of FABP4 expression associated with different rs1054135 genotypes tested by the Mann-Whitney U test.
DISCUSSION
The strong invasiveness of TNBCs is manifested in an early onset of recurrence and metastasis, particularly in the first three years [21]. In our study, 60% of patients in the relapse group suffered disease progression within three years, and the five-year DFS of the total sample was 75.7%. These results are similar to those previously reported for TNBC studies; however, they are significantly worse than those reported for other subtypes [22]. Therefore, the identification of TNBC subgroups relevant to clinical prognosis will aid in the design and administration of individualized treatment plans.
By a two-stage analysis of discovery and validation samples, we identified a novel variant in FABP4 associated with both recurrence risk and DFS of TNBC. In our study, we found that the A allele of rs1054135 could upregulate FABP4 expression in TNBC patients, which was in line with the findings of a previous study that in children with obstructive sleep apnea, the rs1054135 AA genotype was associated with high serum FABP4 levels [23], suggesting a functional relevance of this site.
According to bioinformatics predictions, rs1054135 was a miR-3685 complementary SNP site and a G>A transition at rs1054135 may lead to an increased binding force with miR-3685. Scientists have demonstrated that, in some mRNAs with AU-rich elements (AREs), miRNAs could mediate a direct association of micro-ribonucleoproteins (microRNPs) with the AREs and eventually upregulate translation in some cases [24]. Likewise, variation in rs1054135 may affect the expression of FABP4 through similar molecular mechanisms, while further investigations are needed to validate this speculation.
Adipocyte fatty acid binding protein4 (FABP4) is predominantly expressed in the cytosol of mature adipocytes and reversibly binds long-chain fatty acids. Previous reports have characterized its role in lipid metabolism and transport [25]. In vitro studies showed that cocultivation of several cancer cell lines (ovarian, breast, and colon) with adipocytes induced FABP4 mRNA expression. Controversially, when adding a FABP4 inhibitor to a coculture of ovarian cancer cells and adipocytes, lipid accumulation in the cancer cells and adipocyte-mediated invasion were drastically reduced [26]. Similarly, in our study, stronger immunohistochemical (IHC) staining of FABP4 was observed in adipocytes adjacent to breast tissue, implying that FABP4 may function as a mediator of lipid trafficking, and the expression level of FABP4 maybe an indicator of the regional metabolic level. Moreover, FABP4 was found to be induced by VEGFA and/or the NOTCH pathway in endothelial cells, and inhibition of FABP4 blocks most of the VEGFA effects, suggesting its role in tumour angiogenesis [27]. Nieman KM, et al. reported an up regulation of FABP4 expression in metastatic human ovarian cancer samples compared with primary ovarian tumours; the increased FABP4 levels were shown to fuel rapid tumour growth and support metastasis [28]. Furthermore, previous studies have identified FABP4 as a prognostic marker in breast cancer. Hancke K et al. found that higher serum FABP4 levels were associated with obese breast cancer, as well as greater tumour size and lymph node involvement [29]. A most recent report also showed that FABP4 positivity was associated with significantly shorter DFS and OS in TNBC [30]. However, unlike in our study, tumour tissues instead of stroma were used as the IHC target, and the positive rate of FABP4 was relatively low (2/50), making it a less statistically powerful prognostic biomarker.
Our study found a closed-loop chain between rs1054135, FABP4 expression and TNBC prognosis. Given that FABP4 is a significant medium of fuel supply for tumour growth, and probably involved in tumour angiogenesis, the rs1054135 SNP located in the 3′-UTR of FABP4 may influence patient susceptibility to TNBC recurrence through posttranscriptional regulation of FABP4 expression.
Indeed, the roles of lipid metabolism-related genes and pathways in tumour development have been studied extensively, especially in breast cancer. Several studies indicated that adipose tissue itself is an endocrine organ that could influence tumour growth or differentiation via adipose tissue-derived hormones [31] called adipocytokines, e.g., leptin, resistin, or adiponectin (ApN), most of which showed strong correlations with BMI [32–33]. However, the association between obesity and survival after breast cancer remained controversial for decades until last year; a positive association was demonstrated in meta-analyses of published data [34–35]. In the present study, a positive association between obesity and high recurrence risk was observed for the rs1054135-AA/AG subgroup. This provides additional evidence that body fat content and FABP4 (as key substrates and enzymes of fat metabolism) functioned synergistically when fueling rapid tumour growth and metastasis. Therefore, it is safe to assume that the previous controversy over the association between BMI and breast cancer prognosis maybe related to the distribution difference of the FABP4 genotype among different populations.
Recently, the Women's Intervention Nutrition Study (WINS) revealed that a low-fat diet after diagnosis of early breast cancer can reduce the death rate by 56%for women with both ER- and PR-negative breast cancer [36]. In addition, another large retrospective study reported that statin use was associated with a significant reduction in deaths from breast cancer (aHR = 0.60) [37] and, most importantly, statins were found to suppress the expression of FABP4 by previous basic research [38]. Thus, these findings shed light on the possibility that for obese patients with the rs1054135-AA/AG genotype, a low fat diet and statins could be selectively administered.
To our knowledge, our study is the first to examine the association of TNBC prognosis and SNPs located in the complementary miRNA binding sites of the 3′-UTRs of target genes. Since germline SNP variations are more stable than somatic SNP mutations, the germline SNP prognostic signature may provide more reliable information on individual susceptibility to tumour metastasis and be less likely to be affected by intratumour heterogeneity. Additionally, our findings are important because TNBC patients have fewer immediate therapeutic options, and these patients tend to have more aggressive disease. This study not only demonstrated the significant role of lipid metabolism in the process of TNBC recurrence, but discovered a novel SNP located in the 3′-UTR of FABP4 that acted in concert with BMI and showed a strong association with DFS. Thus, for patients with the rs1054135-AA/AG genotype, low-fat diet intervention and body weight management is strongly recommended. More importantly, these results suggest that cutting off the ‘fuel supply’ may be a promising method for tumours such as TNBC that lack therapeutic targets.
However, despite the aforementioned strengths, we also acknowledge the limitations of this study. TNBC selection was based on immunohistochemistry instead of genomic analysis. Thus, a small proportion of other subtypes may have been involved. However, this confounding factor could hardly restrict the application, given that IHC diagnosis is still the gold standard in clinical practice. The second limitation is the sample size, which may have resulted in our study having limited statistical power. However, a two-step screening and validation process as well as the multiple-testing procedure were used to reduce the false-positive rate, and the validity of our results can be confirmed in future studies. Another limitation is the unequal follow-up time between the discovery and validation cohorts. Given that the primary endpoint of our study was DFS, and that TNBC patients have the highest percentage of early relapse, the relatively longer follow-up time in the discovery cohort was considered mainly due to the long-term observation of patients in the disease-free group. Therefore, it is well-founded to regard the difference in follow-up time between two cohorts as acceptable.
In conclusion, our study identified a lipid metabolism-related gene and an important SNP in the 3′-UTR of FABP4 associated with TNBC prognosis, which may aid in the screening of high-risk patients with TNBC recurrence and the development of novel chemotherapeutic agents.
MATERIALS AND METHODS
Ethics statement
This investigation was conducted in accordance with the ethical standards of the Declaration of Helsinki and following the national and international guidelines and has been approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Hospital.
Study subjects
Tumour tissues and blood samples have been collected from primary breast cancer patients treated in our hospital since 1998, and there are a total of 13,240 blood samples. In the present study, we reviewed all of the pathologically confirmed TNBC cases from this sample library (n=430). Patients with a previous history of cancer (n=4) and insufficient blood samples (n=23) were excluded. Additionally, disease-free survivors with a follow-up time of less than three years were also excluded (n=80). Thus, a total of 323 TNBC patients were included in the final analysis. We artificially designated the date of Jan 1, 2008 a cut-off; patients diagnosed with TNBCs before Jan 1, 2008 were grouped into the discovery cohort (n=161) and those diagnosed after that date were grouped into the validation cohort (n=162). Patients were followed until April 1, 2014 to collect data on clinicopathological features, treatments, and vital status, such as recurrence and death. The DFS time was defined as the time from the date of surgery until the date of the first locoregional recurrence, first distant metastasis, or death from any cause. Patients known to be alive with no evidence of disease progression were censored at the last follow-up date or on April 1, 2014 (whichever came first).
ER (estrogen receptor) and PR (progesterone receptor) status was evaluated based on the IHC results of formalin-fixed, paraffin-embedded breast cancer tissue samples obtained from the patients. A positive ER and PR status was defined by nuclear staining of more than 1% according to guidelines issued by the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) in 2010. Tumours negative for ER, PR, and HER2 were defined as TNBCs. However, there is growing evidence that low-HR-staining tumours (1%-10%) are clinicopathologically more similar to HR-negative than to HR-positive tumours [39]. Therefore, tumours with low and/or focal PR staining were included in our study. The IHC was performed with anti-ER and anti-PR antibodies. To determine the HER2 status, IHC or gene amplification was performed by fluorescence in situ hybridization (FISH).
SNP selection and genotyping
SNP selection was a process taking full advantage of online databases and can be described as follows. First, the top 100 genes known to be differentially expressed in breast cancer were downloaded from the NextBio database (www.nextbio.com), searching using the keyword “breast cancer”. Second, these candidate genes were entered into the Ensembl (http://www.ensembl.org/index.html) and NCBI (http://hapmap.ncbi.nlm.nih.gov/) databases to select SNPs located in the 3′-UTRs of these differentially expressed genes with a minor allele frequency (MAF)≥ 5% in the ethnic Han Chinese people (n=204). Lastly, the MirSNP (http://cmbi.bjmu.edu.cn/mirsnp) publicly available online database was used for the final screening [40]. MirSNP contains a collection of human SNPs in predicted miRNA-mRNA binding sites, and 414,510 SNPs were identified to affect miRNA-mRNA binding through a miRNA target prediction algorithm, miRanda. In other words, only those SNPs with potential effects on miRNA-mRNA binding were included (n=140). Primers and probes were designed using MassARRAY Typer 4.0 software. In addition, 29 SNPs were excluded due to interference with primer binding. Therefore, 111 SNPs were included in the final genotyping.
For purposes of economy and efficiency, genotyping of the combined samples (n=323) was conducted using the MassARRAY MALDI-TOF System (Sequenom Inc., San Diego, CA, USA) at once by the method described in the Sequenom Genotyping Protocol, while association studies for the individual cohorts were analyzed separately. Duplicate samples and negative controls (without DNA) were included for quality assurance of genotyping. Concordance for duplicate samples was 100% for all assays. The analysts who carried out the genotyping were blinded to the group information on each sample.
Immunohistochemistry
IHC staining of FABP4 was performed on formalin-fixed, paraffin-embedded tissue sections. As FABP4 is primarily expressed in the cytosol of mature adipocytes, we chose adipocytes adjacent to tumour tissues as targets. Briefly, 4-μm-thick sections were cut with a microtome, transferred onto adhesive slides, and then dried at 62°C for 15 min. All slides were incubated with primary antibody (FABP4, 1:100, ab92501, Abcam, Cambridge, UK). After applying primary antibodies, the tissues were incubated in blocking solution for 1 h at 37°C. Subsequently, immunodetection was performed using a commercial streptavidin-biotin kit according to the manufacturer's instructions, which involved incubation with biotinylated anti-mouse or anti-rabbit immunoglobulin, followed by peroxidase-labelled streptavidin and 3, 3′-diaminobenzidine chromogenic substrate. The primary antibody incubation step was omitted from the negative control. Finally, the slides were counterstained with Harris haematoxylin.
Integrated optical density (IOD)
Using the Moticcam 2306® 4 tablet (MOTIC Company Ltd., China), cellular membranes of adipocytes adjacent to breast tissues were selected at random from the digitized IHC images and their contours were precisely delineated with an Intuos pen using the selecting tool available within the ImageJ software (Image-Pro Plus 6.0). The contours of cellular membranes were transformed into vectorial masks and saved as TIFF format files. The latter were subjected to an ImageJ algorithm, which computed individual membrane area and associated IODs of the FABP4 staining. The technicians were blinded to group information and SNP data for each sample.
Statistical analyses
The differences in patients' characteristics for study inclusion were assessed by Pearson's χ2 tests, and all P values represent two-sided statistical tests. The continuous variable BMI with a normal distribution was expressed as a mean, and the intergroup difference was tested using the unpaired t-test. As for the IOD, a Shapiro–Wilk analysis was employed to validate the distribution characteristic, and a t-test or Mann-Whitney U test was selected for intergroup difference assessment, as appropriate. A P value of less than 0.05 was considered to indicate significance. The Kaplan-Meier and Cox methods were used to estimate the survival function stratified by genotype of the studied genes. Differences across survival curves were examined using a log-rank test.
For individual SNP analysis, we tested three genetic models (additive, dominant, and recessive) to evaluate the significance of SNPs, and the best-fitting model for each SNP was selected by the smallest P value. Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association of SNPs and risk of recurrence and metastasis were adjusted for age, tumour size (≤2cm and >2 cm), lymph-node involvement (no and yes), histological type, menopausal status (no and yes), vascular invasion (no and yes), breast or ovarian cancer history (no and yes), taxane/anthracycline-based chemotherapy (yes or no), and radiotherapy (no and yes). Given the number of SNPs investigated, the Benjamini-Hochberg false discovery rate (FDR) method was used to assess statistical significance after correction for multiple comparisons. We considered an FDR of <0.05 to be noteworthy [41]. Tests for Hardy–Weinberg equilibrium were conducted. All statistical procedures were conducted using SPSS software (version 19.0) and GraphPad Prism5.
Acknowledgments
We thank all of the patients, resident physicians, and attending physicians in the Department of Medical Oncology at the Chinese Academy of Medical Sciences & Cancer Hospital; without their commitment to tissue donation and sample collection, this project would not have been possible.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This work was funded by the National Natural Science Foundation of China (81202109, 81472753), the Wu Jieping Foundation (320.6750.13297), and the Youth fund of Peking Union Medical College.
REFERENCES
- 1.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 2.Irvin WJ, Jr., Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44:2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 3.Perez EA, Moreno-Aspitia A, Aubrey Thompson E, Andorfer CA. Adjuvant therapy of triple negative breast cancer. Breast Cancer Res Treat. 2010;120:285–291. doi: 10.1007/s10549-010-0736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple-negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–57. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–50. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, Pietenpol JA. TNBCtype: A subtyping tool for triple-negative breast cancer. Cancer Inform. 2012;11:147–156. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, Bischoff J, Harich D, Schlimok G, et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 11.Svoboda M, Sana J, Redova M, Navratil J, Palacova M, Fabian P, Slaby O, Vyzula R. MiR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, Shapiro CL, Huebner K. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One. 2013;8:e55910. doi: 10.1371/journal.pone.0055910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svoboda M, Sana J, Redova M, Navratil J, Palacova M, Fabian P, Slaby O, Vyzula R. MiR-34b is associated with clinical outcomein triple-negative breast cancer patients. Diagn Pathol. 2012;7:31. doi: 10.1186/1746-1596-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200family promotes EMT and invasion in cancer cells. EMBO J. 2008;9:582–589. doi: 10.1038/embor.2008.74. Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, Dorairaj J, Geyda K, Pelletier C, Nallur S, Martens JW, Hooning MJ, Kerin M, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12:377–386. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang R, Schlehe B, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, Volkmann J, Varon R, Weber BH, Niederacher D, Arnold N, Meindl A, Bartram CR, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2010;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Liu W, Jiang W, Lin J, Jiang Y, Li B, Pang D. A miRNA binding site single-nucleotide polymorphism in the 3′-UTR region of the IL23R gene is associated with breast cancer. PLoS One. 2012;7:e49823. doi: 10.1371/journal.pone.0049823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung H, Jeon S, Lee KM, Han S, Song M, Choi JY, Park SK, Yoo KY, Noh DY, Ahn SH, Kang D. Common genetic polymorphisms of microRNA biogenesis pathway genes and breast cancer survival. BMC Cancer. 2012;12:195–207. doi: 10.1186/1471-2407-12-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM. The triple negative paradox: primary tumour chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 22.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, Heikkilä P, Heikkinen T, Nevanlinna H, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhushan B, Khalyfa A, Spruyt K, Kheirandish-Gozal L, Capdevila OS, Bhattacharjee R, Kim J, Keating B, Hakonarson H, Gozal D. Fatty-acid binding protein 4 gene polymorphisms and plasma levels in children with obstructive sleep apnea. Sleep Med. 2011;12:666–71. doi: 10.1016/j.sleep.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 25.Scheja L, Makowski L, Uysal KT, Wiesbrock SM, Shimshek DR, Meyers DS, Morgan M, Parker RA, Hotamisligil GS. Altered insulin secretion associated with reduced lipolytic efficiency in aP2−/− mice. Diabetes. 1999;48:1987–1994. doi: 10.2337/diabetes.48.10.1987. [DOI] [PubMed] [Google Scholar]
- 26.Hertzel AV, Hellberg K, Reynolds JM, Kruse AC, Juhlmann BE, Smith AJ, Sanders MA, Ohlendorf DH, Suttles J, Bernlohr DA. Identification and characterization of a small molecule inhibitor of fatty acid binding proteins. J. Med. Chem. 2009;52:6024–6031. doi: 10.1021/jm900720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harjes U, Bridges E, McIntyre A, Fielding BA, Harris AL. Fatty acid-binding protein 4, a point of convergence for angiogenic and metabolic signaling pathways in endothelial cells. J Biol Chem. 2014;289:23168–76. doi: 10.1074/jbc.M114.576512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, Peter ME, Gwin K, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumour growth. Nat Med. 2011;17:1498–503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hancke K, Grubeck D, Hauser N, Kreienberg R, Weiss JM. Adipocyte fatty acid-binding protein as a novel prognostic factor in obese breast cancer patients. Breast Cancer Res Treat. 2010;119:367–377. doi: 10.1007/s10549-009-0577-9. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Lee Y, Koo JS. Differential expression of lipid metabolism related proteins in different breast cancer subtypes. PLoS One. 2015;10:e0119473. doi: 10.1371/journal.pone.0119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Housa D, Housova J, Vernerova Z, Haluzik M. Adipocytokines and cancer. Physiol Res. 2006;55:233–244. doi: 10.33549/physiolres.930848. [DOI] [PubMed] [Google Scholar]
- 32.Vona-Davis L, Rose DP. Adipokines as endocrine, paracrine, and autocrine factors in breast cancer risk and progression. Endocr Rel Cancer. 2007;14:189–206. doi: 10.1677/ERC-06-0068. [DOI] [PubMed] [Google Scholar]
- 33.Lorincz A, Sukumar S. Molecular links between obesity and breast cancer. Endocr Relat Cancer. 2006;13:279–292. doi: 10.1677/erc.1.00729. [DOI] [PubMed] [Google Scholar]
- 34.Pan H, Gray RG, on behalf of the Early Breast Cancer Trialists' Collaborative Group Effect of obesity in premenopausal ER+ early breast cancer: EBCTCG data on 80,000 patients in 70 trials. Program and abstracts of the 2014 American Society of Clinical Oncology Annual Meeting; May 30-June 3, 2014; Chicago, Illinois. Abstract 503. [Google Scholar]
- 35.Chan DS, Vieira AR, Aune D, Bandera EV, Greenwood DC, McTiernan A, Navarro Rosenblatt D, Thune I, Vieira R, Norat T. Body mass index and survival in women with breast cancer-systematic literature review and meta-analysis of 82 follow-up studies. Ann Oncol. 2014;25:1901–1914. doi: 10.1093/annonc/mdu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chlebowski RT, Blackburn GL. Final survival analysis from the randomized Women's Intervention Nutrition Study (WINS) evaluating dietary intervention as adjuvant breast cancer therapy. Program and abstracts of the San Antonio Breast Cancer Symposium; December 9-13, 2014; San Antonio, Texas. Abstract S5-08. [Google Scholar]
- 37.Wang A, Aragaki AK, Tang JY, Kurian AW, Manson JE, Chlebowski RT, Simon MS, Desai PM, Wassertheil-Smoller S, Liu S, Kritchevsky S, Wakelee HA, et al. Statin use and all-cancer mortality: Prospective results from the Women's Health Initiative. Program and abstracts of the 2015 American Society of Clinical Oncology Annual Meeting; May 29-June 2, 2015; Chicago, Illinois. Abstract 1506. [Google Scholar]
- 38.Takeyama N, Itoh Y, Kitazawa Y, Tanaka T. Altered hepatic mitochondrial fatty acid oxidation and ketogenesis in Endotoxic rats. Am J Physiol. 1990;22:498–505. doi: 10.1152/ajpendo.1990.259.4.E498. [DOI] [PubMed] [Google Scholar]
- 39.Deyarmin B, Kane JL, Valente AL, van Laar R, Gallagher C, Shriver CD, Ellsworth RE. Effect of ASCO/CAP guidelines for determining ER status on molecular subtype. Ann Surg Oncol. 2013;20:87–93. doi: 10.1245/s10434-012-2588-8. [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Zhang F, Li T, Lu M, Wang L, Yue W, Zhang D. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics. 2012;13:661. doi: 10.1186/1471-2164-13-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaffer JP. Controlling the false discovery rate with constraints: the Newman-Keuls test revisited. Biometrical J. 2007;49:136–143. doi: 10.1002/bimj.200610297. [DOI] [PubMed] [Google Scholar]


