Abstract
Maternally expressed gene 3 (MEG3), a long non-coding RNA (lncRNA), is involved in cancer development and metastasis. The objective of the present study was to evaluate whether common single nucleotide polymorphisms (SNPs) in MEG3 could be related with colorectal cancer risk in Chinese. We genotyped six tagSNPs of MEG3 in a colorectal cancer case-control study including 518 cases and 527 control subjects. Multivariate logistic regression analysis was applied to calculate adjusted odds ratios (ORs). We found that MEG3 rs7158663 AA genotype, but not GA genotype, had significant increased colorectal cancer risk, compared with GG genotype (OR = 1.96 and P = 0.006 for AA versus GG, and OR = 1.20 and P = 0.171 for GA versus GG). Further stratified analysis indicated that the increased risk was significantly correlated with individuals with age ≤ 60 and family history of cancer. However, there was no significant association between rs7158663 and colorectal tumor site and stage (P = 0.842 for tumor site, and P = 0.601 for tumor stage). These results demonstrate that genetic variants in MEG3 may contribute to the development and risk of colorectal cancer. Further studies are required to confirm these findings.
Keywords: lncRNA, MEG3, colorectal cancer
INTRODUCTION
Colorectal cancer is the third most commonly diagnosed cancer in the world [1]. According to recent reports, the incidence and mortality of colorectal cancer in China have also been increasing in last two decades [2]. It is well known that colorectal cancer susceptibility is related to multiple environmental factors and genetic alterations, such as genetic mutations or polymorphisms [3, 4]. Nevertheless, the role of genetic polymorphism and colorectal cancer susceptibility still remains unknown.
Long non-coding RNAs (lncRNAs) are non-coding molecules larger than 200 nucleotides lacking significant protein-coding capacity [5]. It has been reported that lncRNAs are involved in diverse functions in carcinogenesis, including chromatin modification, transcription, and posttranscriptional processing [6]. In addition, previous studies have indicated that polymorphisms in lncRNAs may influence the risk of gastric cancer [7, 8]. Recent studies have demonstrated that lncRNA maternally expressed gene 3 (MEG3) is abnormal expressed in various human cancers, such as hepatocellular carcinoma [9, 10], bladder cancer [11], glioma [12], and gastric cancer [13]. Moreover, MEG3 overexpression induced cell proliferation and was associated with the development and progression of colorectal cancer [14]. However, little is known about the single nucleotide polymorphisms (SNPs) in MEG3 and colorectal cancer risk.
On the basis of previous findings mentioned above, as well as the influence of SNPs on MEG3, we hypothesis that genetic variants of MEG3 may modify the development of colorectal cancer. The genetic variants of MEG3 may be associated with the expression of MEG3 and consequently influence susceptibility to colorectal cancer. To test the hypothesis, we carried out an association study between tagging SNPs (tagSNPs) in MEG3 and colorectal cancer risk in a hospital-based colorectal cancer case-control study comprising 518 patients and 527 control subjects from China.
RESULTS
The demographic characteristics of participants are described in Table 1. The average age of the patients was 60.0 years old compared with 59.2 years old in controls, which revealed no statistically difference (P = 0.284). Furthermore, there was no significant difference in sex distribution (P = 0.972) or smoking status (P = 0.292). However, the cases were asked to report a significant high rate of family history of cancer than the controls (P < 0.001). Among 518 patients, the number of cases with colon cancer and rectal cancer were 248 (47.9%) and 270 (52.1%), respectively. The tumor stage for I, II, III, and IV were 38 (7.3%), 214 (41.3%), 179 (34.6%), and 87 (16.8%), respectively.
Table 1. Distribution of characteristics among cases and controls.
| Variable | Cases (n = 518) | Controls (n = 527) | P |
|---|---|---|---|
| N (%) | N (%) | ||
| Age (mean ± SD)a | 60.0 ± 12.4 | 59.2 ± 9.4 | 0.284 |
| Gender | |||
| Male | 320 (61.8) | 325 (61.7) | 0.972 |
| Female | 198 (38.2) | 202 (38.3) | |
| Smoking status | |||
| Non-smokers | 347 (67.0) | 369 (70.0) | 0.292 |
| Smokers | 171 (33.0) | 158 (30.0) | |
| Family history of cancer | |||
| No | 405 (78.2) | 488 (92.6) | < 0.001 |
| Yes | 113 (21.8) | 39 (7.4) | |
| Site | |||
| Colon | 248 (47.9) | ||
| Rectum | 270 (52.1) | ||
| Stage | |||
| I | 38 (7.3) | ||
| II | 214 (41.3) | ||
| III | 179 (34.6) | ||
| IV | 87 (16.8) | ||
SD, standard deviation
The primary information of six SNPs in MEG3 is summarized in Table 2. All SNPs in both cases and controls showed a call rate > 97.0%. The genotype frequencies in controls were in line with the Hardy-Weinberg equilibrium model (P = 0.712 for rs3087918, P = 0.930 for rs11160608, P = 0.812 for rs7158663, P = 0.521 for rs4081134, and P = 0.221 for rs10144253).
Table 2. Association analyses between SNPs in MEG3 and colorectal cancer risk.
| SNP | Position | MAF in casea | MAF in controla | Call rate | HWE in case | HWE in control | OR (95%CI)b | Pb |
|---|---|---|---|---|---|---|---|---|
| rs3087918 | 101297963 | 0.367 | 0.381 | 100.0% | 0.344 | 0.712 | 0.94 (0.79-1.12) | 0.493 |
| rs11160608 | 101313093 | 0.433 | 0.436 | 100.0% | 1.000 | 0.930 | 0.98 (0.82-1.16) | 0.788 |
| rs7158663 | 101319424 | 0.295 | 0.242 | 98.9% | 0.138 | 0.812 | 1.31 (1.08-1.59) | 0.007 |
| rs4081134 | 101321788 | 0.227 | 0.219 | 97.8% | 0.314 | 0.521 | 1.04 (0.84-1.29) | 0.711 |
| rs10144253 | 101325962 | 0.470 | 0.472 | 99.8% | 0.659 | 0.221 | 0.98 (0.82-1.17) | 0.843 |
Minor allele frequency.
Additive model adjusted for age, sex, and smoking status.
As shown in Table 2, the allele frequency of rs7158663 exhibited a significant difference between case and control groups (P = 0.007). The minor allele frequency (MAF) of rs7158663 in cases and controls were 0.295 and 0.242, respectively. However, no significant association with colorectal cancer was identified for other five SNPs (P = 0.493 for rs3087918, P = 0.788 for rs11160608, P = 0.711 for rs4081134, and P = 0.843 for rs10144253). In addition, after adjusting for multiple testing using Bonferroni correction, rs7158663 was still significant (P = 0.035).
We further performed a multivariate logistic regression analysis for rs7158663 by adjusting age, sex, and smoking. As listed in Table 3, rs7158663 AA genotype, but not GA genotype, had a significantly elevated risk of colorectal cancer, compared with GG genotype (P = 0.007) (OR = 1.96 and P = 0.006 for AA versus GG, and OR = 1.20 and P = 0.171 for GA versus GG). For genetic models, rs7158663 was significant between cases and controls in dominant, recessive, and additive models (P = 0.035, 0.012, and 0.007, respectively).
Table 3. Genotype frequencies of MEG3 rs7158663 among cases and controls and their association with colorectal cancer risk.
| Genotype | Cases (n = 516) | Controls (n = 517) | OR (95%CI)a | Pa |
|---|---|---|---|---|
| N (%) | N (%) | |||
| GG | 264 (51.2) | 298 (57.6) | 1.00 | |
| GA | 200 (38.7) | 188 (36.4) | 1.20 (0.92-1.56) | 0.171 |
| AA | 52 (10.1) | 31 (6.0) | 1.96 (1.22-3.17) | 0.006 |
| Dominant | 1.31 (1.02-1.67) | 0.035 | ||
| Recessive | 1.82 (1.14-2.91) | 0.012 | ||
| Additive | 1.31 (1.08-1.59) | 0.007 |
Adjusted for age, sex, and smoking status.
We next evaluated the stratified association of rs7158663 with colorectal cancer risk by age, sex, family history of cancer, and smoking habits (Table 4). We observed that the GA/AA genotype hadan increased risk of colorectal cancer (OR = 1.31, P = 0.035). This increased effect was also more evident in subgroups of age ≤ 60 (OR = 1.71, P = 0.003) and patients with family history (OR = 1.25, P = 0.011). Nevertheless, no significant evidence was found for interaction between rs7158663 and these two factors.
Table 4. Stratified analyses for MEG3 rs7158663 genotypes in cases and controls.
| Variable | GG | GA/AA | OR (95%CI)a | Pa |
|---|---|---|---|---|
| N (case/control) | N (case/control) | |||
| Age (y), median | ||||
| ≤60 | 104/158 | 124/111 | 1.71 (1.20-2.44) | 0.003 |
| >60 | 160/140 | 128/108 | 1.04 (0.74-1.46) | 0.844 |
| Gender | ||||
| Male | 167/177 | 152/142 | 1.15 (0.84-1.57) | 0.396 |
| Female | 97/121 | 100/77 | 1.62 (0.98-2.34) | 0.076 |
| Smoking status | ||||
| Non-smokers | 168/207 | 178/155 | 1.42 (0.96-1.87) | 0.062 |
| Smokers | 96/91 | 74/64 | 1.10 (0.70-1.71) | 0.679 |
| Family history of cancer | ||||
| No | 204/268 | 200/210 | 1.25 (0.96-1.63) | 0.099 |
| Yes | 60/30 | 52/9 | 2.98 (1.28-6.92) | 0.011 |
Adjusted for age, sex, and smoking status.
We evaluated the correlations between rs7158663 and clinical features of colorectal cancer, including tumor site and stage. As shown in Table 5, no significant relationship was observed (P = 0.842 for tumor site, and P = 0.601 for tumor stage).
Table 5. Association analyses between MEG3 rs7158663 genotypes and clinicalpathologic characteristics of cases.
| Variable | GG | GA/AA | OR (95%CI)a | Pa |
|---|---|---|---|---|
| N (%) | N (%) | |||
| Site | ||||
| Colon | 126 (24.4) | 122 (23.6) | 1.00 | |
| Rectum | 138 (26.7) | 130 (25.2) | 1.04 (0.73-1.46) | 0.842 |
| Stage | ||||
| I/II | 126 (24.4) | 125 (24.2) | 1.00 | |
| III/IV | 138 (26.7) | 127 (24.6) | 1.10 (0.78-1.55) | 0.601 |
Adjusted for age, sex, and smoking status.
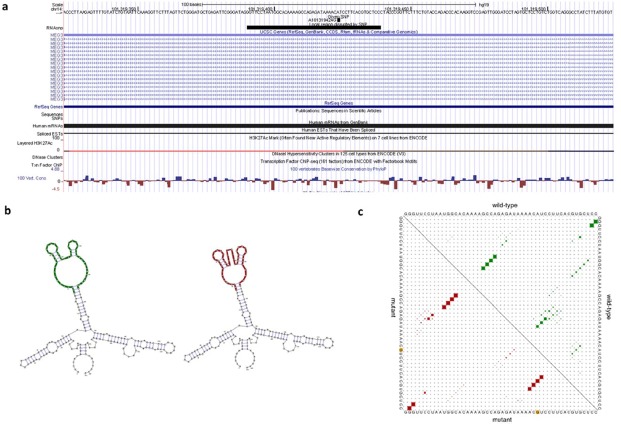
We peformed an in silico analysis of rs7158663 on MEG3 by RNAsnp. As shown in Figure 1, the RNAsnp predicted that rs7158663 G-A allele substitution leaded to a minimum free energy (MFE) change from −53.30 kcal/mol to −55.20 kcal/mol. Moreover, the base pair probabilities were different between rs7158663 G allele and A allele.
Figure 1. The predicting rs7158663 on MEG3 secondary structure.
a. UCSC genome browser view for structure-disruptive rs7158663. b. Minimum free energy structures of rs7158663. c. Dot plot of the global structure.
DISCUSSION
We investigated the association of tagSNPs of MEG3 with colorectal cancer risk in a population of Chinese. We demonstrated, for the first time, that rs7158663 in MEG3 had a strong association with increased risk of colorectal cancer. In addition, the elevated risk was greater for subjects with age ≤ 60 and family history of cancer. These results suggested that rs7158663 may be a susceptibility SNP for colorectal cancer in Chinese populations.
Given MEG3 is a lncRNA that plays critical roles in tumor cellular proliferation, migration and invasion [13, 15, 16], SNPs in MEG3 may affect cell phenotypes and cause the risk of developing cancer. MEG3 is located on chromosome 14q32, a region proposed to contain putative tumor suppressors [17]. Although the regulation of MEG3 and its precise mechanism of action in cancer are still well known, emerging evidence strongly demonstrates that MEG3 functions as a novel lncRNA tumor suppressor [18]. In additional to colorectal normal tissue, MEG3 is highly expressed in brain, lung, and liver [19]. Interestingly, we found that MEG3 rs7158663 is correlated to significantly increased risk of colorectal cancer, which may be explained by the finds from Yin et al. [14]. They found that MEG3 predicts a poor prognosis of colorectal cancer by influencing cell proliferation.
Until now, some studies have indicated that SNPs in lncRNAs could potentially impact various biological processes by influencing biological pathways. For example, Yang et al. first revealed the relationship between lncRNA H19 and gastric cancer; they found an important role for H19 variants in gastric cancer carcinogenesis [8]. Zhang et al. identified an allelic regulation of rs920778 on lncRNA HOTAIR expression, as well as associated with the development and progression of esophageal squamous cell carcinoma [20]. The novel function of rs920778 on HOTAIR was further confirmed in other cancer, including colorectal cancer [21], gastric cancer [22], and breast cancer [23]. The RNAsnp prediction revealed that rs7158663 in MEG3 changed the folding structures of MEG3. Therefore, we speculate that rs7158663 could be a regulatory SNP, which regulate expression of MEG3 and contribute to genetic susceptibility of colorectal cancer.
Several limitations of the present study should be mentioned. Firstly, these findings were based on genetic analysis of a single gene with a relatively small size of colorectal cancer patients, which need to be validated using an independent prospective clinical study. Secondly, our study was based on a hospital-based study; therefore, potentially important sources of selection bias may exist. Thirdly, diet is a major determinant of risk for colorectal cancer [24]; however, our study had incomplete information on dietary consumption. Further studies are needed to determine the association between dietary information and genetic variants in MEG3.
In summary, we identified a genetic susceptibility SNP rs7158663 for colorectal cancer in Chinese. The SNP rs7158663 in MEG3 played a vital role in colorectal carcinogenesis. These data suggest that genetic variants in MEG3 may serve as potential risk factor and targets for colorectal cancer therapy in the further.
MATERIALS AND METHODS
Study subjects
This study consisted of 518 colorectal cancer cases and 527 control subjects from the hospitals of Southeast University Medical College. All cases were patients newly diagnosed with histologically confirmed colorectal adenocarcinomas who were admitted to the hospitals. No patient had received radiotherapy or chemotherapy. Controls were frequency-matched with cases on age and sex who were recruited at the same time period. All controls were unrelated ethnic Han Chinese and these subjects had no history of cancer. All participants provided informed consent after the interview. This research protocol was approved by the Institutional Review Board of Southeast University Medical College.
SNPs selection
We selected the tagSNPs of MEG3 with the MAF > 0.05 in Han Chinese from the 1000 Genome Projects. As a result, six tagSNPs were selected using a pairwise Tagger method with r2 > 0.8 to capture other SNPs, rs10144253.
Genotyping
Genomic DNA was extracted from peripheral blood using the TIANamp Blood DNA kit (Tiangen, China). Genotyping was performed by TaqMan SNP genotyping assay. Real-time TaqMan PCR and genotyping were conducted on an ABI 7500 real-time PCR System (Applied Biosystems, USA). The results of allelic discrimination were analyzed using SDS 2.4 software. For quality control, we included two negative controls (water) and two duplicates in each 96-well plate. Furthermore, about 3% of selected samples were repeated genotyping to confirm the results in a blind fashion.
Statistical analysis
Tests for the Hardy-Weinberg equilibrium in cases and controls were performed by good-of-fit χ2 test. Student's t-test or χ2 test was used to assess the significance of any differences in frequency distributions of demographic variables and genotypes among cases and controls. We estimated the association between genotypes and colorectal cancer risk by odds ratios (ORs) and 95% confidence intervals (CIs) using the logistic regression. The ORs and 95%CIs were further adjusted by for age, sex, and smoking habits. All analyses were two-sided and P < 0.05 was considered significant. All statistical calculations were conducted with SPSS 13.0 software (SPSS Inc., Chicago, IL, USA).
Footnotes
CONFLICTS OF INTERESTS
We declared no conflicts of interests in this study.
REFERENCES
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. Report of incidence and mortality in China cancer registries, 2009. Chinese journal of cancer research [article in Chineese] 2013;2(5):10–21. doi: 10.3978/j.issn.1000-9604.2012.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X, Zhang B, Zheng W. Genetic variants associated with colorectal cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut. 2014;63:326–336. doi: 10.1136/gutjnl-2012-304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nature reviews Genetics. 2009;10:353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 5.Costa FF. Non-coding RNAs: Meet thy masters. BioEssays. 2010;32:599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes & development. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J, Ma G, Chu H, Tong N, Zhu H, Wang M, Qiang F, Zhang Z. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6:31255–31262. doi: 10.18632/oncotarget.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C, Tang R, Ma X, Wang Y, Luo D, Xu Z, Zhu Y, Yang L. Tag SNPs in long non-coding RNA H19 contribute to susceptibility to gastric cancer in the Chinese Han population. Oncotarget. 2015;6:15311–15320. doi: 10.18632/oncotarget.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, Wang P, Sun B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Molecular carcinogenesis. 2016;55:209–19. doi: 10.1002/mc.22270. [DOI] [PubMed] [Google Scholar]
- 11.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Molecular bioSystems. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. Journal of cellular biochemistry. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 13.Yan J, Guo X, Xia J, Shan T, Gu C, Liang Z, Zhao W, Jin S. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Medical oncology. 2014;31:879. doi: 10.1007/s12032-014-0879-6. [DOI] [PubMed] [Google Scholar]
- 14.Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour biology. 2015;36:4851–4859. doi: 10.1007/s13277-015-3139-2. [DOI] [PubMed] [Google Scholar]
- 15.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balik V, Srovnal J, Sulla I, Kalita O, Foltanova T, Vaverka M, Hrabalek L, Hajduch M. MEG3: a novel long noncoding potentially tumour-suppressing RNA in meningiomas. Journal of neuro-oncology. 2013;112:1–8. doi: 10.1007/s11060-012-1038-6. [DOI] [PubMed] [Google Scholar]
- 17.Bando T, Kato Y, Ihara Y, Yamagishi F, Tsukada K, Isobe M. Loss of heterozygosity of 14q32 in colorectal carcinoma. Cancer genetics and cytogenetics. 1999;111:161–165. doi: 10.1016/s0165-4608(98)00242-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. Journal of molecular endocrinology. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. The Journal of clinical endocrinology and metabolism. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062–2067. doi: 10.1093/carcin/bgu103. [DOI] [PubMed] [Google Scholar]
- 21.Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H, Tong N, Chen J, Zhang Z, Wang M. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303–310. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 22.Bayram S, Ulger Y, Sumbul AT, Kaya BY, Rencuzogullari A, Genc A, Sevgiler Y, Bozkurt O, Rencuzogullari E. A functional HOTAIR rs920778 polymorphism does not contributes to gastric cancer in a Turkish population: a case-control study. Familial cancer. 2015;14:561–7. doi: 10.1007/s10689-015-9813-0. [DOI] [PubMed] [Google Scholar]
- 23.Bayram S, Sumbul AT, Batmaci CY, Genc A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour biology. 2015;36:3863–3870. doi: 10.1007/s13277-014-3028-0. [DOI] [PubMed] [Google Scholar]
- 24.Singh PN, Fraser GE. Dietary risk factors for colon cancer in a low-risk population. American journal of epidemiology. 1998;148:761–774. doi: 10.1093/oxfordjournals.aje.a009697. [DOI] [PubMed] [Google Scholar]