Abstract
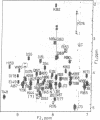
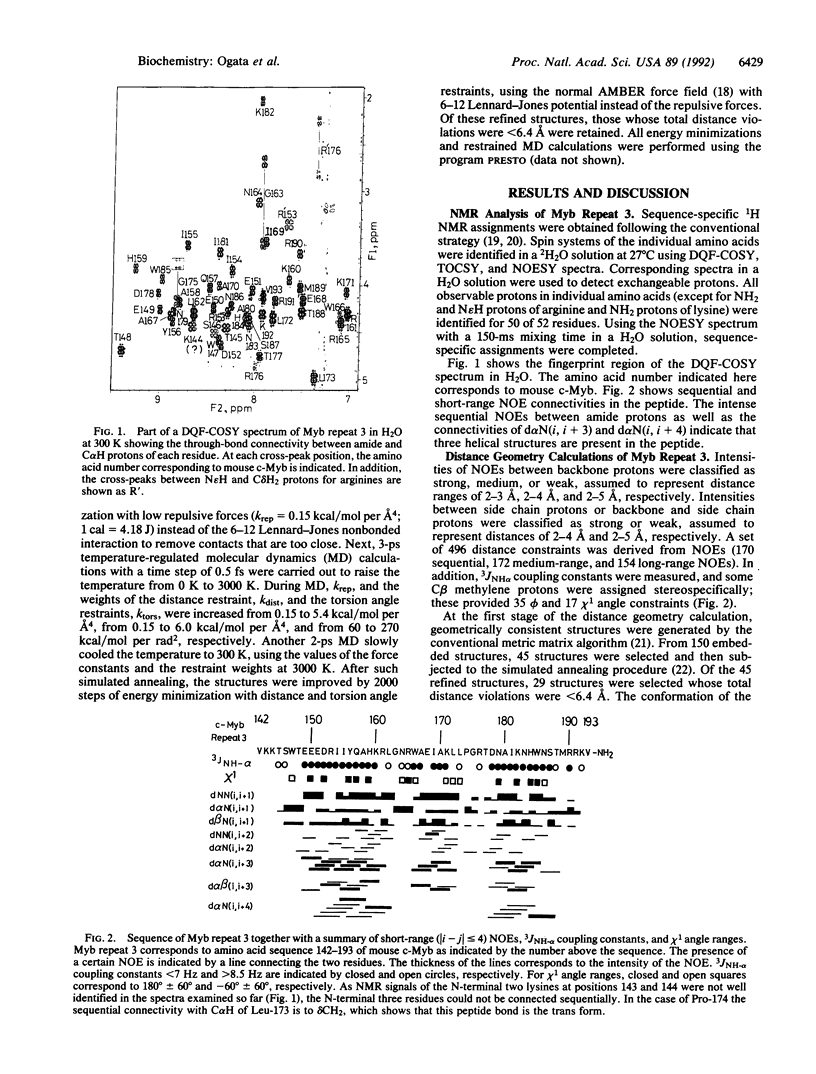
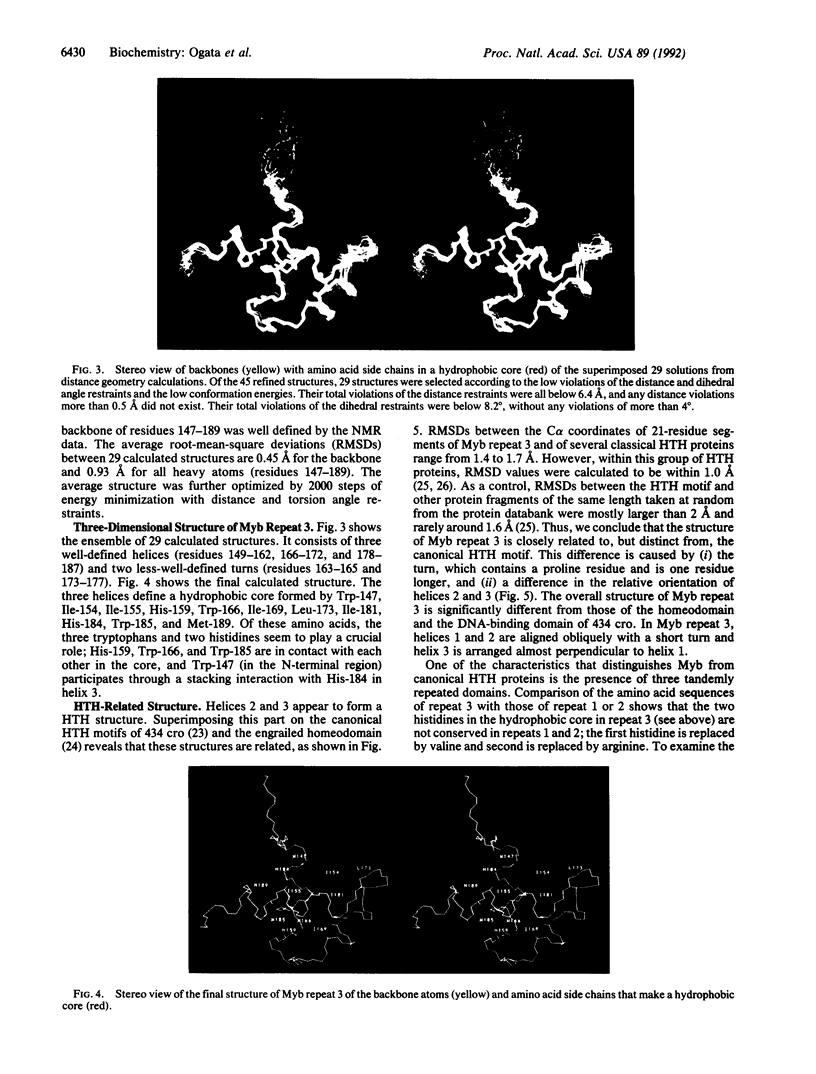
The DNA-binding domain of the c-myb protooncogene product consists of three imperfect tandem repeats of 51 or 52 amino acids, each of which contains three conserved tryptophans, spaced 18 or 19 amino acids apart. The structure of the third repeat, which is essential for sequence-specific DNA binding, has been determined by NMR with distance geometry calculation. It includes three well-defined helices (residues 149-162, 166-172, and 178-187) maintained by a hydrophobic core that includes the three conserved tryptophans, together with two histidines. Helices 2 and 3 form a structure related to but distinct from a canonical helix-turn-helix motif. In particular, the turn between these helices is one amino acid longer than the corresponding turn in bacterial repressors and homeodomains and contains a proline residue. In addition, the architecture of the three helices is different from those of homeodomains and DNA-binding domains of bacterial repressors. Based on the present structure, the binding mode of Myb repeat 3 with a specific DNA is also discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Anton I. A., Frampton J. Tryptophans in myb proteins. Nature. 1988 Dec 22;336(6201):719–719. doi: 10.1038/336719a0. [DOI] [PubMed] [Google Scholar]
- Biedenkapp H., Borgmeyer U., Sippel A. E., Klempnauer K. H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988 Oct 27;335(6193):835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Driscoll P. C., Gronenborn A. M., Beress L., Clore G. M. Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: a study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989 Mar 7;28(5):2188–2198. doi: 10.1021/bi00431a033. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Wand A. J. Main-chain-directed strategy for the assignment of 1H NMR spectra of proteins. Biochemistry. 1987 Sep 22;26(19):5953–5958. doi: 10.1021/bi00393a001. [DOI] [PubMed] [Google Scholar]
- Frampton J., Leutz A., Gibson T., Graf T. DNA-binding domain ancestry. Nature. 1989 Nov 9;342(6246):134–134. doi: 10.1038/342134a0. [DOI] [PubMed] [Google Scholar]
- Gabrielsen O. S., Sentenac A., Fromageot P. Specific DNA binding by c-Myb: evidence for a double helix-turn-helix-related motif. Science. 1991 Sep 6;253(5024):1140–1143. doi: 10.1126/science.1887237. [DOI] [PubMed] [Google Scholar]
- Howe K. M., Reakes C. F., Watson R. J. Characterization of the sequence-specific interaction of mouse c-myb protein with DNA. EMBO J. 1990 Jan;9(1):161–169. doi: 10.1002/j.1460-2075.1990.tb08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C., Sarai A., Sawazaki T., Nakagoshi H., He D. N., Ogata K., Nishimura Y., Ishii S. The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J Biol Chem. 1990 Nov 15;265(32):19990–19995. [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Sippel A. E. The highly conserved amino-terminal region of the protein encoded by the v-myb oncogene functions as a DNA-binding domain. EMBO J. 1987 Sep;6(9):2719–2725. doi: 10.1002/j.1460-2075.1987.tb02565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Ernst R. R., Wüthrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem Biophys Res Commun. 1980 Jul 16;95(1):1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- Mondragón A., Wolberger C., Harrison S. C. Structure of phage 434 Cro protein at 2.35 A resolution. J Mol Biol. 1989 Jan 5;205(1):179–188. doi: 10.1016/0022-2836(89)90374-4. [DOI] [PubMed] [Google Scholar]
- Nakagoshi H., Nagase T., Kanei-Ishii C., Ueno Y., Ishii S. Binding of the c-myb proto-oncogene product to the simian virus 40 enhancer stimulates transcription. J Biol Chem. 1990 Feb 25;265(6):3479–3483. [PubMed] [Google Scholar]
- Ness S. A., Marknell A., Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989 Dec 22;59(6):1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- Nishina Y., Nakagoshi H., Imamoto F., Gonda T. J., Ishii S. Trans-activation by the c-myb proto-oncogene. Nucleic Acids Res. 1989 Jan 11;17(1):107–117. doi: 10.1093/nar/17.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem Biophys Res Commun. 1983 Dec 16;117(2):479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- Saikumar P., Murali R., Reddy E. P. Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8452–8456. doi: 10.1073/pnas.87.21.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H., Kanei-Ishii C., Nagase T., Nakagoshi H., Gonda T. J., Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K., Bishop J. M. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell. 1989 Jul 14;58(1):85–93. doi: 10.1016/0092-8674(89)90405-4. [DOI] [PubMed] [Google Scholar]