Abstract
Purpose
The aldehyde dehydrogenase 2 (ALDH2) gene has been implicated in the development of alcoholic liver cirrhosis (ALC) in East Asians. However, the results are inconsistent. In this study, a meta-analysis was performed to assess the associations between the ALDH2 polymorphism and the risk of ALC.
Materials and Methods
Relevant studies were retrieved by searching PubMed, Web of Science, CNKI, Wanfang and Veipu databases up to January 10, 2015. Pooled odds ratio (OR) and 95% confidence interval (CI) were calculated using either the fixed- or random effects model.
Results
A total of twelve case-control studies included 1003 cases and 2011 controls were included. Overall, the ALDH2 polymorphism was associated with a decreased risk of ALC (*1/*2 vs. *1/*1: OR=0.78, 95% CI: 0.61–0.99). However, in stratification analysis by country, we failed to detect any association among Chinese, Korean or Japanese populations.
Conclusion
The pooled evidence suggests that ALDH2 polymorphism may be an important protective factor for ALC in East Asians.
Keywords: ALDH2, polymorphism, alcoholic liver cirrhosis, meta-analysis
INTRODUCTION
Alcoholic liver disease (ALD) presents as a wide spectrum of liver disease, including fatty liver, acute alcoholic hepatitis, and alcoholic liver cirrhosis (ALC), and hepatocellular carcinoma.1,2 It is well known that almost all alcohol abusers develop hepatic steatosis; however, only a small portion develop signs of liver disease, suggesting that some of the genetic variations are involved in the etiology of ALD.3,4
Aldehyde dehydrogenase 2 (ALDH2), belonging to a low-Km mitochondrial ALDH, is the second enzyme to eliminate most of the acetaldehyde generated during alcohol metabolism. The human ALDH2 gene is located on chromosome 12q24.2 and composed of 13 exons, spanning 46031 bp.5 There are several polymorphism sites in the ALDH2 gene, and the Glu487Lys polymorphism (rs671, also named Glu504Lys, with the glutamate corresponding to *1 allele, and lysine corresponding to *2 allele) has been the most frequently studied. This variant could partially determine blood acetaldehyde concentrations after alcohol consumption: heterozygotes or homozygotes for the ALDH2*2 allele showed 6- and 19-fold higher blood acetaldehyde concentrations than individuals with homozygous common-allele, respectively.6 Furthermore, the variant ALDH2*2 allele is prevalent in East Asian, but is rare in non-Asians.7 Up to now, many studies have investigated the association between the ALDH2 polymorphism and the risk of alcoholic cirrhosis.8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25 However, the results remain controversial. Therefore, in the current study, we conducted a meta-analysis in order to get a robust conclusion about the association between the polymorphism and alcoholic cirrhosis risk among East Asians.
MATERIALS AND METHODS
Search strategy
We searched the electronic literature PubMed, Web of Science, CNKI, Wanfang and Veipu databases for all relevant articles. The last search update was January 10, 2015, using the search terms: "aldehyde dehydrogenase 2 or ALDH2" and "genetic polymorphism or polymorphisms or variant" and "alcoholic liver disease or ALD or alcoholic liver cirrhosis or ALC or cirrhosis". The references of the retrieved literature were also hand-search for additional studies. There was no restriction on time period, sample size, population, language, or type of reports.
Inclusion and exclusion criteria
The studies included must meet the following criteria: 1) evaluate the association between the ALDH2 polymorphism and the risk of ALC, 2) case-control study in design, 3) provide sufficient data for calculation of the odds ratios (ORs) with the corresponding 95% confidence interval (CI). Studies were excluded if one of the following existed: 1) review articles and meta-analysis, 2) studies without the usable data of the ALDH2 genotype, and 3) duplicate publications.
Data extraction
Data were extracted independently by two investigators. The following information was extracted from each study: name of the first author, publication year, country of origin, age, gender, genotyping methods, numbers of genotypes in cases and controls, and evidence of Hardy-Weinberg equilibrium (HWE) in controls. Any encountered discrepancies were resolved by consensus.
Statistical analysis
HWE was evaluated for each study using an internet-based HWE calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). The risk of ALC associated with the ALDH2 polymorphism was estimated for each study by OR and 95% CI. The heterogeneity among the results was assessed by χ2-based Q test as well as the I2 statistic.26 When a significant Q test (p>0.1) or I2<50% indicated homogeneity across studies, the fixed effects model was used,27 or the random effects model was used.28 Then, we performed stratification analyses on country. Analysis of sensitivity was performed to evaluate the stability of the results. The Begg's funnel plot and Egger's regression test were used to assess the publication bias.29,30 All statistical analyses were carried out using the Cochrane Collaboration RevMan 5.2 and STATA package version 12.0 (Stata Corporation, College Station, TX, USA).
RESULTS
Study characteristics
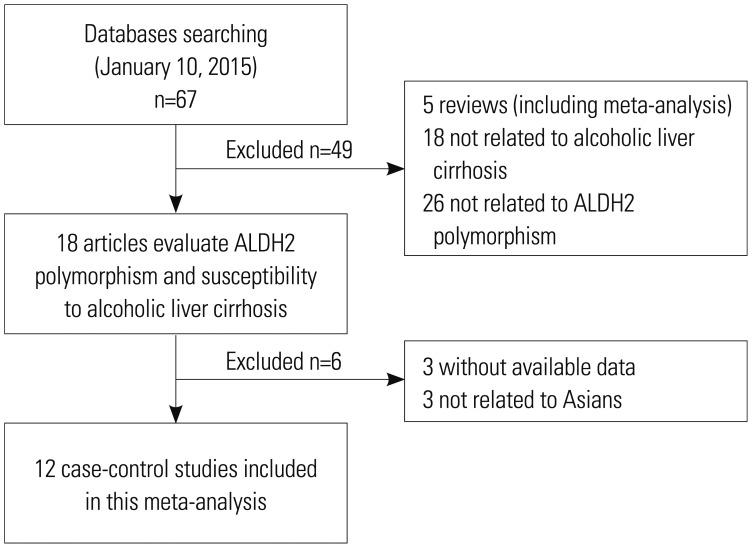
The flow chart of study selection is presented in Fig. 1, a total of 67 articles were identified through database searching. 18 studies8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25 were included based on the inclusion criteria. Because three studies20,21,22 were conducted on non-Asians, they were excluded. Moreover, we excluded three studies because they did not present detailed genotyping information.23,24,25 Therefore, as shown in Table 1, there were 12 case-control studies on ALDH2 polymorphism with 1003 cases and 2011 controls. Of the 12 eligible studies, four studies8,9,10,11 were conducted on Chinese populations, five studies15,16,17,18,19 on Japanese populations and three studies on Korean populations.12,13,14 The distribution of genotypes in the controls was consistent with the HWE for all selected studies, except for two studies.13,19
Fig. 1. The flow chart of study selection based on the inclusion and exclusion criteria. ALDH2, aldehyde dehydrogenase 2.
Table 1. Characteristics of Studies Included in the Meta-Analysis.
| Author | Year | Country | Age (mean±SD) | Gender (M/F) | Genotyping methods | Genotype (case/control) | HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Total | *1/*1 | *1/*2 | |||||
| Chao, et al.8 | 1994 | China | 55.0±3.0 | NR | 27/0 | 50/0 | PCR-directed mutagenesis | 27/50 | 22/44 | 5/6 | 0.652 |
| Chao, et al.9 | 1997 | China | 50.7±12.9 | 49.9±10.6 | 71/4 | 19/0 | PCR-directed mutagenesis | 75/19 | 65/15 | 10/4 | 0.608 |
| Chao, et al.10 | 2000 | China | 50.5±12.6 | 40.8±11.0 | 115/5 | 78/9 | PCR-directed mutagenesis | 116/87 | 98/67 | 18/20 | 0.226 |
| Chao, et al.11 | 2003 | China | 50.0±12.7 | 44.7±9.5 | 150/9 | 35/0 | PCR-directed mutagenesis | 159/35 | 130/28 | 29/7 | 0.511 |
| Kee, et al.12 | 2003 | Korea | NR | NR | NR | NR | PCR-RFLP | 30/12 | 28/9 | 2/3 | 0.621 |
| Kim, et al.13 | 2004 | Korea | 48.8±18.8 | 49.2±20.3 | 19/3 | 44/56 | PCR-RFLP | 22/100 | 17/58 | 5/30 | 0.017 |
| Lee, et al.14 | 2001 | Korea | 52.2±9.3 | 51.7±10.1 | 56/0 | 52/0 | PCR-RFLP | 56/52 | 52/50 | 4/2 | 0.888 |
| Nagata, et al.15 | 1999 | Japan | NR | NR | NR | NR | PCR-SSCP | 27/28 | 25/21 | 2/7 | 0.450 |
| Nagata, et al.16 | 2002 | Japan | NR | NR | NR | 76/68 | PCR-SSCP | 44/29 | 42/26 | 2/3 | 0.769 |
| Yamauchi, et al.17 | 1995 | Japan | 53.0±1.1† | NR | 46/0 | 34/0 | PCR-RFLP | 46/34 | 30/29 | 16/5 | 0.644 |
| Yamauchi, et al.18 | 1995 | Japan | 53.0±1.2† | NR | 42/0 | 34/0 | PCR-RFLP | 42/34 | 30/29 | 12/5 | 0.644 |
| Yokoyama, et al.19 | 2013 | Japan | 55.4±0.5† | 56.5±0.2† | 359/0 | 1543/0 | PCR-RFLP | 359/1543 | 316/1288 | 43/255 | 0.000 |
HWE, Hardy-Weinberg equilibrium; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PCR-SSCP, polymerase chain reaction-singlestrand conformation polymorphism; NR, not reported.
†Mean±SE.
Quantitative data synthesis
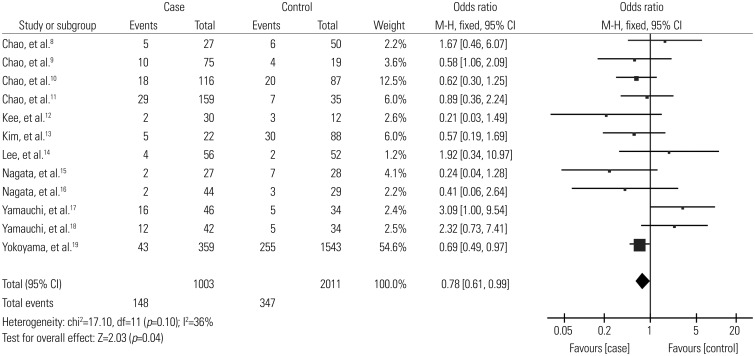
Overall, the ALDH2 polymorphism was associated with a decreased risk of alcoholic cirrhosis (*1/*2 vs. *1/*1: OR=0.78, 95% CI: 0.61–0.99) (Fig. 2).
Fig. 2. Forest plots for the association of ALDH2 and alcoholic cirrhosis risk (*1/*2 vs. *1/*1). ALDH2, aldehyde dehydrogenase 2; CI, confidence interval.
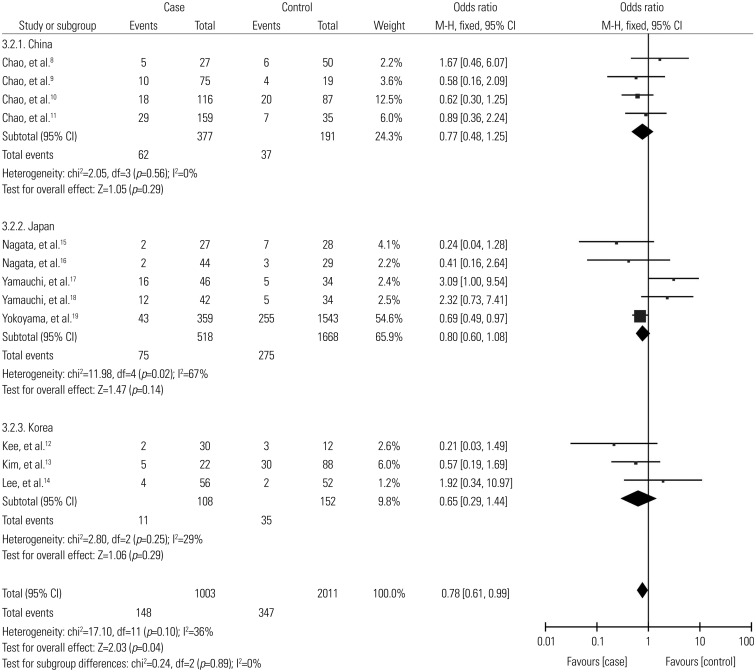
In stratification analyses by country, no significant associations were observed among Chinese (OR=0.77, 95% CI: 0.48–1.25), Korean (OR=0.65, 95% CI: 0.29–1.44), and Japanese populations (OR=0.97, 95% CI: 0.42–2.25) (Fig. 3).
Fig. 3. Subgroup analysis of odds ratios by country for association between ALDH2 polymorphism and risk of alcoholic liver cirrhosis (*1/*2 vs. *1/*1). ALDH2, aldehyde dehydrogenase 2; CI, confidence interval.
Four studies8,9,10,14 reported the alcohol consumption in ALC and control groups. Of which two studies found no significant difference between them, while other studies found that the amount of daily alcohol consumption and/or the duration of alcohol consumption history in ALC group were lower than control group (Table 2). In addition, two studies analysed the usual alcohol consumption between the ALDH2 *1/*1 and ALDH2 *1/*2 genotype in alcoholics. Interestingly, the results were inconsistent. Nagata, et al.16 found that the daily amount of ethanol used was smaller in patients with ALDH2 *1*2 than in patients with ALDH2 *1*1, while the study conducted by Yokoyama, et al.19 showed no significant difference between the ALDH2 genotype groups.
Table 2. Comparison of Alcohol Consumption in Different Groups.
| Author | Year | Country | Group | Alcohol consumption | |
|---|---|---|---|---|---|
| Daily (g) | Duration (yr) | ||||
| Chao, et al.9 | 1997 | China | ALC | 191±105 | 25.3±10.8 |
| Alcoholic without LC | 203±90 | 26.6±10.6 | |||
| Chao, et al.10 | 2000 | China | ALC | 199±118 | 25.6±11.0 |
| Alcoholic without LC | 137±76 | 16.7±9.7 | |||
| Chao, et al.11 | 2003 | China | ALC | 193±113 | 25.4±11.1 |
| Alcoholic without LC | 196±137 | 19.5±8.2 | |||
| Lee, et al.14 | 2001 | Korea | ALC | 165.6±91.8 | 22.7±9.9 |
| Alcoholic without LC | 152.8±99.6 | 22.8±9.8 | |||
ALC, alcoholic liver cirrhosis.
A single study was removed from meta-analysis each time to determine the influence of its individual data sets to the pooled ORs, and the estimated pooled ORs were not materially altered.
Test of heterogeneity
There was no significant heterogeneity for overall comparisons (*1/*2 vs. *1/*1: p=0.10, I2=36%). In the subgroup analysis by country, results were similar among Chinese and Korean populations (p=0.56, I2=0%, p=0.25, I2=29%, respectively), whereas significant heterogeneity between studies was observed in Japanese population (p=0.02, I2=67%).
Publication bias
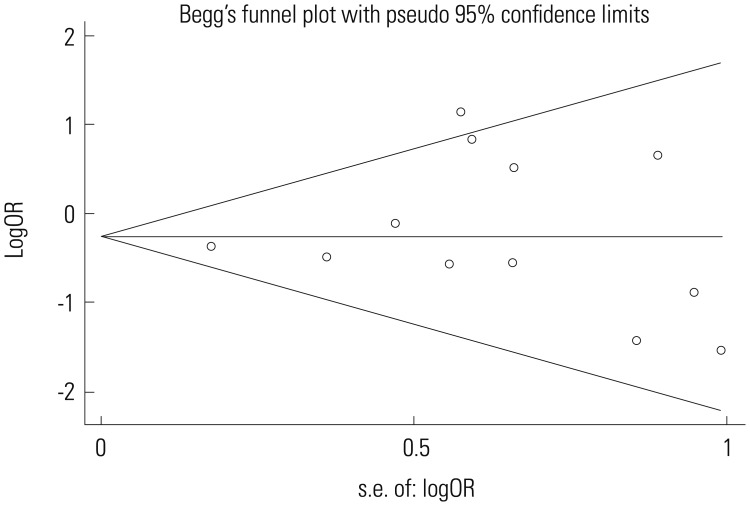
Begg's funnel plot and Egger's test were performed to assess publication bias. As reflected by the funnel plots (Fig. 4) and the corresponding Egger's test, no publication bias was detected in any comparisons (*1/*2 vs. *1/*1: p=0.655).
Fig. 4. Begg's funnel plot of publication bias test for ALDH2 and alcoholic cirrhosis risk (*1/*2 vs. *1/*1). ALDH2, aldehyde dehydrogenase 2; OR, odds ratio.
DISCUSSION
The metabolism of alcohol consists of two steps, it is first catalytically oxidized into acetaldehyde, and then acetaldehyde is metabolized into harmless acetate by ALDH.31 ALDH2 is the major enzyme for acetaldehyde elimination, and its polymorphism determines blood acetaldehyde concentrations after alcohol consumption. It is known that the ALDH2 *2 allele produces a catalytically inactive isozyme, which has greatly reduced or no ability to metabolize acetaldehyde.32 The accumulation of acetaldehyde in the blood and repeated high exposure to acetaldehyde after drinking might contribute to the development of ALC.33 However, the results of studies in this area are inconsistent. Chao, et al.8 reported that the ALDH2 polymorphism may contribute to susceptibility for ALC. Whereas, some studies have reported that no statistically significant association exists between the ALDH2 polymorphism and ALC risk.14,34
Recently, Li, et al.35 conducted a meta-analysis and evaluated the association between ALDH2 polymorphism and the risk of alcoholism and alcohol-induced medical diseases in Asians. The results confirmed the involvement of the human ALDH2 gene in the pathogenesis of alcohol dependence as well as alcohol-induced medical illnesses in East-Asians. However, only 5 studies focusing on ALC were included in the above meta-analysis, due to the limited studies, and further analyses was not conducted. In this study, however, we conducted a comprehensive literature search in different databases (i.e., Web of Science, CNKI, Wanfang and Veipu) and added 8 studies, thus allowing for a larger number of subjects and more precise risk estimation. Thirteen case-control studies included 1025 cases and 2566 controls. In the present study, we found that the ALDH2*2 allele is associated with ALC; individuals with the ALDH2*2/*2 and/or *1/*2 genotype had a lower risk of developing ALC compared to individuals with the ALDH2*/1*1 genotype. The results of our meta-analysis seem to contradict the observations of functional studies of ALDH2, which had suggested that ALDH2 played an important role in the development of ALC. This inconsistency might partly be explained by the fact that ALDH2*1/*2 and *2/*2 subjects with high blood acetaldehyde levels after alcohol consumption could develop intense facial flushing responses with nausea, headache, drowsiness and so on.36 This unpleasant discomfort may prevent people from consuming alcohol and may keep them from developing alcoholism, so that they have much smaller chance to expose to the acetaldehyde, and then decrease the risk of developing ALC.37 In addition, a previous study examined the lifetime drinking history of alcoholics, and the results showed that the ALDH2*1/*1 group experienced the onset of each event in their drinking history, including the onset of habitual, excessive, binge drinking, and alcohol dependence, earlier in life than the ALDH2*1/*2 group.38 Although reactive acetaldehyde is a candidate for the causal agent of the organ injuries in alcoholics, the increased peak blood acetaldehyde levels of alcoholics was found to be inversely correlated with the depressed hepatic ALDH activity.39 Yokoyama, et al.40 found that the levels of blood acetaldehyde in the active ALDH2*1/*1 alcoholics, are comparable with the levels of the inactive heterozygous ALDH2*1/*2 alcoholics with less active ADH1B*1/*1.
In this study, we did not detect a significant publication bias in this meta-analysis, confirming the reliability of our results. As for the heterogeneity, no significant heterogeneity was found in overall comparison, when stratified by country; the heterogeneity was partly decreased or removed among Korean and Chinese populations. However, heterogeneity existed in Japanese population. The above results suggest that the ethnic background might be the source of heterogeneity.
Some limitations should be addressed when explaining the results. First, only published studies were retrieved, publication bias might be possible. Second, our results were based on unadjusted estimates, hence the effect estimates were relatively imprecise. In addition, all studies were conducted in Japan, Korea, and China, which may generate selective bias. More studies focused on other Asians are needed.
In conclusion, the results of this meta-analysis suggest that the ALDH2 polymorphism may be an important protective factor for alcoholic cirrhosis in East Asians.
Footnotes
The authors have no financial conflicts of interest.
References
- 1.Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373:2223–2233. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- 2.O'Shea RS, Dasarathy S, McCullough AJ Practice Guideline Committee of the American Association for the Study of Liver Diseases; Practice Parameters Committee of the American College of Gastroenterology. Alcoholic liver disease. Hepatology. 2010;51:307–328. doi: 10.1002/hep.23258. [DOI] [PubMed] [Google Scholar]
- 3.Monzoni A, Masutti F, Saccoccio G, Bellentani S, Tiribelli C, Giacca M. Genetic determinants of ethanol-induced liver damage. Mol Med. 2001;7:255–262. [PMC free article] [PubMed] [Google Scholar]
- 4.Stickel F, Hampe J. Genetic determinants of alcoholic liver disease. Gut. 2012;61:150–159. doi: 10.1136/gutjnl-2011-301239. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- 6.Mizoi Y, Yamamoto K, Ueno Y, Fukunaga T, Harada S. Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol. 1994;29:707–710. [PubMed] [Google Scholar]
- 7.Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, et al. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88:344–346. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- 8.Chao YC, Liou SR, Chung YY, Tang HS, Hsu CT, Li TK, et al. Polymorphism of alcohol and aldehyde dehydrogenase genes and alcoholic cirrhosis in Chinese patients. Hepatology. 1994;19:360–366. [PubMed] [Google Scholar]
- 9.Chao YC, Young TH, Tang HS, Hsu CT. Alcoholism and alcoholic organ damage and genetic polymorphisms of alcohol metabolizing enzymes in Chinese patients. Hepatology. 1997;25:112–117. doi: 10.1002/hep.510250121. [DOI] [PubMed] [Google Scholar]
- 10.Chao YC, Wang LS, Hsieh TY, Chu CW, Chang FY, Chu HC. Chinese alcoholic patients with esophageal cancer are genetically different from alcoholics with acute pancreatitis and liver cirrhosis. Am J Gastroenterol. 2000;95:2958–2964. doi: 10.1111/j.1572-0241.2000.02328.x. [DOI] [PubMed] [Google Scholar]
- 11.Chao YC, Wang SJ, Chu HC, Chang WK, Hsieh TY. Investigation of alcohol metabolizing enzyme genes in Chinese alcoholics with avascular necrosis of hip joint, pancreatitis and cirrhosis of the liver. Alcohol Alcohol. 2003;38:431–436. doi: 10.1093/alcalc/agg106. [DOI] [PubMed] [Google Scholar]
- 12.Kee JY, Kim MO, You IY, Chai JY, Hong ES, An SC, et al. Effects of genetic polymorphisms of ethanol-metabolizing enzymes on alcohol drinking behaviors. Korean J Hepatol. 2003;9:89–97. [PubMed] [Google Scholar]
- 13.Kim MS, Lee DH, Kang HS, Park HS, Jung S, Lee JW, et al. Genetic polymorphisms of alcohol-metabolizing enzymes and cytokines in patients with alcohol induced pancreatitis and alcoholic liver cirrhosis. Korean J Gastroenterol. 2004;43:355–363. [PubMed] [Google Scholar]
- 14.Lee HC, Lee HS, Jung SH, Yi SY, Jung HK, Yoon JH, et al. Association between polymorphisms of ethanol-metabolizing enzymes and susceptibility to alcoholic cirrhosis in a Korean male population. J Korean Med Sci. 2001;16:745–750. doi: 10.3346/jkms.2001.16.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagata N, Watanabe N, Tsuda M, Tsukamoto H, Matsuzaki S. Relationship between serum levels of anti-low-density lipoproteinacetaldehyde-adduct antibody and aldehyde dehydrogenase 2 heterozygotes in patients with alcoholic liver injury. Alcohol Clin Exp Res. 1999;23(4 Suppl):24S–28S. doi: 10.1111/j.1530-0277.1999.tb04529.x. [DOI] [PubMed] [Google Scholar]
- 16.Nagata N, Hiyoshi M, Shiozawa H, Shiraishi K, Watanabe N, Tsuda M, et al. Assessment of a difference in ALDH2 heterozygotes and alcoholic liver injury. Alcohol Clin Exp Res. 2002;26(8 Suppl):11S–14S. doi: 10.1097/01.ALC.0000027923.82434.7F. [DOI] [PubMed] [Google Scholar]
- 17.Yamauchi M, Maezawa Y, Mizuhara Y, Ohata M, Hirakawa J, Nakajima H, et al. Polymorphisms in alcohol metabolizing enzyme genes and alcoholic cirrhosis in Japanese patients: a multivariate analysis. Hepatology. 1995;22(4 Pt 1):1136–1142. doi: 10.1016/0270-9139(95)90621-5. [DOI] [PubMed] [Google Scholar]
- 18.Yamauchi M, Maezawa Y, Toda G, Suzuki H, Sakurai S. Association of a restriction fragment length polymorphism in the alcohol dehydrogenase 2 gene with Japanese alcoholic liver cirrhosis. J Hepatol. 1995;23:519–523. doi: 10.1016/0168-8278(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama A, Mizukami T, Matsui T, Yokoyama T, Kimura M, Matsushita S, et al. Genetic polymorphisms of alcohol dehydrogenase-1B and aldehyde dehydrogenase-2 and liver cirrhosis, chronic calcific pancreatitis, diabetes mellitus, and hypertension among Japanese alcoholic men. Alcohol Clin Exp Res. 2013;37:1391–1401. doi: 10.1111/acer.12108. [DOI] [PubMed] [Google Scholar]
- 20.Day CP, Bashir R, James OF, Bassendine MF, Crabb DW, Thomasson HR, et al. Investigation of the role of polymorphisms at the alcohol and aldehyde dehydrogenase loci in genetic predisposition to alcohol-related end-organ damage. Hepatology. 1991;14:798–801. doi: 10.1002/hep.1840140509. [DOI] [PubMed] [Google Scholar]
- 21.Frenzer A, Butler WJ, Norton ID, Wilson JS, Apte MV, Pirola RC, et al. Polymorphism in alcohol-metabolizing enzymes, glutathione S-transferases and apolipoprotein E and susceptibility to alcoholinduced cirrhosis and chronic pancreatitis. J Gastroenterol Hepatol. 2002;17:177–182. doi: 10.1046/j.1440-1746.2002.02670.x. [DOI] [PubMed] [Google Scholar]
- 22.García-Bañuelos J, Panduro A, Gordillo-Bastidas D, Gordillo-Bastidas E, Muñoz-Valle JF, Gurrola-Díaz CM, et al. Genetic polymorphisms of genes coding to alcohol-metabolizing enzymes in western Mexicans: association of CYP2E1*c2/CYP2E1*5B allele with cirrhosis and liver function. Alcohol Clin Exp Res. 2012;36:425–431. doi: 10.1111/j.1530-0277.2011.01617.x. [DOI] [PubMed] [Google Scholar]
- 23.Kato S, Tajiri T, Matsukura N, Matsuda N, Taniai N, Mamada H, et al. Genetic polymorphisms of aldehyde dehydrogenase 2, cytochrome p450 2E1 for liver cancer risk in HCV antibody-positive japanese patients and the variations of CYP2E1 mRNA expression levels in the liver due to its polymorphism. Scand J Gastroenterol. 2003;38:886–893. doi: 10.1080/00365520310004489. [DOI] [PubMed] [Google Scholar]
- 24.Ohhira M, Fujimoto Y, Matsumoto A, Ohtake T, Ono M, Kohgo Y. Hepatocellular carcinoma associated with alcoholic liver disease: a clinicopathological study and genetic polymorphism of aldehyde dehydrogenase 2. Alcohol Clin Exp Res. 1996;20(9 Suppl):378A–382A. [PubMed] [Google Scholar]
- 25.Tanaka F, Shiratori Y, Yokosuka O, Imazeki F, Tsukada Y, Omata M. High incidence of ADH2*1/ALDH2*1 genes among Japanese alcohol dependents and patients with alcoholic liver disease. Hepatology. 1996;23:234–239. doi: 10.1053/jhep.1996.v23.pm0008591846. [DOI] [PubMed] [Google Scholar]
- 26.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 27.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 28.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 29.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 30.Egger M, Davey Smith G, Schneider M, Minder C. Bias in metaanalysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eriksson CJ. The role of acetaldehyde in the actions of alcohol (update 2000) Alcohol Clin Exp Res. 2001;25(5 Suppl ISBRA):15S–32S. doi: 10.1097/00000374-200105051-00005. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida A, Hsu LC, Yasunami M. Genetics of human alcohol-metabolizing enzymes. Prog Nucleic Acid Res Mol Biol. 1991;40:255–287. doi: 10.1016/s0079-6603(08)60844-2. [DOI] [PubMed] [Google Scholar]
- 33.Goedde HW, Agarwal DP. Acetaldehyde metabolism: genetic variation and physiological implications. In: Goedde HW, Agarwal DP, editors. Alcoholism: Biomedical and Genetic Aspects. Elmsford: Pergamon Press; 1989. pp. 21–56. [Google Scholar]
- 34.Zintzaras E, Stefanidis I, Santos M, Vidal F. Do alcohol-metabolizing enzyme gene polymorphisms increase the risk of alcoholism and alcoholic liver disease? Hepatology. 2006;43:352–361. doi: 10.1002/hep.21023. [DOI] [PubMed] [Google Scholar]
- 35.Li D, Zhao H, Gelernter J. Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Hum Genet. 2012;131:725–737. doi: 10.1007/s00439-011-1116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enomoto N, Takase S, Yasuhara M, Takada A. Acetaldehyde metabolism in different aldehyde dehydrogenase-2 genotypes. Alcohol Clin Exp Res. 1991;15:141–144. doi: 10.1111/j.1530-0277.1991.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 37.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- 38.Murayama M, Matsushita S, Muramatsu T, Higuchi S. Clinical characteristics and disease course of alcoholics with inactive aldehyde dehydrogenase-2. Alcohol Clin Exp Res. 1998;22:524–527. [PubMed] [Google Scholar]
- 39.Nuutinen H, Lindros KO, Salaspuro M. Determinants of blood acetaldehyde level during ethanol oxidation in chronic alcoholics. Alcohol Clin Exp Res. 1983;7:163–168. doi: 10.1111/j.1530-0277.1983.tb05432.x. [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Yokoyama T. Polymorphisms of alcohol dehydrogenase-1B and aldehyde dehydrogenase-2 and the blood and salivary ethanol and acetaldehyde concentrations of Japanese alcoholic men. Alcohol Clin Exp Res. 2010;34:1246–1256. doi: 10.1111/j.1530-0277.2010.01202.x. [DOI] [PubMed] [Google Scholar]