Abstract
Cell division is a fundamental but complex process that gives rise to two daughter cells. It includes an ordered set of events, altogether called “the cell cycle”, that culminate with cytokinesis, the final stage of mitosis leading to the physical separation of the two daughter cells. Symmetric cell division equally partitions cellular components between the two daughter cells, which are therefore identical to one another and often share the same fate. In many cases, however, cell division is asymmetrical and generates two daughter cells that differ in specific protein inheritance, cell size, or developmental potential. The budding yeast Saccharomyces cerevisiae has proven to be an excellent system to investigate the molecular mechanisms governing asymmetric cell division and cytokinesis. Budding yeast is highly polarized during the cell cycle and divides asymmetrically, producing two cells with distinct sizes and fates. Many components of the machinery establishing cell polarization during budding are relocalized to the division site (i.e., the bud neck) for cytokinesis. In this review we recapitulate how budding yeast cells undergo polarized processes at the bud neck for cell division.
Keywords: Cytokinesis, Budding yeast, Septins, Actomyosin ring, Mitotic exit network, Formins
Introduction
Cell division is a fundamental but complex process that gives rise to two daughter cells. It includes an ordered set of events altogether called “the cell cycle” that culminates in cytokinesis, the final stage of mitosis leading to the physical separation of the two daughter cells. Symmetric cell division equally partitions cellular components between the two daughter cells, which are therefore identical to one another and often share the same fate. In many cases, however, cell division is asymmetrical and generates two daughter cells that differ in specific protein inheritance, cell size, or developmental potential [1–3]. An extensively studied example of asymmetric division is that adopted by stem cells, which give rise to one daughter cell that maintains its stemness and self-renewing potential while the other differentiates. The balance between self-renewal and differentiation is at the basis of tissue homeostasis and, not surprisingly, perturbing this delicate equilibrium can steer hyperproliferation and cancer [4–6].
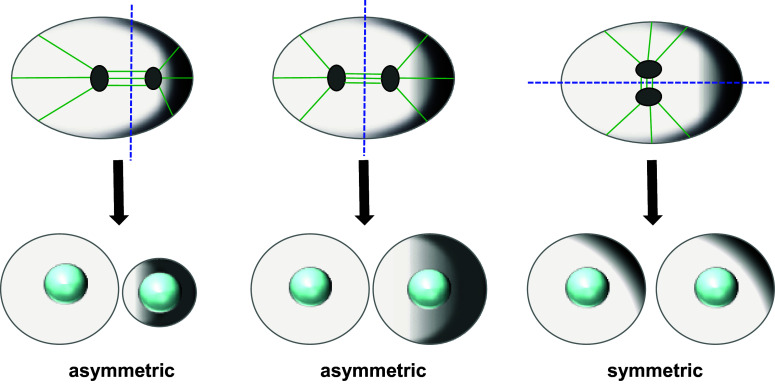
Asymmetric cell divisions arise from special cellular architectures that make cells polarized, with a basal and apical side or a front and a rear. Cell polarity, however, is an intrinsic property of all types of cells and refers to spatial differences in shape, size and function of the cell. Depending on how the mitotic spindle and the cytokinetic furrow are positioned relative to the polarity axis the ensuing cell division is either symmetric or asymmetric [7] (Fig. 1).
Fig. 1.
Spindle positioning relative to the polarity axis determines the outcome of cell division. The cartoon depicts a polarized cell, where a gradient of polarized factors increases from left to right (gray shadow). Depending on spindle positioning, which dictates the position of the cleavage furrow, cell division will be asymmetric or symmetric
Remarkably, positioning of the cytokinetic furrow is always coupled to spindle positioning (reviewed in [8]). Indeed, in many eukaryotic cells the mitotic spindle provides two non-mutually exclusive furrow-specifying signals [9, 10], one that originates from the spindle asters and the other from the central spindle, i.e., the region where polar microtubules interdigitate, which dictate positioning of the cleavage plane halfway between the two spindle poles (reviewed in [11]). Nonetheless, in some organisms, such as budding yeasts, where the site of cell division is defined early during the cell cycle and before spindle assembly, specific surveillance mechanisms delay the onset of cytokinesis until the spindle is properly positioned [12, 13]. Thus, cell polarity, spindle positioning and cytokinesis must be carefully orchestrated to ensure the successful physical separation of the two daughter cells, independently of whether cell division is symmetric or asymmetric.
The budding yeast Saccharomyces cerevisiae has proven to be an excellent system to investigate the molecular mechanisms governing cell polarity and cytokinesis. Budding yeast is highly polarized during the cell cycle and divides asymmetrically, producing two cells with distinct sizes and fates. Indeed, a bud emerges from the mother cell at the G1/S transition and keeps growing in size until cytokinesis, when it gives rise to a daughter cell. At this stage the mother cell is normally bigger than its daughter and progressively ages, while its daughter retains full lifespan [14]. Furthermore, mother and daughter cell undergo distinct transcriptional programs that allow, for instance, mating type switching to occur only in the mother cell, while expression of cell wall hydrolytic enzymes is restricted to the daughter cell [15, 16]. Strikingly, many components of the machinery establishing cell polarization during budding are relocalized to the bud neck (the constriction between mother and daughter cell where cytokinesis takes place) later on during the cell cycle for cytokinesis.
Besides these notable features, tractable genetics, powerful biochemistry, proteomics and cell biology approaches make yeast an attractive model for studying the intricate events governing asymmetric cell division, based on the precedent that fundamental principles in the control of cell division were discovered in budding yeast and proved fully applicable to higher eukaryotes.
In this review we recapitulate how budding yeast cells undergo polarized processes at the bud neck for cell division.
Cell polarization
The ability to polarize is a fundamental property of all types of cells, being crucial for numerous cellular processes such as proliferation, differentiation and development. Simple unicellular eukaryotes, bacteria, cells of multicellular invertebrates or vertebrates are polarized. This results in an extraordinary diversity in the shapes of polarized cells that have been optimized for specialized cell functions, such as the ability to communicate over long distances (neurons), to provide barriers that regulate ion homeostasis between different biological compartments (epithelia), and to unevenly distribute cellular components to daughter cells upon cell division.
At first glance, this diversity of cell shapes and functions suggests that each cell type might have evolved completely different ways to generate cell polarity that distinguishes, for example, budding yeast from a multi-cellular epithelium. Surprisingly, while the final organization of polarized cells is diverse, the basic toolbox of proteins and core mechanisms responsible for polarization are conserved from yeast to humans [17]. Indeed, a common theme in the establishment of a site of polarization is the recruitment of specific lipids and proteins at a given position of the cell surface by membrane traffic along the cytoskeleton. Polarized distribution of macromolecules is achieved by delivery and fusion of vesicles with the plasma membrane (exocytosis), as well as by endocytic internalization and recycling of the molecules that diffuse laterally along the membrane. Signaling proteins, such as Rho-like GTPases (e.g. Cdc42 and Rho1) and Rab-like GTPases are then responsible for the reorganization of the cytoskeleton necessary to polarize the cell surface [18].
Defects in cell polarity can lead to cancer formation and metastasis. For instance, the ability of transformed epithelial cells to disseminate to distant organs is linked to a mesenchymal transition where their apico-basal polarity is lost [19, 20].
Since much of the cellular machinery that contributes to establishing and maintaining epithelial cell polarity is evolutionary conserved, dissecting polarity establishment in simple models, such as yeasts, has been invaluable to understand the basic principles of this process and its derangement during cancer progression.
Polarized growth in the budding yeast S. cerevisiae
The budding yeast S. cerevisiae undergoes highly polarized cell growth throughout its life cycle and follows a stereotypical pattern of growth and division called budding [21, 22] (Fig. 2). Cells first select a site for bud emergence on the basis of cortical landmarks laid in relation to the previous division. Then, an axis of polarity directed toward this site is established by recruitment of signaling molecules. The established site then organizes a cytoskeletal framework targeting secretion for bud emergence. Further cell growth at this stage is mostly restricted to the bud, while the mother cell orchestrates the duplication and segregation of its organelles. Cells then undergo mitosis and cytokinesis, during which polarized secretion is directed to the bud neck to add new membrane and lay down the septum that separates mother and daughter cells.
Fig. 2.
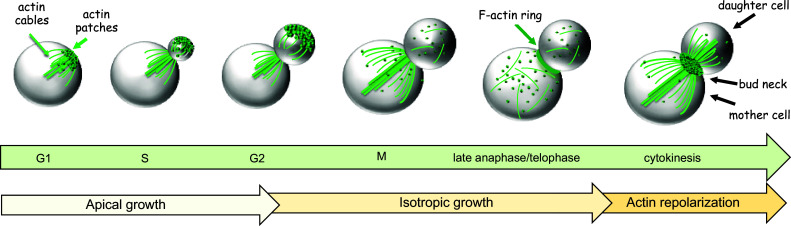
Organization of the actin cytoskeleton during budding yeast cell cycle. The cartoon illustrates budding yeast cells at different cell cycle stages and the distribution of actin structures (patches, cables and contractile F-ring) therein. Depending on whether actin organization is polarized, cell growth can be apical (directed towards the tip of the bud) or isotropic (with the bud expanding in all directions). After a transient depolarization of actin patches and cables in mitosis, the actin cytoskeleton repolarizes in telophase to bring about cytokinesis. See text for details
Most aspects of polarized growth in S. cerevisiae arise from the polarization of the actin cytoskeleton [21–24]. Filamentous actin structures (F actin) comprise of (1) actin patches, (2) actin cables and (3) the cytokinetic actin ring.
Actin patches are mobile and discrete F actin rich bodies that are nucleated by the Arp2/3 complex and represent sites of endocytosis. Many endocytic proteins are indeed linked to the Arp2/3 for formation, maturation or scission of the actin patches (reviewed in [25–27]).
Actin cables are linear F actin bundles that act as “tracks” to guide the delivery of secretory vesicles towards the site of growth [28, 29]. They are anchored at discrete regions of the cell cortex (such as the nascent bud site or the bud neck) and radiate through the rest of the cell, underlying the cell cortex [28–30]. Actin cables are nucleated by the conserved diaphanous-related-formins (DRFs, below in detail) Bni1 (Bud-neck involved) and Bnr1 (BNI1-related) [31–33].
The cytokinetic F actin ring assembles at the bud neck, contracts and disassembles [34, 35]. This is closely followed by cell wall addition (i.e., septum formation) between the dividing cells. The two formins Bni1 and Bnr1 are collectively required for F actin ring assembly and contraction, with Bni1 playing a prominent role [34, 35].
As mentioned above, the very first step towards cytokinesis is the selection of an incipient bud site, which is chosen relative to cortical landmarks remaining from the previous cell division [36–38]. Excellent reviews can be found in the literature on this topic [23, 39–41], which therefore will not be treated here. Once the presumptive bud site has been selected in late G1, the actin cytoskeleton becomes highly polarized (Fig. 2). Cortical patches concentrate at the location of the new bud while actin cables emanate from this site. As a bud emerges, cortical patches initially cluster at its tip, while cables nucleated at the bud tip extend into the mother cell. This configuration supports trafficking required for bud growth from the tip (apical growth). Later on, patches and cables redistribute randomly within the bud, while cables in the mother cell still extend from the bud neck. During this phase, bud growth continues by expansion in all directions (isotropic growth). Finally, when cells exit mitosis actin repolarization occurs at the bud neck to support cytokinesis; the contractile F actin ring assembles and actin cables direct secretion towards the division site for septum formation (Fig. 2). Actin patches also concentrate at the mother and daughter side of the bud neck [28, 42], presumably for endocytic internalization and/or recycling of cytokinetic factors.
Rho GTPases in the establishment of cell polarity
Rho GTPases are conserved proteins belonging to the Ras superfamily of small G proteins that are regarded as molecular switches, as they can oscillate between an inactive GDP-bound state and an active GTP-bound state [43, 44]. Like all G proteins, Rho GTPases are endowed with intrinsic GTPase activity. The rate-limiting step in GTPase activation is the release of GDP aided by guanine nucleotide exchange factors (GEFs), which allow GTPase-binding to GTP that in turn is present in the cells at higher concentrations than GDP. Conversely, GTP hydrolysis can be stimulated by GTPase-activating proteins GAPs) that shift the balance to the inactive state of the GTPase [45, 46]. Other regulators of Rho GTPases include the guanine nucleotide dissociation inhibitors (GDIs) that can lock the GTPase in the GDP- or GTP-bound form, as well as extract it from the membrane, thereby preventing its GEF-mediated activation [47].
Establishment of cell polarity in budding yeast requires the Rho GTPase Cdc42, which accumulates at the presumptive bud site through a process involving its GEF Cdc24 and the scaffold protein Bem1 that bridges Cdc24 to the Cdc42 effector Cla4, thereby generating a positive feedback loop that clusters Cdc42 to a single cortical site [48–50]. Cdc42 is further concentrated to a focused vertex by recycling mechanisms that include the GDI Rdi1 as well as the opposing activities of exo- and endocytosis [51, 52]. Once concentrated at a single focus, active Cdc42 organizes the actin cytoskeleton and septins to promote polarized secretion and cell growth (see below). Known Cdc42 effectors include the partially redundant PAK (p21-activated kinases) Cla4 and Ste20, which play major redundant roles in actin and septin organization [53–58], the formin Bni1 ([59], see below), the proteins Gic1 and Gic2, which promote septin recruitment and formin activity ([60–63], see below) and the Sec3 component of the exocyst complex, which plays essential role in exocytosis through vesicle targeting and docking to the plasma membrane [64, 65].
Besides Cdc42, budding yeast has five additional Rho GTPases that are named Rho1-5. Like Cdc42, Rho1 is essential for cell viability and plays a major role in cytokinesis through assembly of the cytokinetic contractile ring and the division septum (see below). Its effectors include formins [66, 67], the glucan synthase Fks1 [68], protein kinase C (Pkc1, [69, 70]) and the exocyst subunit Sec3 [71]. In contrast, Rho2-5 are dispensable for survival of yeast cells and their respective roles are ill-defined, although Rho3 and Rho4 share an essential role in the establishment of cell polarity and have been collectively implicated in formin activation [59, 66].
Formins as key regulators of cell polarity and cytokinesis
Formins are large multi-domain proteins found in plants, fungi and mammals. Although their number is highly variable in different organisms, formin structure and function are highly conserved [72, 73].
In budding yeast there are two arrays of actin cables, one polarized toward the bud cortex and the other toward the mother-bud neck, that are nucleated by the two formins, Bni1 and Bnr1 [31, 32, 67, 74–76]. In budding yeast, neither one of the two formins is essential, but cell viability requires at least one of them [66], suggesting that they share at least one essential function. However, Bni1 and Bnr1 clearly also play distinct cellular roles, which is highlighted by their different patterns of cellular localization and respective mutant phenotypes [77–80]. From bud emergence to mitotic exit Bnr1 resides at the bud neck, where it is relatively static and nucleates actin cables extending into the mother cell [76, 77, 81]. In contrast, Bni1 localizes throughout most of the cell cycle to the bud tip, where it nucleates actin cables, and to the bud neck immediately before cytokinesis, when it replaces Bnr1, helping to form the contractile actomyosin ring (CAR) [77, 82]. At the bud tip, Bni1 binds to several components of the polarisome (i.e., Spa2, Pea2 and Bud6, [31, 83]), a protein complex involved in cell polarity that localizes at sites of polarized growth [84]. Two motifs named SBD (Spa2-binding domain) and BBD (Bud6-binding domain) (Fig. 3) have been mapped in the middle and C terminal region of Bni1, respectively, linking Bni1 to the polarisome [78, 79, 83]. The Spa2–Bni1 binding is required for Bni1 localization at the bud tip and for proper regulation of actin architecture. Indeed, in the absence of Spa2, Bni1 gets redistributed to the cytosol instead of localizing at bud cortical sites [83]. In contrast, loss of Bud6 has a minor impact on Bni1 localization [31]. However, a C terminal fragment of Bud6 can stimulate the actin-polymerizing activity of Bni1, as supported by in vitro experiments that led to the proposal that Bud6 acts as a nucleation-promoting factor [79, 85]. Interestingly, Bud6 can also enhance in vitro actin nucleation by the other formin Bnr1 when assisted by the Bil1 protein (Bud6-interacting ligand) [86], in agreement with previous data showing that the C terminal part of Bud6 participates in formin-dependent actin cable organization in vivo [78].
Fig. 3.
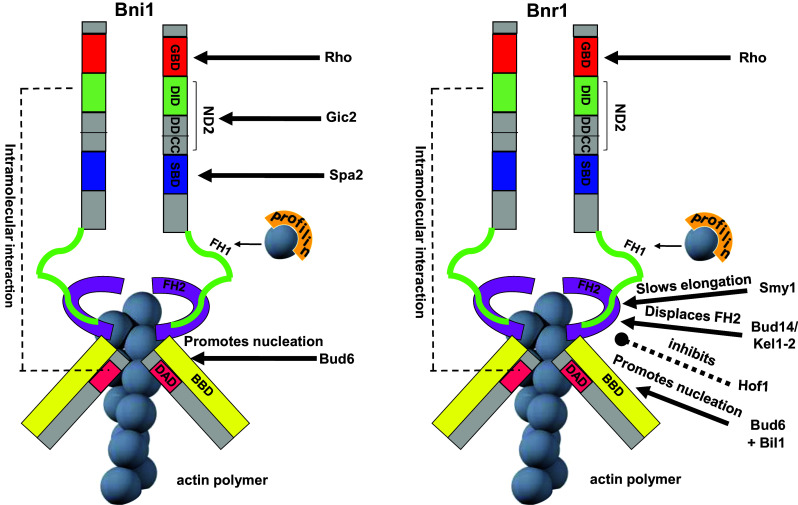
Structural organization of the yeast formins Bni1 and Bnr1. Formins form a doughnut-shaped dimer that encircles the nascent actin filament during its elongation. The main interactors and regulators of each formin are depicted. See text for details. GBD GTPase-binding domain, DID diaphanous inhibitory domain, DD dimerization domain, CC coiled coil, SBD Spa2-binding domain, FH1/2 formin homology domain, DAD diaphanous auto-regulatory domain, BBD Bud6-binding domain
Formin recruitment to and release from membranes, as well as its actin-nucleating activity, involve additional formin modifications and/or binding partners that tune their function. The formin homology domain 1 (FH1, Fig. 3) is a proline-rich motif that binds profilin, among other proteins. Profilin is an actin-binding protein that recruits actin monomers to the active region of formin when bound to FH1 [66, 74], thereby stimulating formin-induced actin polymerization [32, 87]. The second formin homology domain (FH2, Fig. 3) lies next to FH1 and is required to form a doughnut-shaped formin dimer that encircles the nascent actin filament during its elongation [88]. Consistently, the dimeric architecture of formin is relevant for its actin nucleation activity [87, 89].
Formins have two additional important regulatory motifs, the DID (Diaphanous Inhibitory Domain) and the DAD (Diaphanous Auto-regulatory Domain) (Fig. 3) that were originally identified in the Drosophila formin Diaphanous [90]. DID and DAD can interact with one another within the same formin molecule, thereby locking formin in a close inhibited state [91, 92]. Other formin domains can relieve this autoinhibition to promote an open and active state. Among them, the GBD (GTPase binding) and the ND2 (N-terminal) domains (Fig. 3) bind to Rho GTPases and the Gic2 protein, respectively, and also control Bni1 localization at the bud tip [62, 93–95]. The GBD is located next to the DID domain, and its binding to Rho relieves formin autoinhibition by hindering DID interaction with DAD [94, 96–100]. Whether formin binding to a Rho GTPase is always necessary for its activation is unclear, especially since other domains can also relieve autoinhibition.
To add additional layers of complexity to formin regulation, other cofactors modulate the biological function of formins. Tropomyosins, which are master regulators of muscle cells contraction, also play key roles in non-muscle cells by controlling actin dynamics and cell migration [101, 102]. In budding yeast, they control assembly and stability of actin cables [103]. Additionally, tropomyosins promote formin-mediated formation of contractile ring assembly in fission yeast [104], raising the possibility that they might play a similar function also in budding yeast.
Yet another formin regulator is the kinesin-like myosin-passenger protein Smy1, which acts as a Bnr1 damper in vitro and in vivo without affecting Bni1 [105, 106]. Smy1 slows down the elongation rate of Bnr1-mediated actin polymerization by direct binding to Bnr1. Accordingly, cells lacking Smy1 show extremely long actin cables with prominent defects in their architecture [105]. Recently, Smy1, Bnr1 and the myosin V motor protein Myo2 that delivers Smy1 to formin have been involved in an “antenna mechanism” that senses and controls the length of actin cables [107].
Another set of formin tuners act in parallel with the ones listed above to ensure proper actin cable architecture. A complex formed by Bud14 and the Kelch-domain proteins Kel1 and Kel2, which are involved in cell polarity and morphogenesis, senses the length of actin cables and eventually displaces Bnr1 from actin filaments [108–110]. Bud14 does not suppress Bnr1 actin-polymerizing activity but rather the permanence of formin on actin, thereby attenuating the elongation rate of actin filaments.
The F-BAR protein Hof1, which controls septin organization and septum deposition ([81, 111, 112], see below), inhibits the actin-nucleating activity of Bnr1 both in vitro and in vivo [113], thereby tuning the architecture of the actin cable network. Conversely, Bnr1 is somehow activated in vivo by septins (see below) and the septin-associated kinase Gin4 [114].
Finally, formin activity is likely controlled by post-translational modifications. For instance, phosphorylation of Bni1 by the Prk1 kinase unleashes its autoinhibition [115]. In mating cells, Bni1 phosphorylation by the Fus3 MAP kinase is important for its localization to and assembly of actin cables [116], while dephosphorylation of both Bni1 and Bnr1 by Cdc14, as well as Bnr1 dephosphorylation by protein phosphatase 1 (the PP1 Glc7), seems to trigger the replacement of Bnr1 with Bni1 during mitotic exit [117, 118]. Finally, a truncated variant of Bni1 was recently shown to be ubiquitylated in vivo by the Rsp5 E3-ubiquitin ligase and subsequently degraded to reorganize the actin cytoskeleton under stress conditions and wound healing [119].
Actin polarization at the bud neck for cytokinesis
Just before cell division, the actin cytoskeleton repolarizes to the bud neck (Fig. 2). Thus, actin structures (actin cables, actin patches and the F actin ring) converge at the cell division site. In particular, actin cables rearrange to be polarized towards the bud neck and ensure that membrane trafficking will bring secretory vesicles and proteins to the cytokinesis site to bring about membrane closure. This remarkable reorganization of the actin cytoskeleton is driven by inactivation of mitotic cyclin B-CDK complexes at mitotic exit [42, 120] (see “The mitotic exit network”). Approximately at the same time, many polarity factors, such as the Cdc42 and Rho1 GTPases, polarisome components (e.g. Bni1, Spa2 and Bud6) and the exocyst complex, translocate from the bud tip to the bud neck, thus contributing to the actin rearrangements accompanying this transition [76, 81, 82, 84, 121–123]. Most likely, the relocalization of some of these proteins occurs to reinforce Bni1-dependent polymerization of actin cables and ring at the bud neck. While Rho1 presumably directly promotes local Bni1 activation for F actin ring assembly (see below, [124]), it has not been established if Cdc42 directly activates Bni1 for actin polymerization prior to cytokinesis. However, inactivation of the redundant PAK kinases Ste20 and Cla4, which are known Cdc42 effectors, during mitosis abolishes actin repolarization at the bud neck [55], suggesting that Cdc42 might have an indirect role in formin activation at cytokinesis. In turn, Ste20, and perhaps Cla4, might regulate Bni1 activity through direct phosphorylation [54, 125]. Interestingly, although Cdc42 persists at the bud neck until cytokinesis has been accomplished, the levels of active GTP-bound Cdc42 decrease at cytokinesis [123, 126, 127]. Inhibition of Cdc42, and Ste20 downstream to Cdc42, is in turn important for efficient recruitment of cytokinesis factors to the bud neck and proper cell division [128]. Consistent with the antagonism between Cdc42 and Rho1/RhoA that has been shown in many eukaryotic systems, the polo kinase Cdc5 is required for Cdc42 inhibition while promoting the recruitment of Rho1 to the neck through phosphorylation of one of its guanine nucleotide exchange factors (GEFs) [129, 130].
While Spa2 mediates localization of the formin Bni1 at the bud tip [83], it is dispensable for its redistribution to the bud neck at cytokinesis [82]. However, Spa2 and Bud6 might contribute to formin-dependent actin polymerization. Although the precise mechanism by which Bni1 relocates from the bud tip to the neck is still elusive, the Cdc14 phosphatase and Bni1 phosphorylation were shown to be involved in this process [117]. A good candidate for promoting Bni1 recruitment to the bud neck is the exocyst complex. The exocyst is a conserved protein complex made by eight subunits (Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70 and Exo84) that tethers exocytic vesicles to the plasma membrane during secretion [131–133]. Inactivation of the exocyst complex through temperature-sensitive mutations leads to disappearance of actin cables and a general depolarization of actin, which suggests that the exocyst regulates actin dynamics [134, 135]. Strikingly, in fission yeast Sec3 is essential for the localization of the formin For3 (the fission yeast counterpart of Bni1) at the plasma membrane where the two proteins interact [136]. Importantly, the distribution and/or activity of the exocyst complex is controlled by both Cdc42 [64, 137, 138] and Rho1 [71], presumably in a reciprocal manner depending on the cell cycle stage, suggesting yet another possible route through which Rho-like GTPases might control formin activity.
The septin ring
The septin ring and cytokinesis
Studies in budding yeast and mammalian cells indicate that septins act as scaffold to recruit cytokinetic factors to the site of cell division (reviewed in [139, 140]). Septins were first discovered in budding yeast through a genetic screen for mutants defective in cell division [141] and are cytoskeletal GTP-binding proteins that form oligomeric complexes that can in turn self-organize in higher-order structures, such as filaments and rings. Although septins have been found at the division site in most cell types examined so far, the extent to which they contribute to cytokinesis varies from one organism to another. For example, in the fission yeast S. pombe septins appear at the division plane only after the cytokinetic ring has fully assembled [142, 143] and their deletion causes only a mild cell separation defect [144, 145]. In stark contrast, several of S. cerevisiae septins are essential for viability and cytokinesis.
The five septins expressed in vegetative budding yeast cells (Cdc3, Cdc10, Cdc11, Cdc12 and Shs1) form hetero-octamers composed by two copies of the core Cdc10, Cdc3 and Cdc12 subunits and two copies of the alternative septins Cdc11 and Shs1 arranged in palindromic linear rods [57, 146, 147]. The rods collide on the plasma membrane to join end-to-end in non-polar filaments [148] that in turn organize in a ring. Recent data showed that the non-essential yeast septin, Shs1, curves septin filament bundles into rings in vitro and promotes proper septin organization at the bud neck in vivo [147].
The bud neck protein Bni5 had been identified as multicopy suppressor of septin mutants [149] and has been recently shown to crosslink septin filaments in vitro [150], likely providing structural stability to the septin ring in vivo. Bni5 directly interacts with the septins Cdc11 and Shs1, as well as with Myo1, mediating its recruitment to the division site throughout most of the cell cycle until cytokinesis [151–153].
Septins associate with membranes, and in particular with positively charged phosphoinositides, such as phosphatidylinositol-4,5-diphosphate (PIP2) [154], through a highly conserved polybasic region at the N terminus [155, 156]. In budding yeast PIP2 is enriched in membrane areas of polarized growth and the bud neck [157] and it stimulates formation and organization of septin filaments that are in turn essential for cell viability [154, 158].
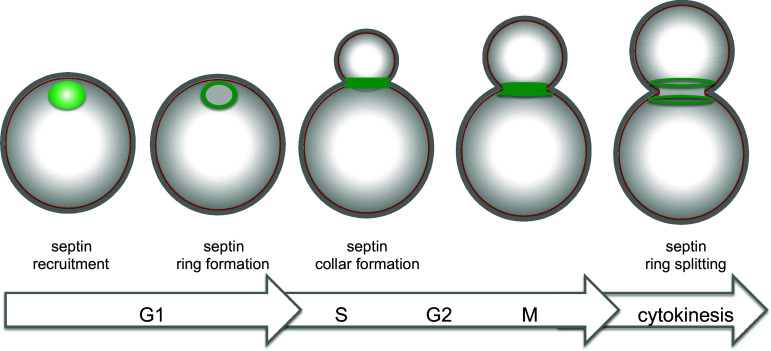
Septins are first recruited to the presumptive bud site as unorganized septin clouds or patches, which are then rapidly transformed into a cortical septin ring in late G1. Fluorescence Recovery After Photobleaching (FRAP) experiments indicate that septin structures prior to bud emergence are highly dynamic. At the time of bud emergence the septin ring expands into a rigid hourglass-like structure referred to as septin collar, which spans the whole bud neck and scaffolds many cytokinetic factors [159, 160]. Immediately prior to cytokinesis the collar splits into two distinct rings that sandwich the contractile actomyosin ring (CAR, see below) and are highly dynamic (reviewed in [139, 140]) (Fig. 4). The physiological relevance of ring splitting for cytokinesis has yet to be elucidated. However, coincident with or immediately after septin-ring splitting the CAR constricts in between the split septin rings and the cleavage furrow ingress, bringing about deposition of the primary septum [161, 162]. Remarkably, septin ring splitting is accompanied by a striking change in septin arrangement that was revealed by polarized fluorescence microscopy. Indeed, while septin filaments inside the collar are arranged in parallel arrays aligned along the mother-bud axis, they are found rotated by 90° in split septin rings [163, 164]. How these observations can be reconciled with earlier electron microscopy (EM) studies showing that septin rings are made by circumferential septin filaments encircling the bud neck [165] has been subjected to extensive debate. Recent data obtained by platinum-replica EM and correlative light/EM suggest that the early septin collar is made by double septin filaments oriented along the mother-bud axis, while later on during mitosis it acquires orthogonally oriented circumferential septin filaments that confer a gauze-like appearance to the structure [166]. During septin ring splitting the septin double filaments are somehow depolymerized, leaving two parallel rings of filaments around the bud neck. Interestingly, although the myosin II Myo1 and the non-essential septin Shs1 are not strictly required for septin collar formation, they seem to affect the overall organization of the mature septin collar [166].
Fig. 4.
Main steps in budding yeast cytokinesis. See text for details
The Cdc42 GTPase is essential for septin recruitment to the presumptive bud site [63, 167] and cycles of Cdc42 GTP-binding and hydrolysis are required for septin collar formation [159, 168]. Among the known Cdc42 effectors, the paralogous membrane proteins Gic1 and Gic2 seem to play a crucial role in septin recruitment. Accordingly, septin deposition and budding mostly fail in gic1 gic2 double mutants at high temperatures [63]. Gic1 localizes at the presumptive bud site and bud tip at early stages of the cell cycle and at the bud neck later on [63], and it has been recently shown to bundle and crosslink septin filaments in vitro, thereby stabilizing them [169]. Surprisingly, the inactive GDP-bound form of Cdc42 was found to bind directly septin filaments and to depolymerize them when present at high concentrations, while creating lateral crosslinks between septin filaments at low concentrations. On the basis of these and other observations it has been proposed that the initial recruitment of septin octamers to the future bud site is promoted by Cdc42-GDP itself; afterwards, septin filaments get bundled and stabilized by Gic1, presumably bound to Cdc42-GTP [169]. Once a polarized cap of septin filaments has been formed at the presumptive bud site, the septin ring is sculpted by polarized exocytosis that creates a hole in the middle of the cap [170]. At cytokinesis, Cdc42 accumulates at the bud neck where it might induce septin depolymerisation [169], to favor recycling of septin octamers for the following cell cycle [171]. Intriguingly, septins inhibit Cdc42 activity at the bud neck in a negative feedback loop through GTPase-activating proteins, which promote the Cdc42 GDP-bound inactive state [170]. Of note, according to the model proposed by Sadian et al. [169] this event could stimulate, at the same time, further recruitment of septin octamers.
Direct binding to septin filaments and activation of Gic1 and Gic2 are not the only function of Cdc42 in septin regulation. Another Cdc42 effector, the PAK kinase Cla4, directly phosphorylates the septin Cdc10 in vitro and in vivo, thereby preserving the integrity of septin architecture [57]. Furthermore, the formin Bni1, which is another effector of Cdc42 (see above), contributes to septin ring formation along with Cla4 [56].
Anillin is a multi-domain cytokinesis protein that in metazoans interacts with a plethora of partners, including actin, myosin, septins and formins, among many others (reviewed in [172]). In budding yeast, the anillin-like protein Bud4 associates with septins in mitosis and colocalizes with the septin ring until after the next G1 phase, to then disappear at the onset of budding [173, 174]. Bud4 is required to stabilize the septin ring during splitting [162, 175], similar to the fission yeast anillin Mid2 [142, 143]. However, the mechanism underlying this stabilization has yet to be discovered, as well as the physiological relevance of septin ring splitting. Indeed, BUD4 deletion causes mild cytokinesis defects that get more pronounced in sensitized mutant backgrounds [176].
The Rho1 GTPase, besides playing a pivotal role in CAR assembly [124](see below), has been recently shown to stabilize septins during their recruitment to the presumptive bud site through activation of its effector protein kinase C (Pkc1). Pkc1 in turn modulates the turnover at the bud neck of the F-BAR protein Syp1 through direct phosphorylation [177]. Syp1 is an endocytic protein that has been implicated in timely septin deposition and in stimulating septin ring dynamics through an unknown mechanism [177, 178]. Remarkably, Syp1 is recruited to the presumptive bud site in G1, at the same time as septins, and forms a ring that surrounds and is larger than the septin ring. After budding the Syp1 ring is thus found asymmetrically located on the mother side of the septin collar [177]. This peculiar spatial arrangement of Syp1 relative to septins is highly reminiscent of the coordination between secretion and endocytosis occurring during the establishment of cell polarity in G1, where endocytosis corrals at the membrane a vertex of active exocytosis for bud emergence [51]. Thus, altogether these data raise the possibility that timely accumulation of septins at the future bud site in late G1 might be facilitated by endocytic recycling. Interestingly, the EH domain-containing protein Ede1, which is a major partner of Syp1 for endocytosis [179, 180], has been recently implicated in cytokinesis [181].
Other functions of the yeast septin ring
Besides being necessary for cytokinesis, the budding yeast septin ring has been implicated in several other polarized processes. Some of them are intimately linked to the second main function of septins, in addition to scaffolding, as cortical barriers to prevent the free diffusion of membrane proteins between different compartments (reviewed in [182, 183]). For instance, the septin ring hampers the diffusion to the mother cell of the mitotic exit regulator Lte1, normally localized in the bud cortex, thereby ensuring the proper coupling between correct spindle positioning and mitotic exit [184]. The septin ring also restricts to the bud the accumulation of the machinery responsible for the asymmetric localization of certain mRNAs [185, 186]. Additionally, the septin ring segregates some membrane proteins of the endoplasmic reticulum (ER) to a specific cell compartment, without influencing the distribution of ER luminal proteins that instead remain freely diffusible [187]. Finally, the septin ring contributes to proper spindle positioning early in mitosis [188] and promotes mitotic entry by scaffolding at the bud neck the machinery responsible for the degradation of the Wee1-like kinase Swe1, which inactivates mitotic cyclin-dependent kinases (CDKs) by inhibitory phosphorylation [189–192].
Septin post-translational modifications
Septins are targeted by many post-translational modifications, such as phosphorylation, acetylation, sumoylation and ubiquitination that likely modulate the state transitions of the septin ring during the cell cycle. Several protein kinases, such as Cla4, the Nim (never in mitosis)-related kinases Gin4, Kcc4 and Hsl1, as well as the Elm1 kinase, localize at the bud neck in a septin-dependent manner and promote septin collar formation and stabilization (reviewed in [139, 140, 193]). Specifically, Cla4 phosphorylates several septins in vitro [57] and is regulated by Elm1 [194], while Gin4 phosphorylates Shs1. Gin4 is phosphorylated and activated by Elm1, which together with Cla4 also promotes its recruitment to septins [190, 195, 196]. G1 cyclin-dependent kinases (CDKs) have also been implicated in septin phosphorylation, and CDK-dependent phosphorylation of Shs1 was shown to enhance its interaction with Gin4 [197, 198]. Although the extensive level of phosphorylation and the large number of kinases involved have hampered so far the functional dissection of this post-translational modification, as well as the assessment of the precise role of the Hsl1 and Kcc4 kinases in septin regulation, altogether septin phosphorylation seems to accompany septin ring stabilization. Consistently, the protein phosphatase PP2A bound to the Rts1 regulatory subunit reverses phosphorylation of at least Shs1 and contributes to timely septin disassembly after cytokinesis [199].
Several septins were found to be sumoylated in mitosis by the Siz1 and Siz2 Sumo-ligases [200, 201]. A mutant lacking the major sumoylation sites in Cdc3, Cdc11 and Shs1 displays prominent defects in septin ring disassembly at the end of mitosis. However, the lack of a similar phenotype in siz1 siz2 double mutants or in ubc9 temperature-sensitive mutant that affects the only Sumo-conjugating enzyme leaves open the possibility that the sumoylated lysines of septins might be targeted by other post-translational modifications, namely acetylation or ubiquitination [201].
The septins Cdc11 and Shs1 have been recently shown to be ubiquitinated by the Dma1 and Dma2 E3 ubiquitin ligases [202], which had been previously involved in septin ring stability [203, 204]. However, the role of septin ubiquitination remains to be established.
Finally, septin acetylation by the NuA4 and Esa1 lysine acetyltransferases was found to stabilize the septin collar [205]. Intriguingly, in mutants affecting septin acetylation septin complexes contain actin, suggesting that interaction of septins with the actin cytoskeleton might be deleterious for septin collar stability. Along the same line, in mammalian interphase cells, where septins colocalize with actin in long linear bundles or arcs, actin depolymerisation by cytochalasin D treatment leads to the formation of septin rings [206].
Although septin filaments are apolar and the septin collar is symmetric [207], several proteins that are recruited to the bud neck in a septin-dependent manner localize asymmetrically on the septin collar. For instance, the PP1 phosphatase Glc7 associated to its regulatory subunit Bni4, the F-BAR protein Syp1, the kinase Gin4 and the sumo-ligases Siz1 and Siz2 localize on the mother side of the septin collar, while the kinase Kcc4 is restricted to its bud side [201, 208, 209]. How this asymmetry is generated is unclear but it might rely on specific posttranslational modifications and in turn generate asymmetric septin modifications.
Assembly of the actomyosin ring
In many eukaryotic organisms, including budding yeast, cytokinesis involves a contractile actomyosin ring (CAR) made by the motor protein myosin II and actin filaments. The CAR assembles at the site of cell division and drives furrow ingression (reviewed by [210]). In S. cerevisiae the CAR is coordinated with and guides the formation of the primary septum, which involves the addition of a new cell wall between the dividing cells (see “Constriction of the actomyosin ring”). In the absence of a functional CAR budding yeast cells fail to invaginate the plasma membrane during cytokinesis but in some strain backgrounds they can eventually survive thanks to the formation of aberrant remedial septa [34, 211–213].
CAR assembly in budding yeast is a sequential order of events that starts in late G1 with the septin-dependent recruitment to the presumptive bud site of the single myosin type II heavy chain Myo1, along with its regulatory light chain Mlc2, [214, 215]. In mitosis, at a time when the essential myosin light chain Mlc1 appears at the bud neck, Myo1 interacts also with Mlc1. In telophase Mlc1 promotes the accumulation to the bud neck of the IQGAP protein Iqg1, which is in turn essential for recruiting filamentous actin (F actin) to the CAR, as well as for a second wave of Myo1 targeting to the neck that further increases its local levels [151, 214, 216–220].
The two formins Bni1 and Bnr1 are also essential for the engagement of F actin at the CAR [112, 124], although their exact relationship with Iqg1 has not been established. Furthermore, formins have been recently shown to contribute to the accumulation of Mlc1 at the bud neck during cytokinesis [221], suggesting an additional mechanism by which they could participate in CAR assembly. In principle, many of the formin regulators described above (see “Rho GTPases in the establishment of cell polarity”) could contribute to timely CAR assembly to various extents.
In S. pombe recruitment of formins to the medial cortex, where cytokinesis occurs, is partly mediated by the cytokinetic protein Cdc15, which contains an F-BAR domain (F-BAR: ‘FCH and BAR’, where FCH = Fes/CIP4 Homology and BAR = Bin-Amphiphysin-Rvs) to bind membranes and is a key regulator of CAR assembly and stability [222–226]. Its counterpart in budding yeast, called Hof1 (homologue of fifteen), interacts and partially constricts with the CAR but is thought to be dispensable for CAR function. Rather, it has been implicated in primary septum formation (see below) [81, 111, 112, 227, 228]. However, it has been recently reported that Hof1 functions redundantly with the yeast amphiphysin Rvs167, which also contains a BAR domain, in promoting F actin assembly at the CAR [229], suggesting that it might have a conserved role in CAR formation.
The precise function of the Mlc2/Myo1 complex early in the cell cycle is unknown, but has been proposed to stimulate the retrograde flow of cargos on actin cables [230]. In late G1 Myo1 recruitment to the neck depends on septins and the septin-binding protein Bni5 (see below) and is characterized by high turnover [34, 149, 152, 231]. As the cell cycle proceeds, Myo1 remains localized at the bud neck until F actin is recruited around anaphase to form the CAR [34, 35]. Shortly before cytokinesis Myo1 levels further increase at the bud neck through a mechanism that involves Mlc1 and Iqg1, and Myo1 becomes immobile at the neck where it acts as scaffold for the cytokinetic machinery [151, 231].
As already mentioned, the CAR consists of actin filaments nucleated by formins, which are in turn activated by the Rho1 GTPase [67, 112, 124]. Indeed, Rho1 is essential for assembly of the F actin ring [124], similar to RhoA in many eukaryotic organisms (reviewed in [232]). Rho1 is recruited to the division site through a major mechanism involving its GEFs (Rom1, Rom2 and Tus1) and a distinct backup mechanism depending on interaction between the C terminus of Rho1 and acidic phospholipids [129]. Furthermore, the polo kinase Cdc5 is necessary for Rho1 and Bni1 localization at the bud neck, probably through direct phosphorylation of the GEFs Rom2 and Tus1 [130].
The exact arrangement of actin filaments inside the ring and the precise mechanism by which budding yeast formins contribute to the assembly/contraction of the cytokinetic ring have not been fully elucidated. In fission yeast it has been suggested that pre-existing actin cables might coalesce into the cytokinetic actin ring [233]. However, this model does not seem to apply to budding yeast, where cells with F actin rings are mostly devoid of actin cables, suggesting that the two structures compete with one another for formin-dependent polymerization [124].
Likely, both formins Bnr1 and Bni1 can promote the assembly of the actin ring at the budding yeast bud neck since both form actin cables and localize at the bud neck, albeit in a mutually exclusive manner, during CAR assembly. Consistently, lack of either formin does not affect CAR formation, whereas inactivation of both, as well as inactivation of tropomyosins and profilin, disrupts actin recruitment to the CAR [112, 124].
Strikingly, at the onset of cytokinesis, concomitant with CAR contraction, Bnr1 leaves the bud neck through a process that appears to be linked to its dephosphorylation [117, 118], thus empowering Bni1 as a prominent player for CAR contraction. Accordingly, in bni1 null mutant cells the actin ring still forms but often fails to contract, while this is not the case for bnr1∆ mutants [112]. Thus, although Bni1 and Bnr1 seem to play overlapping roles in actin ring formation, their coordinated interplay is important for proper ring contraction, possibly through the interaction with other polarized factors involved in cytokinesis.
The exact function of the IQGAP Iqg1 in CAR assembly has not been fully understood. Mammalian IQGAP can crosslink actin filaments [234] and has been proposed to act as scaffold for the actin assembly machinery [235, 236], while budding yeast Iqg1 can bind actin through its N terminal calponin-homology domain (CHD), suggesting that it might directly recruit actin to the CAR [217, 218]. Consistently, IQG1 overexpression causes premature actin ring formation [217]. How Iqg1 and formins cooperate to assemble the actin ring is unclear. Mammalian IQGAP interacts physically with the formin Dia1 and is required for its proper localization [237]. Similarly, C. albicans Iqg1 associates with both formins Bni1 and Bnr1 and promotes efficient recruitment of Bni1 to the bud neck [238]. Thus, Iqg1 might on one side organize actin filaments polymerized by formins and on the other favor efficient formin activity.
Constriction of the actomyosin ring
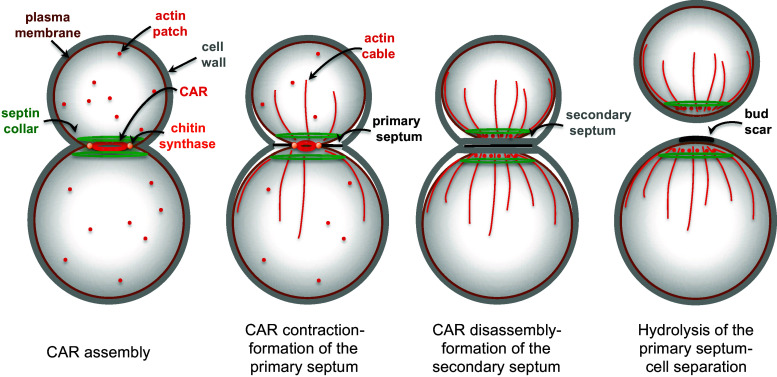
Shortly after its complete formation the CAR constricts (Fig. 5). In many organisms, CAR constriction during cytokinesis is thought to drive invagination of the overlying plasma membrane inward generating the force to cleave the cell in two (reviewed by [210]). In budding yeast CAR also drives membrane deposition through vesicle targeting and contributes to formation of the primary septum. The mechanism of CAR constriction has originally been inferred from that by which actomyosin generates force in the striated muscle, which stems from the sliding of bipolar myosin filaments along actin filaments that are organized in regular antiparallel arrays [239, 240]. However, this model does not seem to apply to budding yeast CAR. Indeed, Myo1 levels progressively decrease as the CAR constricts [162, 241], while they would be expected to remain constant if a sliding mechanism fully accounted for constriction. Furthermore, unlike in other organisms, the motor domain of Myo1 is not strictly required for CAR constriction and cytokinesis [151, 242, 243], while the rest of the protein is essential in most strain backgrounds and necessary for actin assembly in the CAR [34, 213, 244]. In agreement with these observations, CAR contraction in S. cerevisiae has been recently shown to be mainly driven by actin depolymerisation promoted by the cofilin Cof1. Actin depolymerisation by Cof1 synergizes with the motor activity of Myo1 to promote fast CAR constriction, with the latter mechanism playing a less prominent role than the former [243]. Since the action of cofilin can be stimulated by actin crosslinking to generate contractile stress [245], the IQGAP Iqg1 has been proposed to play such a role during yeast cytokinesis [243]. Consistently, deletion of the C terminal GTPase-activating protein-related domain of Iqg1 prevents CAR constriction without affecting CAR assembly [218], while the phosphorylation-deficient mutants of IQG1 slow down CAR constriction, while advancing CAR formation [238, 246]. Interestingly, although Iqg1 is necessary for CAR assembly and constriction, its ubiquitin-dependent degradation mediated by the anaphase-promoting complex is important for CAR disassembly after cytokinesis [241].
Fig. 5.
The septin ring during budding yeast cell cycle. Sequential stages of septin organization during the cell cycle of budding yeast
Around the time of CAR constriction the myosin V Myo2, which transports post-Golgi vesicles along actin cables, and the exocyst complex, which tethers secretory vesicles to the plasma membrane, get recruited to the CAR to promote delivery of membrane and essential cargoes to the division site [120, 151, 162, 199, 231]. The essential myosin light chain Mlc1 is required for Myo2 tethering to the bud neck [247], thereby coordinating CAR formation with membrane trafficking. Remarkably, interfering with membrane traffic at the bud neck through mutations affecting Myo2 or exocyst subunits leads to CAR destabilization during constriction, without affecting CAR assembly [120]. Similarly, loss of the chitin synthase Chs2, which is required for primary septum formation and is transported to the bud neck through Myo2- and exocyst-mediated secretion (see below), affects the stability of the CAR during contraction [120, 212], suggesting that CAR constriction, secretion and septation are intimately connected processes (Fig. 5).
Septum formation
Contractile actomyosin ring constriction in budding yeast is coupled to the centripetal deposit of a primary septum that physically separates the two daughter cells. The primary septum is a chitin disk deposited by the action of the chitin synthase 2 (Chs2, Fig. 5). Chs2 is synthesized in G2/M and accumulates in the endoplasmic reticulum (ER) until the end of mitosis [248]. Inactivation of mitotic CDKs or Chs2 dephosphorylation by the Cdc14 phosphatase triggers the translocation of the chitin synthase Chs2 from the ER to the bud neck [249–251] (see also “The mitotic exit network”). As mentioned above, Chs2 is a cargo of the exocyst complex and delivered to the bud neck through Myo2-dependent transport along actin cables. Then, Chs2 persists at the bud neck to form the primary septum in coordination with CAR contraction [120, 212, 248] (Fig. 5). Once this process is accomplished, Chs2 is removed from the neck by endocytosis and transferred to the vacuole for degradation [252].
The F-BAR protein Hof1 forms a ternary complex with the cytokinetic proteins Inn1 and Cyk3 to couple CAR contraction with membrane ingression and primary septum deposition. Mutants affecting the Hof1–Inn1–Cyk3 complex exhibit various degrees of cytokinesis defects and Inn1, but not Hof1 and Cyk3, is essential for cell viability [111, 112, 227, 253–257]. Furthermore, overexpression of HOF1 and CYK3 efficiently rescues the cytokinetic defects of iqg1 mutant cells without restoring a CAR [254]. The Hof1–Inn1–Cyk3 complex is thought to be mainly involved in cytokinesis by promoting primary septum formation, most likely by activating the chitin synthase Chs2 [181, 251, 255, 258]. Consistently, Hof1 interacts physically with Chs2 and stabilizes it at the bud neck during CAR constriction, while overexpression of HOF1 or CYK3 rescues the cytokinetic defects of hypomorphic, but not null, chs2 mutants [111, 256]. In addition, mutants affecting the Hof1–Inn1–Cyk3 complex fail to undergo centripetal and symmetric CAR contraction, resulting in CAR destabilization during constriction similar to chs2 mutant cells [120, 212, 227, 228, 257].
Shortly after the primary septum starts being assembled, cells deposit a secondary septum composed of glucans (polymers of glucose) and mannoproteins (heavily glycosylated cell wall proteins bearing abundant mannose sugars) on each side of the chitin disk (Fig. 5). Synthesis of 1,3-beta-linked glucans, which confer most of the rigidity to the yeast cell wall, is accounted for by the redundant 1,3-beta-glucan synthases Fks1 and Fks2, which in turn are effectors of the Rho1 GTPase (reviewed in [259]), while synthesis of mannoproteins requires a mannosyltransferase complex (reviewed in [260]. Formation of the secondary septum also involves directed secretion [199]. Strikingly, actin cables are oriented toward the bud neck and actin patches cluster at the bud neck during this process enforcing polarized vesicle traffic (Fig. 5). Finally, chitin synthase 3 (Chs3) also contributes to deposition of the secondary septum [261]. In contrast to deletion of CHS2, which abolishes primary septum formation and causes severe cytokinesis failure, deletion of CHS3, either alone or in combination with that of the third chitin synthase Chs1, does not cause obvious cytokinetic defects [251, 261, 262]. However, chs2 mutant cells can survive thanks to the deposition of aberrant remedial septa mostly made by Chs3 that fill up the intercellular space [261, 263]. Remarkably, remedial septa are also built in the absence of CAR assembly, such as in myo1∆ mutants, and therefore represent a major backup cytokinetic mechanism and a resource for cells to rapidly adapt to these adverse conditions [211].
Upon completion of a primary and secondary septum, the cell wall between mother and daughter cell is degraded by hydrolytic enzymes, such as the chitinase Cts1 [264] and several glucanases, including Dse4 and Egt2 [265, 266], thereby allowing cell separation (Fig. 5). Transcription of the genes responsible for cell wall digestion is driven by the Ace2 transcription factor and occurs only at the M to G1 transition of the cell cycle [267], thereby contributing to ensure proper timing of cell separation. Strikingly, the Ace2-dependent transcriptional program driving expression of most hydrolytic enzymes is restricted to the bud, thus explaining why after cell division a birth scar of undigested cell wall is only visible in the mother cells [15].
The mitotic exit network
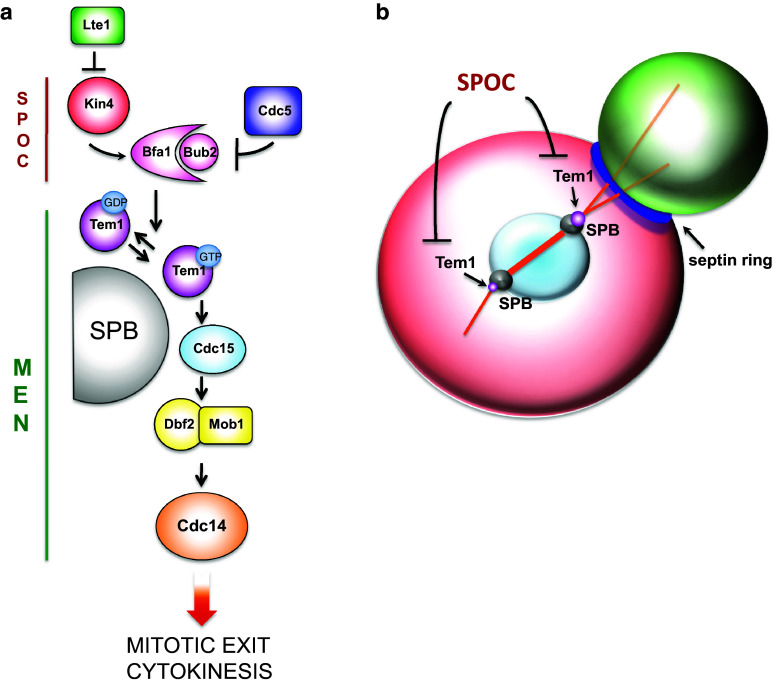
The budding yeast Mitotic Exit Network (MEN), is an essential kinase cascade that is similarly organized to the fission yeast Septation Initiation Network and the metazoan Hippo pathway (reviewed in [268–270]). MEN plays a crucial role in actin repolarization at the end of mitosis, as well as in cytokinesis, and comprises an upstream GTPase (Tem1), its effector Cdc15 kinase, the Mob1-Dbf2 kinase and the Cdc14 phosphatase. The polo kinase Cdc5 activates Tem1 by dampening the activity of the two component GTPase-activating protein Bub2-Bfa1, which keeps a large pool of Tem1 inactive until telophase (reviewed in [271]). Other upstream regulators, such as the polo kinase Cdc5, the bud-localized cortical protein Lte1 and the mother cell-specific Kin4 kinase, modulate Tem1 activation especially in relation to spindle positioning and nuclear division (Fig. 6a). Tem1 and MEN are indeed the targets of the Spindle Position Checkpoint (SPOC), which keeps Tem1 inhibited until the spindle elongates properly along the mother-bud polarity axis in anaphase, thereby preventing mitotic exit and cytokinesis in case of spindle misalignment (reviewed in [12, 13]; Fig. 6b). Intriguingly, proper mitochondrial inheritance from the mother to the daughter cells is required for the function of MEN in cytokinesis [272], suggesting that budding yeast cells keep MEN activity in standby until a balanced set of chromosomes and organelles have been segregated to the bud.
Fig. 6.
The mitotic exit network (MEN) and its regulation by the spindle position checkpoint (SPOC). a MEN signaling takes place mostly at SPBs, where the GTPase Tem1 in its active GTP-bound state promotes recruitment and activation of the Cdc15 protein kinase, which in turn recruits the Dbf2-Mob1 kinase complex that ultimately activates the Cdc14 phosphatase, thereby triggering mitotic exit and cytokinesis. Tem1 is kept inactive by the GTPase-activating protein Bfa1-Bub2 that can be inhibited by the polo kinase Cdc5, whose activity is counteracted by the kinase Kin4 in the mother cell. In turn, the Lte1 protein, which is localized specifically in the bud, restrains Kin4 in the mother compartment. b The MEN inhibitor Kin4 and the MEN activator Lte1 are spatially segregated in the mother and bud compartment, respectively (Kin4 red, Lte1 green). As long as an SPB has not moved into the bud, Tem1 and MEN are kept inactive, thereby coupling spindle positioning and nuclear division with mitotic exit
The core MEN actors are thought to work in a linear cascade (Tem1 > Cdc15 > Mob-Dbf2 > Cdc14), although feedback controls by Cdc14 on various MEN components have been discovered [273–275]. MEN signaling for mitotic exit (i.e., inactivation of mitotic CDKs) occurs at spindle pole bodies (SPBs, i.e., the budding yeast microtubule-organizing centers) by recruitment of MEN components to the SPB scaffold Nud1 (reviewed in [268, 276]; Fig. 6).
The Cdc14 phosphatase promotes mitotic exit in two ways, i.e., by inhibiting mitotic cyclinB-CDKs and by reversing CDK-driven phosphorylation events [277]. MEN is strictly required for its full activation in telophase (reviewed in [278]). Thus, one reason for MEN being involved in cytokinesis is linked to inactivation of CDKs, which otherwise prevent cytokinesis in many eukaryotic systems (reviewed in [8]). For instance, in budding yeast CDK inactivation in telophase is required for actin repolarization and targeted secretion at the bud neck [42, 120]. Many MEN proteins, however, relocalize from SPBs to the bud neck upon mitotic exit, suggesting the existence of additional direct roles in cytokinesis regulation [130, 279–285]. This is indeed the case. Although to date the list of MEN-regulated cytokinesis proteins is likely incomplete, we know several examples of cytokinesis factors that are phosphorylated by MEN kinases or dephosphorylated by Cdc14 (see below). Furthermore, inactivation of MEN proteins in conditions that allow mitotic exit prevents CAR contraction [161, 250]. It is interesting to note that the fission yeast Septation Initiation Network, while being similarly organized to the MEN and involving orthologous proteins, is specifically required for cytokinesis and dispensable for mitotic exit [268, 286].
Although MEN has been clearly involved in CAR constriction and cytokinesis (see below), its role in CAR assembly is controversial. Some MEN mutants have been reported to fail recruiting F actin to the CAR at restrictive temperature [35, 246, 280, 287], while others were shown to proficiently assemble an apparently functional actin ring [112, 130, 282]. Since activation of the Cdc14 phosphatase is required for actin ring formation [246], it is quite surprising that the upstream MEN factors are dispensable for this process. Likely, the ability of some MEN mutants to assemble a functional CAR is ascribable to an incomplete inactivation of the corresponding MEN proteins.
An important target of Cdc14 in CAR assembly is the IQGAP Iqg1 (see “The septin ring”). Iqg1 is phosphorylated in vivo by mitotic CDKs both in S. cerevisiae [288, 289] and in C. albicans [238] and is dephosphorylated by Cdc14 [246]. Mutating the CDK-dependent phosphorylation sites of Iqg1 to non-phosphorylatable alanines leads to premature CAR assembly before anaphase, thus recapitulating the phenotype of cells that either overexpress CDC14 or have reduced levels of mitotic CDKs [238, 246, 290, 291]. Additionally, expression of non-phosphorylatable Iqg1 rescues the inability of cdc14 mutant cells to assemble the F actin ring at restrictive temperature [246], suggesting that Iqg1 is a crucial MEN target in this process.
The chitin synthase Chs2 is phosphorylated by mitotic CDKs and dephosphorylated by Cdc14 to promote its timely relocalization from the ER to the bud neck [249]. Cdc14 has also been shown to interact with and dephosphorylate both formins Bni1 and Bnr1. In cdc14 and cdc15 mutants Bni1 fails to localize to the bud neck, whereas CDC14 overexpression in metaphase displaces Bnr1 from the CAR while recruiting Bni1 [117]. Furthermore, Inn1 recruitment to the neck and activity is likely regulated by CDK-dependent phosphorylation and subsequent dephosphorylation mediated by Cdc14 [181, 250, 255], although the exact mechanism underlying this control remains to be defined. Finally, high CDK activity inhibits the daughter-specific transcriptional program responsible for the expression of the septum-degrading enzymes at the end of cytokinesis through phosphorylation of the transcription factor Ace2, while Cdc14 reverses inhibition [15, 16, 267, 292, 293]. Additional potential cytokinesis targets of mitotic CDKs and/or Cdc14 have been recently identified and hold promises for exciting discoveries in the future [117, 181].
Although activation of the Cdc14 phosphatase appears to be the main function of MEN in cytokinesis [181, 291], upstream MEN factors might contribute directly to this process. For instance, the MEN kinase Dbf2 directly phosphorylates Chs2 and likely stimulates its removal from the CAR by endocytosis [251]. Furthermore, Dbf2-dependent phosphorylation promotes activation of the Chs2 regulatory complex Hof1–Inn1–Cyk3 in several ways. On one hand, MEN contributes to the efficient recruitment of Chs2, Hof1, Inn1 and Cyk3 to the bud neck even independently of mitotic exit [250]. On the other, Hof1 phosphorylation by Dbf2 dissociates it from the septin ring and relocalizes it to the CAR [228].
Concluding remarks
The last 20 years have witnessed a blooming of papers addressing the mechanisms that regulate cytokinesis. The budding yeast S. cerevisiae remains an outstanding model system to study this process, as many of the basic principles underlying cytokinesis are conserved in more complex eukaryotes.
In spite of cutting-edge technologies that have considerably improved the resolution of cytokinetic events and the enormous efforts by researchers in the field, many important questions await an answer, such as how precisely the CAR is organized and what contributes to its contraction, how formins promote CAR assembly, how CAR constriction is coupled to membrane addition, what drives the splitting of the septin ring, which are the critical targets of mitotic CDKs and MEN in cell division, and so forth.
One major obstacle to the progress of our knowledge in this field is the redundancy and intertwinings of cytokinetic pathways and proteins involved, which often makes the contribution of each hard to assess. Nevertheless, we can therefore expect in the years to come exciting discoveries that will shed light on such a fascinating and intricate process providing, hopefully, a complete and detailed picture of cytokinesis.
Acknowledgments
We apologize to all the authors whose works could not be cited here due to the vastity of the subject. We thank M. Granata, L. Merlini, G. Rancati, S. Rincon, A. Sanchez-Diaz and M. Segal for critical reading of the manuscript. Work in S. Piatti’s lab is supported by the Fondation pour la Recherche Médicale (Grant DEQ 20150331740), the Ligue Nationale contre le Cancer and the Fondation ARC (Grant PJA 20141201926).
References
- 1.Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135(9):1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 3.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132(4):583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8(6):462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- 5.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441(7097):1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 6.Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23(23):2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11(4):365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 8.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131(5):847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436(7051):731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- 10.Dechant R, Glotzer M. Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev Cell. 2003;4(3):333–344. doi: 10.1016/S1534-5807(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 11.Oliferenko S, Chew TG, Balasubramanian MK. Positioning cytokinesis. Genes Dev. 2009;23(6):660–674. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 12.Caydasi AK, Ibrahim B, Pereira G. Monitoring spindle orientation: spindle position checkpoint in charge. Cell Div. 2010;5:28. doi: 10.1186/1747-1028-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraschini R, Venturetti M, Chiroli E, Piatti S. The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans. 2008;36(Pt 3):416–420. doi: 10.1042/BST0360416. [DOI] [PubMed] [Google Scholar]
- 14.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183(4677):1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 15.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107(6):739–750. doi: 10.1016/S0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 16.Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, Weiss EL. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6(8):e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422(6933):766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71(1):48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macara IG, McCaffrey L. Cell polarity in morphogenesis and metastasis. Philos Trans R Soc Lond B Biol Sci. 2013;368(1629):20130012. doi: 10.1098/rstb.2013.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muthuswamy SK, Xue B. Cell polarity as a regulator of cancer cell behavior plasticity. Annu Rev Cell Dev Biol. 2012;28:599–625. doi: 10.1146/annurev-cellbio-092910-154244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113(Pt 4):571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 22.Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 2000;113(3):365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- 23.Martin SG, Arkowitz RA. Cell polarization in budding and fission yeasts. FEMS Microbiol Rev. 2014;38(2):228–253. doi: 10.1111/1574-6976.12055. [DOI] [PubMed] [Google Scholar]
- 24.Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70(3):605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 26.Goode BL, Eskin JA, Wendland B. Actin and endocytosis in budding yeast. Genetics. 2015;199(2):315–358. doi: 10.1534/genetics.112.145540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smythe E, Ayscough KR. Actin regulation in endocytosis. J Cell Sci. 2006;119(Pt 22):4589–4598. doi: 10.1242/jcs.03247. [DOI] [PubMed] [Google Scholar]
- 28.Adams AE, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae . J Cell Biol. 1984;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143(7):1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- 30.Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces . J Cell Biol. 1984;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4(1):42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- 32.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4(8):626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 33.Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae . Mol Cell Biol. 1996;16(4):1857–1870. doi: 10.1128/MCB.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142(5):1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140(2):355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65(7):1203–1212. doi: 10.1016/0092-8674(91)90015-Q. [DOI] [PubMed] [Google Scholar]
- 37.Park HO, Bi E, Pringle JR, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci USA. 1997;94(9):4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park HO, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365(6443):269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- 39.Bi E, Park HO. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191(2):347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5(2):179–186. doi: 10.1016/S1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- 41.Howell AS, Lew DJ. Morphogenesis and the cell cycle. Genetics. 2012;190(1):51–77. doi: 10.1534/genetics.111.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120(6):1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 44.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 45.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 46.Schweins T, Wittinghofer A. GTP-binding proteins. Structures, interactions and relationships. Curr Biol. 1994;4(6):547–550. doi: 10.1016/S0960-9822(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 47.Dransart E, Olofsson B, Cherfils J. RhoGDIs revisited: novel roles in Rho regulation. Traffic. 2005;6(11):957–966. doi: 10.1111/j.1600-0854.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 48.Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276(10):7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- 49.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139(4):731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol. 2008;18(22):1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jose M, Tollis S, Nair D, Sibarita JB, McCusker D. Robust polarity establishment occurs via an endocytosis-based cortical corralling mechanism. J Cell Biol. 2013;200(4):407–418. doi: 10.1083/jcb.201206081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slaughter BD, Das A, Schwartz JW, Rubinstein B, Li R. Dual modes of cdc42 recycling fine-tune polarized morphogenesis. Dev Cell. 2009;17(6):823–835. doi: 10.1016/j.devcel.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9(15):1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 54.Goehring AS, Mitchell DA, Tong AH, Keniry ME, Boone C, Sprague GF., Jr Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol Biol Cell. 2003;14(4):1501–1516. doi: 10.1091/mbc.E02-06-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holly SP, Blumer KJ. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae . J Cell Biol. 1999;147(4):845–856. doi: 10.1083/jcb.147.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae . Mol Biol Cell. 2004;15(12):5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol. 2004;164(5):701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss EL, Bishop AC, Shokat KM, Drubin DG. Chemical genetic analysis of the budding-yeast p21-activated kinase cla4p [In Process Citation] Nat Cell Biol. 2000;2(10):677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- 59.Dong Y, Pruyne D, Bretscher A. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J Cell Biol. 2003;161(6):1081–1092. doi: 10.1083/jcb.200212040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown JL, Jaquenoud M, Gulli MP, Chant J, Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11(22):2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen GC, Kim YJ, Chan CS. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae . Genes Dev. 1997;11(22):2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen H, Kuo CC, Kang H, Howell AS, Zyla TR, Jin M, Lew DJ. Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p. Mol Biol Cell. 2012;23(19):3814–3826. doi: 10.1091/mbc.E12-05-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17(3):1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Bi E, Novick P, Du L, Kozminski KG, Lipschutz JH, Guo W. Cdc42 interacts with the exocyst and regulates polarized secretion. J Biol Chem. 2001;276(50):46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X, Orlando K, He B, Xi F, Zhang J, Zajac A, Guo W. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol. 2008;180(1):145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae . EMBO J. 1997;16(10):2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae . EMBO J. 1996;15(22):6060–6068. [PMC free article] [PubMed] [Google Scholar]
- 68.Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin DE, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science. 1996;272(5259):279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 69.Kamada Y, Qadota H, Python CP, Anraku Y, Ohya Y, Levin DE. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271(16):9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 70.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae . EMBO J. 1995;14(23):5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo W, Tamanoi F, Novick P. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat Cell Biol. 2001;3(4):353–360. doi: 10.1038/35070029. [DOI] [PubMed] [Google Scholar]
- 72.Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M. Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol Biol Evol. 2008;25(12):2717–2733. doi: 10.1093/molbev/msn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11(1):62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 74.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276(5309):118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 75.Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4(3):260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- 76.Pruyne D, Gao L, Bi E, Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol Biol Cell. 2004;15(11):4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18(5):1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Delgehyr N, Lopes CS, Moir CA, Huisman SM, Segal M. Dissecting the involvement of formins in Bud6p-mediated cortical capture of microtubules in S. cerevisiae . J Cell Sci. 2008;121(Pt 22):3803–3814. doi: 10.1242/jcs.036269. [DOI] [PubMed] [Google Scholar]
- 79.Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280(30):28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- 80.Wen KK, Rubenstein PA. Differential regulation of actin polymerization and structure by yeast formin isoforms. J Biol Chem. 2009;284(25):16776–16783. doi: 10.1074/jbc.M109.006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae . J Biol Chem. 1998;273(43):28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- 82.Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, Irie K, Takai Y. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae . Mol Cell Biol. 2001;21(3):827–839. doi: 10.1128/MCB.21.3.827-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p–Bni1p–Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae . Mol Biol Cell. 1998;9(5):1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18(7):4053–4069. doi: 10.1128/MCB.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22(21):4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL. Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. J Cell Biol. 2013;201(4):595–611. doi: 10.1083/jcb.201212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15(2):896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116(5):711–723. doi: 10.1016/S0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 89.Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42(2):486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- 90.Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120(12):3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 91.Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276(4):2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- 92.Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280(8):6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- 93.Gould CJ, Maiti S, Michelot A, Graziano BR, Blanchoin L, Goode BL. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21(5):384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lammers M, Rose R, Scrima A, Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 2005;24(23):4176–4187. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wallar BJ, Stropich BN, Schoenherr JA, Holman HA, Kitchen SM, Alberts AS. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281(7):4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- 96.Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13(15):1335–1340. doi: 10.1016/S0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 97.Liu W, Sato A, Khadka D, Bharti R, Diaz H, Runnels LW, Habas R. Mechanism of activation of the Formin protein Daam1. Proc Natl Acad Sci USA. 2008;105(1):210–215. doi: 10.1073/pnas.0707277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nezami AG, Poy F, Eck MJ. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14(2):257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 99.Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435(7041):513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 100.Seth A, Otomo C, Rosen MK. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol. 2006;174(5):701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lees JG, Bach CT, O’Neill GM. Interior decoration: tropomyosin in actin dynamics and cell migration. Cell Adhes Migr. 2011;5(2):181–186. doi: 10.4161/cam.5.2.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vindin H, Gunning P. Cytoskeletal tropomyosins: choreographers of actin filament functional diversity. J Muscle Res Cell Motil. 2013;34(3–4):261–274. doi: 10.1007/s10974-013-9355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57(2):233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- 104.Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20(8):2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chesarone-Cataldo M, Guerin C, Yu JH, Wedlich-Soldner R, Blanchoin L, Goode BL. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev Cell. 2011;21(2):217–230. doi: 10.1016/j.devcel.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eskin JA, Rankova A, Johnston AB, Alioto SL, Goode BL. Common formin-regulating sequences in Smy1 and Bud14 are required for the control of actin cable assembly in vivo. Mol Biol Cell. 2016;27:828–837. doi: 10.1091/mbc.E15-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mohapatra L, Goode BL, Kondev J. Antenna mechanism of length control of actin cables. PLoS Comput Biol. 2015;11(6):e1004160. doi: 10.1371/journal.pcbi.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16(2):292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gould CJ, Chesarone-Cataldo M, Alioto SL, Salin B, Sagot I, Goode BL. Saccharomyces cerevisiae Kelch proteins and Bud14 protein form a stable 520-kDa formin regulatory complex that controls actin cable assembly and cell morphogenesis. J Biol Chem. 2014;289(26):18290–18301. doi: 10.1074/jbc.M114.548719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Philips J, Herskowitz I. Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae . J Cell Biol. 1998;143(2):375–389. doi: 10.1083/jcb.143.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol Biol Cell. 2013;24(9):1305–1320. doi: 10.1091/mbc.E12-11-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae . Mol Biol Cell. 2000;11(2):593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Graziano BR, Yu HY, Alioto SL, Eskin JA, Ydenberg CA, Waterman DP, Garabedian M, Goode BL. The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol Biol Cell. 2014;25(11):1730–1743. doi: 10.1091/mbc.E14-03-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buttery SM, Kono K, Stokasimov E, Pellman D. Regulation of the formin Bnr1 by septins anda MARK/Par1-family septin-associated kinase. Mol Biol Cell. 2012;23(20):4041–4053. doi: 10.1091/mbc.E12-05-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang J, Neo SP, Cai M. Regulation of the yeast formin Bni1p by the actin-regulating kinase Prk1p. Traffic. 2009;10(5):528–535. doi: 10.1111/j.1600-0854.2009.00893.x. [DOI] [PubMed] [Google Scholar]
- 116.Matheos D, Metodiev M, Muller E, Stone D, Rose MD. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J Cell Biol. 2004;165(1):99–109. doi: 10.1083/jcb.200309089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem. 2011;286(7):5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Orii M, Kono K, Wen HI, Nakanishi M. PP1-dependent formin Bnr1 dephosphorylation and delocalization from a cell division site. PLoS One. 2016;11(1):e0146941. doi: 10.1371/journal.pone.0146941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 2012;150(1):151–164. doi: 10.1016/j.cell.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 120.VerPlank L, Li R. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol Biol Cell. 2005;16(5):2529–2543. doi: 10.1091/mbc.E04-12-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8(4):729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Finger FP, Hughes TE, Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92(4):559–571. doi: 10.1016/S0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- 123.Meitinger F, Richter H, Heisel S, Hub B, Seufert W, Pereira G. A safeguard mechanism regulates Rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biol. 2013;11(2):e1001495. doi: 10.1371/journal.pbio.1001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tolliday N, VerPlank L, Li R. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol. 2002;12(21):1864–1870. doi: 10.1016/S0960-9822(02)01238-1. [DOI] [PubMed] [Google Scholar]
- 125.Annan RB, Lee AY, Reid ID, Sayad A, Whiteway M, Hallett M, Thomas DY. A biochemical genomics screen for substrates of Ste20p kinase enables the in silico prediction of novel substrates. PLoS One. 2009;4(12):e8279. doi: 10.1371/journal.pone.0008279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol. 2007;179(7):1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ziman M, Preuss D, Mulholland J, O’Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4(12):1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol. 2013;202(2):231–240. doi: 10.1083/jcb.201301090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yoshida S, Bartolini S, Pellman D. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23(7):810–823. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, Pellman D. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313(5783):108–111. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- 131.Heider MR, Gu M, Duffy CM, Mirza AM, Marcotte LL, Walls AC, Farrall N, Hakhverdyan Z, Field MC, Rout MP, Frost A, Munson M. Subunit connectivity, assembly determinants and architecture of the yeast exocyst complex. Nat Struct Mol Biol. 2016;23(1):59–66. doi: 10.1038/nsmb.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae . EMBO J. 1996;15(23):6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 133.TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae . J Cell Biol. 1995;130(2):299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aronov S, Gerst JE. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J Biol Chem. 2004;279(35):36962–36971. doi: 10.1074/jbc.M402068200. [DOI] [PubMed] [Google Scholar]
- 135.Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol Biol Cell. 1997;8(8):1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jourdain I, Dooley HC, Toda T. Fission yeast sec3 bridges the exocyst complex to the actin cytoskeleton. Traffic. 2012;13(11):1481–1495. doi: 10.1111/j.1600-0854.2012.01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Adamo JE, Moskow JJ, Gladfelter AS, Viterbo D, Lew DJ, Brennwald PJ. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J Cell Biol. 2001;155(4):581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Roumanie O, Wu H, Molk JN, Rossi G, Bloom K, Brennwald P. Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J Cell Biol. 2005;170(4):583–594. doi: 10.1083/jcb.200504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2010;21(3):141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol. 2008;9(6):478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- 141.Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69(2):265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 142.Berlin A, Paoletti A, Chang F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160(7):1083–1092. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tasto JJ, Morrell JL, Gould KL. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160(7):1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.An H, Morrell JL, Jennings JL, Link AJ, Gould KL. Requirements of fission yeast septins for complex formation, localization, and function. Mol Biol Cell. 2004;15(12):5551–5564. doi: 10.1091/mbc.E04-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martin-Cuadrado AB, Morrell JL, Konomi M, An H, Petit C, Osumi M, Balasubramanian M, Gould KL, Del Rey F, de Aldana CR. Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol Biol Cell. 2005;16(10):4867–4881. doi: 10.1091/mbc.E04-12-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bertin A, McMurray MA, Grob P, Park SS, Garcia G, 3rd, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105(24):8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Garcia G, 3rd, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol. 2011;195(6):993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bridges AA, Zhang H, Mehta SB, Occhipinti P, Tani T, Gladfelter AS. Septin assemblies form by diffusion-driven annealing on membranes. Proc Natl Acad Sci USA. 2014;111(6):2146–2151. doi: 10.1073/pnas.1314138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee PR, Song S, Ro HS, Park CJ, Lippincott J, Li R, Pringle JR, De Virgilio C, Longtine MS, Lee KS. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae . Mol Cell Biol. 2002;22(19):6906–6920. doi: 10.1128/MCB.22.19.6906-6920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Patasi C, Godocikova J, Michlikova S, Nie Y, Kacerikova R, Kvalova K, Raunser S, Farkasovsky M. The role of Bni5 in the regulation of septin higher-order structure formation. Biol Chem. 2015;396(12):1325–1337. doi: 10.1515/hsz-2015-0165. [DOI] [PubMed] [Google Scholar]
- 151.Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191(7):1333–1350. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Finnigan GC, Booth EA, Duvalyan A, Liao EN, Thorner J. The carboxy-terminal tails of septins Cdc11 and Shs1 recruit myosin-II binding factor Bni5 to the bud neck in Saccharomyces cerevisiae . Genetics. 2015;200(3):843–862. doi: 10.1534/genetics.115.176503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schneider C, Grois J, Renz C, Gronemeyer T, Johnsson N. Septin rings act as a template for myosin higher-order structures and inhibit redundant polarity establishment. J Cell Sci. 2013;126(Pt 15):3390–3400. doi: 10.1242/jcs.125302. [DOI] [PubMed] [Google Scholar]
- 154.Bertin A, McMurray MA, Thai L, Garcia G, 3rd, Votin V, Grob P, Allyn T, Thorner J, Nogales E. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404:711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Casamayor A, Snyder M. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol Cell Biol. 2003;23(8):2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang J, Kong C, Xie H, McPherson PS, Grinstein S, Trimble WS. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9(24):1458–1467. doi: 10.1016/S0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- 157.Garrenton LS, Stefan CJ, McMurray MA, Emr SD, Thorner J. Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci USA. 2010;107(26):11805–11810. doi: 10.1073/pnas.1005817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McMurray MA, Bertin A, Garcia G, 3rd, Lam L, Nogales E, Thorner J. Septin filament formation is essential in budding yeast. Dev Cell. 2011;20(4):540–549. doi: 10.1016/j.devcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Caviston JP, Longtine M, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell. 2003;14(10):4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4(3):345–357. doi: 10.1016/S1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 161.Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114(Pt 7):1379–1386. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- 162.Wloka C, Nishihama R, Onishi M, Oh Y, Hanna J, Pringle JR, Krauss M, Bi E. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol Chem. 2011;392(8–9):813–829. doi: 10.1515/BC.2011.083. [DOI] [PubMed] [Google Scholar]
- 163.Demay BS, Bai X, Howard L, Occhipinti P, Meseroll RA, Spiliotis ET, Oldenbourg R, Gladfelter AS. Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J Cell Biol. 2011;193(6):1065–1081. doi: 10.1083/jcb.201012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443(7110):466–469. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- 165.Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69(3):717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ong K, Wloka C, Okada S, Svitkina T, Bi E. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun. 2014;5:5698. doi: 10.1038/ncomms6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cid VJ, Adamikova L, Sanchez M, Molina M, Nombela C. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 2001;147(Pt 6):1437–1450. doi: 10.1099/00221287-147-6-1437. [DOI] [PubMed] [Google Scholar]
- 168.Gladfelter AS, Bose I, Zyla TR, Bardes ES, Lew DJ. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156(2):315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sadian Y, Gatsogiannis C, Patasi C, Hofnagel O, Goody RS, Farkasovsky M, Raunser S. The role of Cdc42 and Gic1 in the regulation of septin filament formation and dissociation. eLife. 2013;2:e01085. doi: 10.7554/eLife.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Okada S, Leda M, Hanna J, Savage NS, Bi E, Goryachev AB. Daughter cell identity emerges from the interplay of cdc42, septins, and exocytosis. Dev Cell. 2013;26(2):148–161. doi: 10.1016/j.devcel.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McMurray MA, Thorner J. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr Biol. 2008;18(16):1203–1208. doi: 10.1016/j.cub.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Piekny AJ, Maddox AS. The myriad roles of anillin during cytokinesis. Semin Cell Dev Biol. 2010;21(9):881–891. doi: 10.1016/j.semcdb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 173.Kang PJ, Angerman E, Jung CH, Park HO. Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J Cell Sci. 2012;125(Pt 16):3840–3849. doi: 10.1242/jcs.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sanders SL, Herskowitz I. The BUD4 protein of yeast, required for axial budding, is localized to the mother/BUD neck in a cell cycle-dependent manner. J Cell Biol. 1996;134(2):413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu H, Guo J, Zhou YT, Gao XD. The anillin-related region of Bud4 is the major functional determinant for Bud4’s function in septin organization during bud growth and axial bud site selection in budding yeast. Eukaryot Cell. 2015;14(3):241–251. doi: 10.1128/EC.00268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eluere R, Varlet I, Bernadac A, Simon MN. Cdk and the anillin homolog Bud4 define a new pathway regulating septin organization in yeast. Cell Cycle. 2012;11(1):151–158. doi: 10.4161/cc.11.1.18542. [DOI] [PubMed] [Google Scholar]
- 177.Merlini L, Bolognesi A, Juanes MA, Vandermoere F, Courtellemont T, Pascolutti R, Seveno M, Barral Y, Piatti S. Rho1- and Pkc1-dependent phosphorylation of the F-BAR protein Syp1 contributes to septin ring assembly. Mol Biol Cell. 2015;26(18):3245–3262. doi: 10.1091/mbc.E15-06-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Qiu W, Neo SP, Yu X, Cai M. A novel septin-associated protein, Syp1p, is required for normal cell cycle-dependent septin cytoskeleton dynamics in yeast. Genetics. 2008;180(3):1445–1457. doi: 10.1534/genetics.108.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28(20):3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20(22):4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kuilman T, Maiolica A, Godfrey M, Scheidel N, Aebersold R, Uhlmann F. Identification of Cdk targets that control cytokinesis. EMBO J. 2014;34(1):81–96. doi: 10.15252/embj.201488958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16(4):493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 183.Saarikangas J, Barral Y. The emerging functions of septins in metazoans. EMBO Rep. 2011;12(11):1118–1126. doi: 10.1038/embor.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Castillon GA, Adames NR, Rosello CH, Seidel HS, Longtine MS, Cooper JA, Heil-Chapdelaine RA. Septins have a dual role in controlling mitotic exit in budding yeast. Curr Biol. 2003;13(8):654–658. doi: 10.1016/S0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 185.Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci USA. 2003;100(20):11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290(5490):341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 187.Luedeke C, Frei SB, Sbalzarini I, Schwarz H, Spang A, Barral Y. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J Cell Biol. 2005;169(6):897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kusch J, Meyer A, Snyder MP, Barral Y. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 2002;16(13):1627–1639. doi: 10.1101/gad.222602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.King K, Jin M, Lew D. Roles of Hsl1p and Hsl7p in Swe1p degradation: beyond septin tethering. Eukaryot Cell. 2012;11(12):1496–1502. doi: 10.1128/EC.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae . Mol Cell Biol. 2000;20(11):4049–4061. doi: 10.1128/MCB.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.McMillan JN, Longtine MS, Sia RA, Theesfeld CL, Bardes ES, Pringle JR, Lew DJ. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol. 1999;19(10):6929–6939. doi: 10.1128/MCB.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae . Mol Cell Biol. 1999;19(10):7123–7137. doi: 10.1128/MCB.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Versele M, Thorner J. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 2005;15(8):414–424. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sreenivasan A, Kellogg D. The elm1 kinase functions in a mitotic signaling network in budding yeast. Mol Cell Biol. 1999;19(12):7983–7994. doi: 10.1128/MCB.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Asano S, Park JE, Yu LR, Zhou M, Sakchaisri K, Park CJ, Kang YH, Thorner J, Veenstra TD, Lee KS. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J Biol Chem. 2006;281(37):27090–27098. doi: 10.1074/jbc.M601483200. [DOI] [PubMed] [Google Scholar]
- 196.Mortensen EM, McDonald H, Yates J, 3rd, Kellogg DR. Cell cycle-dependent assembly of a Gin4-septin complex. Mol Biol Cell. 2002;13(6):2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Egelhofer TA, Villen J, McCusker D, Gygi SP, Kellogg DR. The septins function in G1 pathways that influence the pattern of cell growth in budding yeast. PLoS One. 2008;3(4):e2022. doi: 10.1371/journal.pone.0002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tang CS, Reed SI. Phosphorylation of the septin cdc3 in g1 by the cdc28 kinase is essential for efficient septin ring disassembly. Cell Cycle. 2002;1(1):42–49. [PubMed] [Google Scholar]
- 199.Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305(5682):393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- 200.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147(5):981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106(6):735–744. doi: 10.1016/S0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 202.Chahwan R, Gravel S, Matsusaka T, Jackson SP. Dma/RNF8 proteins are evolutionarily conserved E3 ubiquitin ligases that target septins. Cell Cycle. 2013;12(6):1000–1008. doi: 10.4161/cc.23947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fraschini R, Bilotta D, Lucchini G, Piatti S. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol Biol Cell. 2004;15(8):3796–3810. doi: 10.1091/mbc.E04-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Merlini L, Fraschini R, Boettcher B, Barral Y, Lucchini G, Piatti S. Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. PLoS Genet. 2012;8(4):e1002670. doi: 10.1371/journal.pgen.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mitchell L, Lau A, Lambert JP, Zhou H, Fong Y, Couture JF, Figeys D, Baetz K. Regulation of septin dynamics by the Saccharomyces cerevisiae lysine acetyltransferase NuA4. PLoS One. 2011;6(10):e25336. doi: 10.1371/journal.pone.0025336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kinoshita M, Field CM, Coughlin ML, Straight AF, Mitchison TJ. Self- and actin-templated assembly of mammalian septins. Dev Cell. 2002;3(6):791–802. doi: 10.1016/S1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 207.Vrabioiu AM, Mitchison TJ. Symmetry of septin hourglass and ring structures. J Mol Biol. 2007;372(1):37–49. doi: 10.1016/j.jmb.2007.05.100. [DOI] [PubMed] [Google Scholar]
- 208.Kozubowski L, Larson JR, Tatchell K. Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae . Mol Biol Cell. 2005;16(8):3455–3466. doi: 10.1091/mbc.E04-09-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Longtine MS, Fares H, Pringle JR. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143(3):719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307(5716):1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 211.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135(5):879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci. 2002;115(Pt 2):293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- 213.Tolliday N, Pitcher M, Li R. Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol Biol Cell. 2003;14(2):798–809. doi: 10.1091/mbc.E02-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Luo J, Vallen EA, Dravis C, Tcheperegine SE, Drees B, Bi E. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae . J Cell Biol. 2004;165(6):843–855. doi: 10.1083/jcb.200401040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Watts FZ, Shiels G, Orr E. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 1987;6(11):3499–3505. doi: 10.1002/j.1460-2075.1987.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Boyne JR, Yosuf HM, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113(Pt 24):4533–4543. doi: 10.1242/jcs.113.24.4533. [DOI] [PubMed] [Google Scholar]
- 217.Epp JA, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7(12):921–929. doi: 10.1016/S0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- 218.Shannon KB, Li R. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol Biol Cell. 1999;10(2):283–296. doi: 10.1091/mbc.10.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shannon KB, Li R. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr Biol. 2000;10(12):727–730. doi: 10.1016/S0960-9822(00)00539-X. [DOI] [PubMed] [Google Scholar]
- 220.Tian C, Wu Y, Johnsson N. Stepwise and cooperative assembly of a cytokinetic core complex in Saccharomyces cerevisiae . J Cell Sci. 2014;127(Pt 16):3614–3624. doi: 10.1242/jcs.153429. [DOI] [PubMed] [Google Scholar]
- 221.Feng Z, Okada S, Cai G, Zhou B, Bi E. MyosinII heavy chain and formin mediate the targeting of myosin essential light chain to the division site before and during cytokinesis. Mol Biol Cell. 2015;26(7):1211–1224. doi: 10.1091/mbc.E14-09-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe . J Cell Biol. 2003;162(5):851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82(3):435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- 224.Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192(6):1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe . Mol Biol Cell. 2006;17(7):3254–3266. doi: 10.1091/mbc.E05-11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Willet AH, McDonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol. 2015;208(4):391–399. doi: 10.1083/jcb.201411097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143(7):1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25(8):875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Nkosi PJ, Targosz BS, Labib K, Sanchez-Diaz A. Hof1 and Rvs167 have redundant roles in actomyosin ring function during cytokinesis in budding yeast. PLoS One. 2013;8(2):e57846. doi: 10.1371/journal.pone.0057846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Huckaba TM, Lipkin T, Pon LA. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J Cell Biol. 2006;175(6):957–969. doi: 10.1083/jcb.200609155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wloka C, Vallen EA, The L, Fang X, Oh Y, Bi E. Immobile myosin-II plays a scaffolding role during cytokinesis in budding yeast. J Cell Biol. 2013;200(3):271–286. doi: 10.1083/jcb.201208030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15(12):651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 233.Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419(6902):82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- 234.Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K. Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J Biol Chem. 1997;272(47):29579–29583. doi: 10.1074/jbc.272.47.29579. [DOI] [PubMed] [Google Scholar]
- 235.Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15(12):2997–3005. [PMC free article] [PubMed] [Google Scholar]
- 236.Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, Kroschewski R. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282(1):426–435. doi: 10.1074/jbc.M607711200. [DOI] [PubMed] [Google Scholar]
- 237.Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178(2):193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Li CR, Wang YM, Wang Y. The IQGAP Iqg1 is a regulatory target of CDK for cytokinesis in Candida albicans. EMBO J. 2008;27(22):2998–3010. doi: 10.1038/emboj.2008.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Huxley H, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173(4412):973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 240.Huxley HE. The mechanism of muscular contraction. Science. 1969;164(3886):1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 241.Tully GH, Nishihama R, Pringle JR, Morgan DO. The anaphase-promoting complex promotes actomyosin-ring disassembly during cytokinesis in yeast. Mol Biol Cell. 2009;20(4):1201–1212. doi: 10.1091/mbc.E08-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lord M, Laves E, Pollard TD. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol Biol Cell. 2005;16(11):5346–5355. doi: 10.1091/mbc.E05-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mendes Pinto I, Rubinstein B, Kucharavy A, Unruh JR, Li R. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev Cell. 2012;22(6):1247–1260. doi: 10.1016/j.devcel.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Rahal R, Amon A. The Polo-like kinase Cdc5 interacts with FEAR network components and Cdc14. Cell Cycle. 2008;7(20):3262–3272. doi: 10.4161/cc.7.20.6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zumdieck A, Kruse K, Bringmann H, Hyman AA, Julicher F. Stress generation and filament turnover during actin ring constriction. PLoS One. 2007;2(8):e696. doi: 10.1371/journal.pone.0000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Miller DP, Hall H, Chaparian R, Mara M, Mueller A, Hall MC, Shannon KB. Dephosphorylation of Iqg1 by Cdc14 regulates cytokinesis in budding yeast. Mol Biol Cell. 2015;26(16):2913–2926. doi: 10.1091/mbc.E14-12-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wagner W, Bielli P, Wacha S, Ragnini-Wilson A. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 2002;21(23):6397–6408. doi: 10.1093/emboj/cdf650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zhang G, Kashimshetty R, Ng KE, Tan HB, Yeong FM. Exit from mitosis triggers Chs2p transport from the endoplasmic reticulum to mother-daughter neck via the secretory pathway in budding yeast. J Cell Biol. 2006;174(2):207–220. doi: 10.1083/jcb.200604094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chin CF, Bennett AM, Ma WK, Hall MC, Yeong FM. Dependence of Chs2 ER export on dephosphorylation by cytoplasmic Cdc14 ensures that septum formation follows mitosis. Mol Biol Cell. 2012;23(1):45–58. doi: 10.1091/mbc.E11-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Meitinger F, Petrova B, Lombardi IM, Bertazzi DT, Hub B, Zentgraf H, Pereira G. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci. 2010;123(Pt 11):1851–1861. doi: 10.1242/jcs.063891. [DOI] [PubMed] [Google Scholar]
- 251.Oh Y, Chang KJ, Orlean P, Wloka C, Deshaies R, Bi E. Mitotic exit kinase Dbf2 directly phosphorylates chitin synthase Chs2 to regulate cytokinesis in budding yeast. Mol Biol Cell. 2012;23(13):2445–2456. doi: 10.1091/mbc.E12-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135(3):597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Jendretzki A, Ciklic I, Rodicio R, Schmitz HP, Heinisch JJ. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol Genet Genomics. 2009;282(4):437–451. doi: 10.1007/s00438-009-0476-0. [DOI] [PubMed] [Google Scholar]
- 254.Korinek WS, Bi E, Epp JA, Wang L, Ho J, Chant J. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr Biol. 2000;10(15):947–950. doi: 10.1016/S0960-9822(00)00626-6. [DOI] [PubMed] [Google Scholar]
- 255.Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, Bi E. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae . J Cell Biol. 2009;185(6):995–1012. doi: 10.1083/jcb.200903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Onishi M, Ko N, Nishihama R, Pringle JR. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol. 2013;202(2):311–329. doi: 10.1083/jcb.201302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Sanchez-Diaz A, Marchesi V, Murray S, Jones R, Pereira G, Edmondson R, Allen T, Labib K. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat Cell Biol. 2008;10(4):395–406. doi: 10.1038/ncb1701. [DOI] [PubMed] [Google Scholar]
- 258.Devrekanli A, Foltman M, Roncero C, Sanchez-Diaz A, Labib K. Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J Cell Sci. 2012;125(Pt 22):5453–5466. doi: 10.1242/jcs.109157. [DOI] [PubMed] [Google Scholar]
- 259.Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189(4):1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae . Microbiol Mol Biol Rev. 2006;70(2):317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Duran A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114(1):111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Sburlati A, Cabib E. Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae . J Biol Chem. 1986;261(32):15147–15152. [PubMed] [Google Scholar]
- 263.Cabib E, Schmidt M. Chitin synthase III activity, but not the chitin ring, is required for remedial septa formation in budding yeast. FEMS Microbiol Lett. 2003;224(2):299–305. doi: 10.1016/S0378-1097(03)00477-4. [DOI] [PubMed] [Google Scholar]
- 264.Kuranda MJ, Robbins PW. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae . J Biol Chem. 1991;266(29):19758–19767. [PubMed] [Google Scholar]
- 265.Baladron V, Ufano S, Duenas E, Martin-Cuadrado AB, del Rey F, Vazquez de Aldana CR. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae . Eukaryot Cell. 2002;1(5):774–786. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kovacech B, Nasmyth K, Schuster T. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol Cell Biol. 1996;16(7):3264–3274. doi: 10.1128/MCB.16.7.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Mazanka E, Weiss EL. Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol Biol Cell. 2010;21(16):2809–2820. doi: 10.1091/mbc.E10-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2(11):815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- 269.Hergovich A, Hemmings BA. Hippo signalling in the G2/M cell cycle phase: lessons learned from the yeast MEN and SIN pathways. Semin Cell Dev Biol. 2012;23(7):794–802. doi: 10.1016/j.semcdb.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Krapp A, Gulli MP, Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol. 2004;14(17):R722–R730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 271.Scarfone I, Piatti S. Coupling spindle position with mitotic exit in budding yeast: the multifaceted role of the small GTPase Tem1. Small GTPases. 2015;6(4):1–6. doi: 10.1080/21541248.2015.1109023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Garcia-Rodriguez LJ, Crider DG, Gay AC, Salanueva IJ, Boldogh IR, Pon LA. Mitochondrial inheritance is required for MEN-regulated cytokinesis in budding yeast. Curr Biol. 2009;19(20):1730–1735. doi: 10.1016/j.cub.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Jaspersen SL, Morgan DO. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol. 2000;10(10):615–618. doi: 10.1016/S0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- 274.Konig C, Maekawa H, Schiebel E. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J Cell Biol. 2010;188(3):351–368. doi: 10.1083/jcb.200911128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Pereira G, Manson C, Grindlay J, Schiebel E. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J Cell Biol. 2002;157(3):367–379. doi: 10.1083/jcb.200112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Pereira G, Schiebel E. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr Opin Cell Biol. 2001;13(6):762–769. doi: 10.1016/S0955-0674(00)00281-7. [DOI] [PubMed] [Google Scholar]
- 277.Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk- dependent phosphorylation. Mol Cell. 1998;2(6):709–718. doi: 10.1016/S1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 278.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 279.Bembenek J, Kang J, Kurischko C, Li B, Raab JR, Belanger KD, Luca FC, Yu H. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle. 2005;4(7):961–971. doi: 10.4161/cc.4.7.1798. [DOI] [PubMed] [Google Scholar]
- 280.Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J Cell Sci. 2000;113(Pt 19):3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- 281.Hwa Lim H, Yeong FM, Surana U. Inactivation of mitotic kinase triggers translocation of MEN components to mother-daughter neck in yeast. Mol Biol Cell. 2003;14(11):4734–4743. doi: 10.1091/mbc.E03-04-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21(20):6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20(1):286–298. doi: 10.1128/MCB.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Xu S, Huang HK, Kaiser P, Latterich M, Hunter T. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol. 2000;10(6):329–332. doi: 10.1016/S0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]
- 285.Yoshida S, Toh-e A. Regulation of the localization of Dbf2 and mob1 during cell division of Saccharomyces cerevisiae . Genes Genet Syst. 2001;76(2):141–147. doi: 10.1266/ggs.76.141. [DOI] [PubMed] [Google Scholar]
- 286.Simanis V. The mitotic exit and septation initiation networks. J Cell Sci. 2003;116(Pt 21):4261–4262. doi: 10.1242/jcs.00777. [DOI] [PubMed] [Google Scholar]
- 287.Corbett M, Xiong Y, Boyne JR, Wright DJ, Munro E, Price C. IQGAP and mitotic exit network (MEN) proteins are required for cytokinesis and re-polarization of the actin cytoskeleton in the budding yeast, Saccharomyces cerevisiae . Eur J Cell Biol. 2006;85(11):1201–1215. doi: 10.1016/j.ejcb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 288.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325(5948):1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10(7):676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Naylor SG, Morgan DO. Cdk1-dependent phosphorylation of Iqg1 governs actomyosin ring assembly prior to cytokinesis. J Cell Sci. 2014;127(Pt 5):1128–1137. doi: 10.1242/jcs.144097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sanchez-Diaz A, Nkosi PJ, Murray S, Labib K. The mitotic exit network and Cdc14 phosphatase initiate cytokinesis by counteracting CDK phosphorylations and blocking polarised growth. EMBO J. 2012;31(17):3620–3634. doi: 10.1038/emboj.2012.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bidlingmaier S, Weiss EL, Seidel C, Drubin DG, Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae . Mol Cell Biol. 2001;21(7):2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Brace J, Hsu J, Weiss EL. Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis. Mol Cell Biol. 2011;31(4):721–735. doi: 10.1128/MCB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]