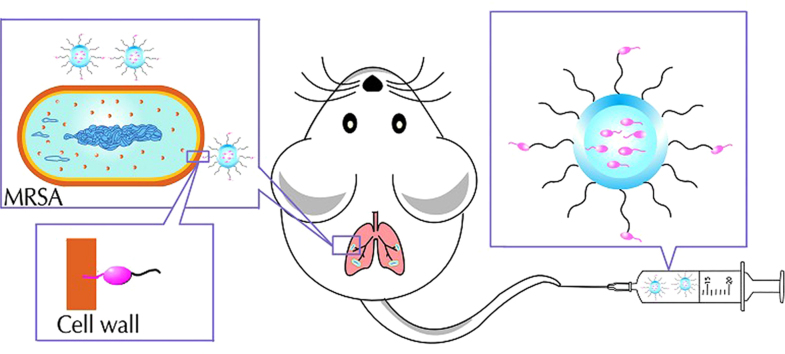
Abstract
Considering that some antibacterial agents can identify the outer structure of pathogens like cell wall and/or cell membrane, we explored a self-enhanced targeted delivery strategy by which a small amount of the antibiotic molecules were modified on the surface of carriers as targeting ligands of certain bacteria while more antibiotic molecules were loaded inside the carriers, and thus has the potential to improve the drug concentration at the infection site, enhance efficacy and reduce potential toxicity. In this study, a novel targeted delivery system against methicillin-resistant Staphylococcus aureus (MRSA) pneumonia was constructed with daptomycin, a lipopeptide antibiotic, which can bind to the cell wall of S. aureus via its hydrophobic tail. Daptomycin was conjugated with N-hydroxysuccinimidyl–polyethylene glycol–1,2-distearoyl-sn-glycero-3-phosphoethanolamine to synthesize a targeting compound (Dapt–PEG–DSPE) which could be anchored on the surface of liposomes, while additional daptomycin molecules were encapsulated inside the liposomes. These daptomycin-modified, daptomycin-loaded liposomes (DPD-L[D]) showed specific binding to MRSA as detected by flow cytometry and good targeting capabilities in vivo to MRSA-infected lungs in a pneumonia model. DPD-L[D] exhibited more favorable antibacterial efficacy against MRSA than conventional PEGylated liposomal daptomycin both in vitro and in vivo. Our study demonstrates that daptomycin-modified liposomes can enhance MRSA-targeted delivery of encapsulated antibiotic, suggesting a novel drug delivery approach for existing antimicrobial agents.
KEY WORDS: Daptomycin, Liposome, Targeted drug delivery, Lung infection, Staphylococcus aureus
Graphical abstract
Daptomycin, as targeting molecules conjugated with NHS–PEG3400–DSPE, were modified on the surface of liposomes with more daptomycin molecules encapsulated. The specific targeting effect and promising therapeutic efficacy of the Dapt–PEG–DSPE-modified liposome against methicillin-resistant Staphylococcus aureus has been investigated, both in vitro and in vivo. This study may provide a novel drug delivery approach for treatment of staphylococcal infections.
1. Introduction
“Magic bullets”, put forward by Paul Ehrlich, was first used to refer to compounds that had special affinity for pathogens and destructive power over them1. This concept has led to the development of a number of nano-sized drug carriers modified by targeting ligands, referred to as “active targeting drug delivery systems” and widely used in tumor therapy. Considering the increasing difficulties in developing new antibacterial agents and the limitations of the existing antibiotics, including toxicity to normal tissues, drug resistance, and rapid degradation and clearance, novel drug delivery approaches are urgently required to address these problems. Unlike the relatively high similarity between cancer cells and normal tissue cells, there are huge differences between prokaryotic cells (pathogens) and eukaryotic cells (hosts), especially the outer part of bacteria providing an “Achilles׳ Heel” for recognition and tracking. By attaching a targeting moiety such as an antibody, an aptamer or peptides to the surface of a nano-carrier, these drug delivery agents can selectively bind to microorganisms or infected cells and release drug payloads to kill or inhibit the growth of the microorganisms, thereby significantly enhancing the therapeutic efficacy of existing antimicrobial therapies2. However, the current used ligands for targeting pathogen is very limited, with some suffereing from in vivo stability and high cost, which necessitates the design or development of new ligands.
It is worth noting that there are many antimicrobial agents that target the bacterial cell wall and/or cell membrane. For some of them corresponding targets exist on the surface of bacterial cells allowing specific binding. There is a possibility, therefore, that those cell wall– and membrane-active agents may work as targeting molecules for bacteria rather than as a direct inhibitor when used at a very low dose. For example, vancomycin, a glycopeptide antibiotic which can bind to Gram-positive bacteria via a five-point hydrogen bond interaction with the d-Ala–d-Ala moieties of N-acetylmuramic acid and N-acetylglucosamine peptide subunits in the cell wall, was used as a molecular recognition agent for Staphylococcus aureus in vitro3. UBI29-41, a human antimicrobial peptide fragment, was conjugated to chloramphenicol for the targeted therapy of bacterial infections4. These findings suggest that some antibiotic agents may have potential to be targeting ligands of pathogens, in addition to their anti-bacterial properties.
Daptomycin, a novel cyclic lipopeptide antibiotic, has been proven to be a successful and less toxic alternative to vancomycin in the treatment of multidrug resistant Gram-positive pathogens5. It has a unique chemical structure containing a 13-member amino acid cyclic lipopeptide (hydrophilic core) with a decanoyl side chain (lipophilic tail). Daptomycin exerts its antibacterial effect via binding to the cell wall of Gram-positive bacteria, resulting in perturbation and depolarization of the cell membrane6. Currently, Cubicin®, an injectable solution, is the only approved formulation of daptomycin. Therefore, there is a growing interest in the development of new formulations and delivery methods to further enhance the in vivo performance of daptomycin7, 8.
Liposomes as nano-carriers have been widely used for antimicrobial drug delivery, including polymyxin B liposomes9, benzyl penicillin liposomes10 and teicoplanin liposomes11. Liposomes can encapsulate both hydrophilic and hydrophobic compounds, increasing the drug solubility. They provide better pharmacokinetics and biodistribution versus nonencapsulated drug, and enhance activity against both extracellular and intracellular pathogens12. Conventional liposomes can be further designed with active targeting property by surface modification to better target and selectively bind to microorganisms, and thereby improve drug accumulation at the infected sites, reduce adverse effects, decrease drug toxicity, and increase efficacy.
Herein, we propose a self-enhanced targeted delivery strategy: daptomycin molecules, modified as targeting molecules by conjugation with N-Hydroxysuccinimidyl (NHS)–polyethylene glycol (MW3400 Da, PEG3400)–1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE), were modified and placed on the surface of liposomes in which more daptomycin molecules were encapsulated. The methicillin-resistant S. aureus (MRSA) binding capability of these daptomycin-modified liposomes was measured in vitro. Mouse models of MRSA hematogenous pulmonary infection were used to further evaluate the targeting efficacy and curative effects in vivo.
2. Materials and methods
2.1. Materials
Daptomycin was purchased from Meilun Biotech Co. (Dalian, China). NHS–PEG3400–DSPE was supplied by Nanocs Corp. (New York, NY, USA). Soybean phospholipids (SPC) were from Aladdin Bio-Chem Technology Co., Ltd. (Shanghai, China). m-Polyethylene glycol2000 (mPEG2000)–DSPE was obtained from AVT pharmaceuticals Co., Ltd. (Shanghai, China). 1,1′-Dioctadecyl-3,3,3,3-tetramethylindotricarbocyanineiodide (DiR) was supplied by Invitrogen Corp. (Life Technologies, Carlsbad, CA, USA). Fluoresceinamine isomer II (FAM) was from Sigma-Aldrich (St. Louis, MO, USA). The other reagents were all of analytical grade.
MRSA252 was incubated in tryptic soy broth (Nissui Pharmaceuticals, Japan) at 37 °C. Six-week-old male/female Kunming (KM) mice weighing 20±2 g were purchased from the experimental animal center of Third Military Medical University (Chongqing, China) and kept in regulated conditions. All animal experiments were performed in accordance with the guidelines approved by the ethics committee of the College of Pharmaceutical Sciences, Southwest University, China.
2.2. Targeting compound synthesis and characterization
For the synthesis of the targeting compound Dapt–PEG–DSPE, daptomycin was conjugated with NHS–PEG3400–DSPE by an amide reaction13. Daptomycin and NHS–PEG3400–DSPE were dissolved in newly distilled dimethyl fumarate (DMF) at a molar ratio of 1.2:1 and the pH of the solution was adjusted to 8.0–8.5 with an appropriate amount of triethylamine, followed by stirring continuously for 72 h at 25 °C. The reaction endpoint was confirmed by HPLC and the product was purified by dialysis (MW cut off 3500 Da) against distilled water for 48 h. Finally, the product solution was lyophilized. To confirm the conjugation of daptomycin with NHS–PEG3400–DSPE, the final product was assayed by a matrix-assisted laser desorption/ionization time-of-flight MS (MALDI-TOF-MS, Autoflex speed, Bruker, Germany).
2.3. Preparation of liposomes
Daptomycin-loaded liposomes modified by Dapt–PEG–DSPE (DPD-L[D]) were prepared by a thin-film hydration method. Briefly, SPC:cholesterol:mPEG2000–DSPE:Dapt–PEG–DSPE (73.5:24.5:1.5:0.5, molar ratio) was dissolved in methanol. To form a lipid film, the mixture was evaporated to remove methanol and mixed with physiological saline containing daptomycin (drug:lipids=1:10, w/w) and sonicated. The liposomal size was controlled by a miniextruder and the liposomes were purified by Sepharose CL-4B chromatography to remove free drug. PEGylated liposomal daptomycin (mPEG-L[D], SPC:cholesterol:mPEG2000–DSPE=73.5:24.5:2, molar ratio; daptomycin:lipids=1:10, w/w) and Dapt–PEG–DSPE-modified blank liposomes (DPD-Lip; SPC:cholesterol:mPEG2000–DSPE:Dapt–PEG–DSPE=73.5:24.5:1.5:0.5, molar ratio) were prepared by the same method as DPD-L[D].
A preparation of liposomes loading with fluorescent reagents was prepared following a procedure identical to that used for Dapt-loaded liposomes, with daptomycin replaced by FAM dispersed in solution or by DiR dissolved in methanol, respectively.
2.4. Characterization of liposomes
The particle size and polydispersity index of the liposomes was measured by dynamic light scattering using a Zetasizer Nano ZS (Malvern, Worcestershire, UK). The entrapment efficiency of DPD-L[D] was determined by gel filtration. The crude liposomes prior to chromatographic purification were divided into two equal parts. One part was purified using the Sephadex CL-4B column and the eluent collected. The other was diluted into the same volume of the eluent above. The two samples were then treated with 10% (v/v) Triton X-100 for demulsification. Encapsulation efficiency (EE) was calculated as follows: EE (%)=(W/W0)×100, where W and W0 are the daptomycin content in purified liposomes and total drug in dispersion, respectively. The content of daptomycin was measured by high performance liquid chromatography (HPLC).
2.5. Determination of the minimum inhibitory concentration (MIC)
The MIC of DPD-L[D], mPEG-L[D], and free daptomycin were determined by the broth micro-dilution method in 96-well plates14. Since daptomycin is a calcium-dependent antibiotic, the MICs were measured by using Mueller–Hinton II broth with 50 μg/mL calcium ions15. Each preparation was added to 100 μL Mueller–Hinton II broth at sequential 2-fold dilutions, with concentrations in the range of 64–0.0625 μg/mL. A 100 μL MRSA252 aliquot (106 CFU/mL) was added to each well. The plates were incubated at 37 °C for 24 h. The MIC was designated as the lowest concentration of daptomycin at which there was no visible and measurable MRSA growth after 24 h of incubation. Each experiment was performed in triplicate.
2.6. In vitro targeting efficacy
Flow cytometry (ACEA Novo Cyte, 2060R, Shanghai, China) was used to measure the specific binding between Dapt–PEG–DSPE-modified FAM-loaded liposomes (DPD-L[FAM]) and bacteria. PEGylated liposomal FAM (mPEG-L[FAM]) and free FAM to MRSA252 were also evaluated in parallel. The mixture of DPD-Lip (Dapt–PEG–DSPE-modified blank liposome) and free FAM were used as controls. MRSA252 was incubated in Mueller-Hinton II broth with shaking overnight to logarithmic phase growth at 37 °C. Bacteria were collected by centrifugation at 3000×g for 10 min and resuspended in fresh medium to the desired density (108 CFU/mL). FAM-loaded liposomes were mixed with bacterial suspension at a 1:9 (v/v) ratio and incubated at 37 °C for 1 and 2 h. Samples then were centrifuged (3000×g, 10 min) to collect the bacterial cells and rinsed with PBS 3 times, then measured by flow cytometry. The bacterial suspension served as a blank control. The velocity of flow was 14 μL/min, and the diameter of the sample flow orifice was 7.7 μm. Bacterially-associated FAM was excited with an argon laser (488 nm), and fluorescence was detected at 530 nm.
2.7. Establishment and evaluation of MRSA252 hematogenous pneumonia model
Animal models were established by the method previously reported16, 17. Briefly, MRSA252 in the logarithmic phase were collected by centrifugation at 3000×g for 10 min and resuspended in sterilized normal saline and washed three times. Subsequently, the suspension was warmed to 45 °C. The bacterial suspension was mixed with 4% (w/v) molten agar in the proportion of 1:1 at 45 °C. One milliliter of the agar-bacterium suspension was rapidly injected into 49 mL of rapidly stirred ice-bath-chilled sterile saline via a 1 mL syringe with 26-gauge needle, and agar beads formed. The final concentration of agar was 0.04% (w/v), and the final number of bacteria was 2×109 CFU/mL.
A murine model of hematogenous pneumonia infection was generated by injecting 250 μL suspension containing bacteria-loaded agar beads into the tail vein of KM mice. As a contrast, 250 μL bacteria suspension (2×109 CFU/mL) was injected. Six infected mice in each group were killed to get lung, liver, kidney and spleen under aseptic conditions at 1, 3 and 7 days post-injection. The tissue was weighed and homogenized for bacteria counts, suspended in sterile saline and cultured quantitatively by serial dilutions on MH IIA plates at 37 °C. The number of colony forming units (CFU) was counted 24 h later.
2.8. In vivo optical imaging
Mice (normal or pneumonic) were intravenously administered Dapt–PEG–DSPE-modified DiR-loaded liposomes (DPD-L[DiR]) or PEGylated liposomal DiR (mPEG-L[DiR]). To detect DiR fluorescence, an in vivo imaging system (FX-Pro, BRUKER) with a 730 nm excitation filter and an emission filter at 790 nm were used. Hair was removed from the mice using 8% (w/v) sodium sulfide solution. Mice were scanned at 2, 4, 8 and 12 h. Meanwhile, different organs (heart, liver, spleen, lung, kidney and brain) were removed for ex vivo imaging.
2.9. Biodistribution study
Seventy-two KM-infected mice were randomly assigned to three groups: treated with DPD-L[D], mPEG-L[D], or free daptomycin via intravenous administration. The dosage in each group was equivalent to a 25 mg/kg dose of daptomycin. The mice were killed with removal of liver, spleen, lung and kidney at predetermined time intervals (0.5, 4, 8 and 24 h). The tissues were weighed and homogenized in physiological saline. Daptomycin was extracted from the tissue samples by a mixed solvent of acetonitrile and methanol (1:1, v/v). The supernatant was transferred and then dried by a stream of nitrogen gas. The residue was redissolved in ultrapure water and further analyzed by HPLC. For HPLC detection of daptomycin, the mobile phase was composed of acetonitrile and 0.5% NH4H2PO4 at a volume ratio of 40:60. The mobile phase was delivered at a flow rate of 0.7 mL/min and kept at 30 °C. Column eluate was monitored at 221 nm.
2.10. In vivo pharmacodynamics effects
This study included five groups: three treatment groups with 30 pneumonic mice (n=10 per group) with each group receiving a single dose treatment via tail vein with one of the preparations: DPD-L[D], mPEG-L[D] and saline. The dosage of DPD-L[D] or mPEG-L[D] was equivalent to a 25 mg/kg dose of daptomycin which is only half of the reported dosage18. Herein, Dapt–PEG–DSPE was calculated as an equivalent amount of daptomycin in the preparation of DPD-L[D]. Meanwhile, normal mice received an injection in the tail vein of either saline or a suspension containing agar beads without bacteria. The survival rate of the mice and body weight changes were monitored for 10 days. Lung samples were obtained from pneumonic mice administered DPD-L[D], mPEG-L[D] or saline at the 3rd day post-injection for histopathological evaluation and bacterial number counts.
A preliminary safety assessment of the daptomycin-loaded liposomes treatment was conducted via studying serum biochemical parameters, including alanine aminotransferase, aspartate aminotransferase, blood urea nitrogen, creatinine phosphokinase, creatinine and uric acid, using a fully automated biochemistry analyser (TMS-1024i, BOEKI, Japan).
2.11. Data analysis
Survival data are presented using Kaplan–Meier plots and were analyzed using a log-rank test. Student׳s t-test was employed for comparing two groups, and the one-way analysis of variance (ANOVA) was used for multiple group comparisons. A value of P<0.05 was considered statistically significant.
3. Results and discussion
3.1. Preparation and characterization of liposomes
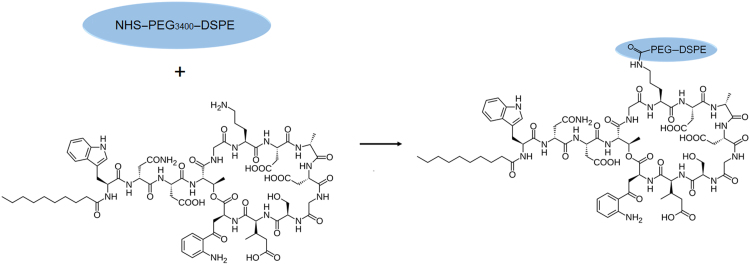
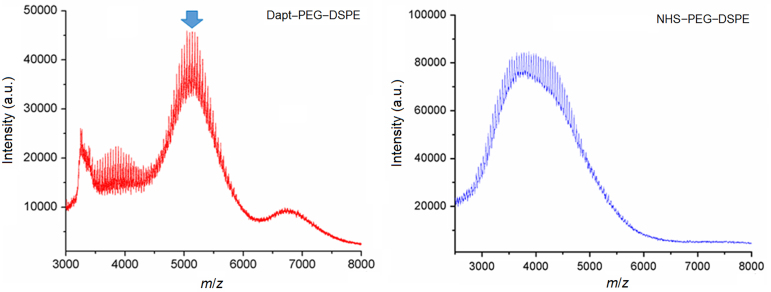
The hydrophilic core of daptomycin contains an active amino group that can react with succinimide, so the targeting compound can be synthesized by conjugation of daptomycin (MW 1620.68 Da) with NHS–PEG3400–DSPE in DMF through a nucleophilic substitution reaction which forms the amide bond19 (Scheme 1). As shown in Fig. 1, the experimental MW of NHS–PEG3400–DSPE and Dapt–PEG–DSPE determined by MALDI-TOF-MS were around 3700 and 5300 Da, respectively. These values were in accordance with the theoretical MW and confirmed the identity of the synthesis product.
Scheme 1.
Model for the synthesis of Dapt–PEG–DSPE (DPD).
Figure 1.
The MALDI-TOF mass spectrometry analysis of Dapt–PEG–DSPE (DPD) and NHS–PEG3400–DSPE. The arrow indicates the peak corresponding to the targeting compound. The molecular weight of DPD was approximately 5300 Da.
Since S. aureus ranges from 0.9–1.6 µm in diameter20, much smaller than eukaryotic cells, control of the liposomal particle size may be necessary. The characteristics of liposomes modified with Dapt–PEG–DSPE or mPEG–DSPE are summarized in Table 1. As shown, all liposomes had a uniform particle size less than 100 nm, with a narrow distribution. Approximately 90% of the daptomycin can be loaded into liposomes. In addition, the stability of drug-loaded liposomes was evaluated at 25 °C. There were no obvious size changes and no drug leakage detected over one month (Table 2).
Table 1.
Characterization of liposomes.
| Liposomes | Z-average (nm) | PDI | Entrapment efficiency (%) |
|---|---|---|---|
| DPD-L[D] | 94.60±1.12 | 0.24±0.12 | 91.85±2.16 |
| mPEG-L[D] | 93.88±2.15 | 0.22±0.08 | 93.73±1.47 |
| DPD-L[FAM] | 91.41±2.02 | 0.22±0.06 | 27.56±1.06 |
| mPEG-L[FAM] | 90.23±1.42 | 0.26±0.09 | 28.21±0.63 |
| DPD-L[DiR] | 88.25±1.33 | 0.21±0.05 | 88.79±2.06 |
| mPEG-L[DiR] | 86.72±0.95 | 0.19±0.03 | 90.21±1.54 |
Data are expressed as mean±SD, n=3.
Table 2.
Stability evaluation of DPD-L[D] at 25±0.3 °C.
| Time (day) | Z-average (nm) | PDI | Drug leakage (%) |
|---|---|---|---|
| 0 | 94.60±1.12 | 0.24±0.12 | –a |
| 7 | 93.92±1.91 | 0.25±0.08 | 0.35±0.13 |
| 15 | 98.24±2.73 | 0.29±0.02 | 0.60±0.21 |
| 30 | 100.79±2.38 | 0.32±0.06 | 1.45±0.16 |
Data are expressed as mean±SD, n = 3.
Not applicable.
3.2. In vitro MRSA targeting and pharmacodynamics
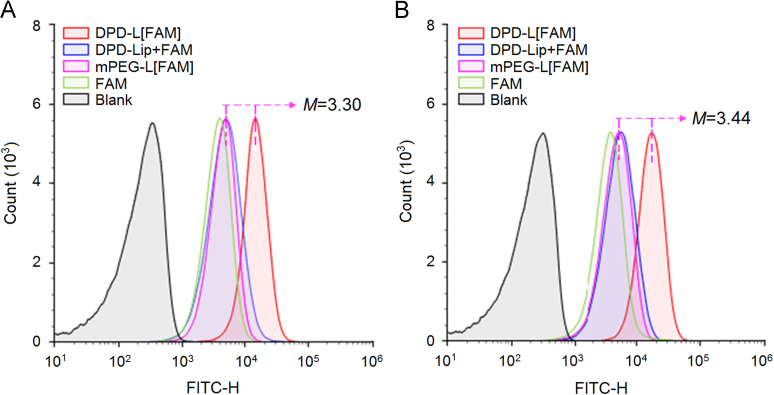
The uptake of Dapt–PEG–DSPE-modified liposomes by MRSA was detected by flow cytometry, and water-soluble FAM served as fluorescence probe (Fig. 2). There was a significant difference in fluorescence intensity between the DPD-L[FAM] group and that of mPEG-L[FAM] group, the former was 3.30- and 3.44-fold higher than the latter after incubation for 1 and 2 h, respectively. No obvious increase in fluorescence signal was observed in the treatment group by the physical mixture of DPD-Lip and FAM when compared with that of mPEG-L[FAM] group, supporting the potential of daptomycin as a targeting ligand to mediate liposomal drug delivery to the targeted bacteria.
Figure 2.
Flow cytometric measurement of the interaction of bacteria with different formulations at 1 h (A) and 2 h (B).
The broth micro-dilution method was used to determine the MIC of each formulation. The results showed MICs of free daptomycin, mPEG-L[D] and DPD-L[D] against MRSA252 of 0.5, 0.5, and 0.25 μg/mL, respectively. It is worth noting that Dapt–PEG–DSPE was calculated as an equivalent amount of daptomycin in the preparation of DPD-L[D]. Therefore, the above results primarily illustrate that the placement of a fraction of daptomycin on the surface enhanced the anti-bacterial efficacy of liposomal daptomycin.
3.3. Establishment of S. aureus hematogenous pneumonia model
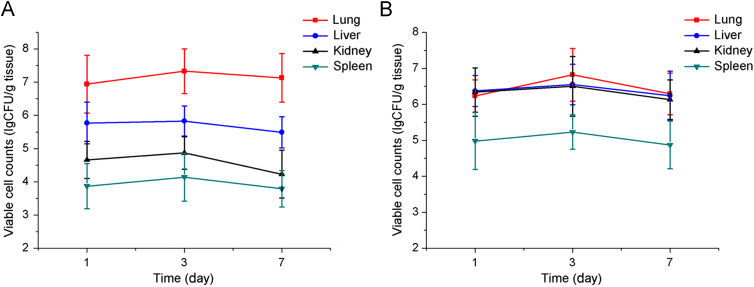
In this study, pneumonia models were constructed via intravenous administration of bacteria embedded in agar beads18. It is well accepted that larger microspheres (greater than 20 μm) will be effectively accumulated in lung via the mechanical trapping effect of pulmonary capillaries after intravenous injection21. Accordingly, it can be expected that bacteria-loaded agar beads in the micrometer range may be concentrated in lung leading to pneumonia. We also have established an animal model simply by giving a bacterial suspension prepared in saline for comparison, and time-dependent changes in the number of viable MRSA252 in the major organs are shown in Fig. 3. The mice injected with MRSA252 enmeshed in agar beads is designated group A, and the mice infected by a bacterial suspension designated group N. The bacterial number in the lungs was markedly higher than that of other organs in group A (Fig. 3A) 1, 3 or 7 days post-infection (P<0.01) and was more than 10-fold higher than that in the livers. There were no marked differences in the bacterial numbers between the lung, liver and kidneys of group N (Fig. 3B). These findings indicate that the bacteria-loaded agar bead technique increased infection efficiency and selectivity to lung, confirming that a murine model of hematogenous pneumonia infection has been established.
Figure 3.
The bacterial loads in the major organs of pneumonia-model mice established by different approach. The mice were injected with MRSA252 enmeshed in agar beads (A) or infected by bacteria suspension (B). Data are expressed as mean±SD, n=6.
3.4. Evaluation of targeting ability and biodistribution in vivo
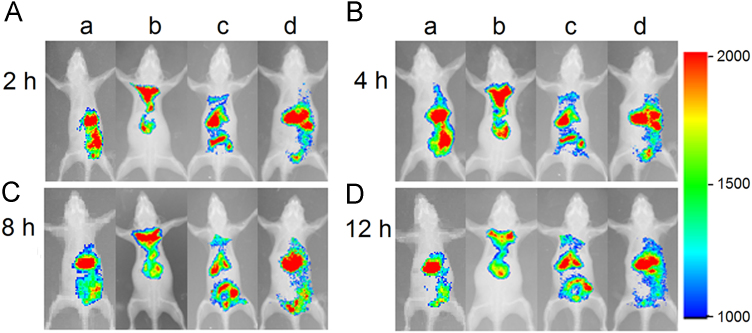
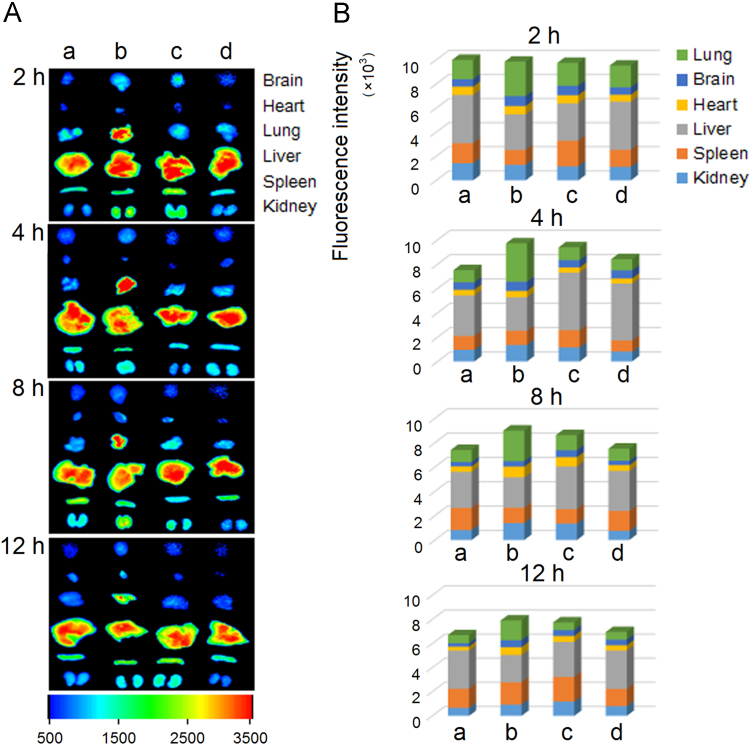
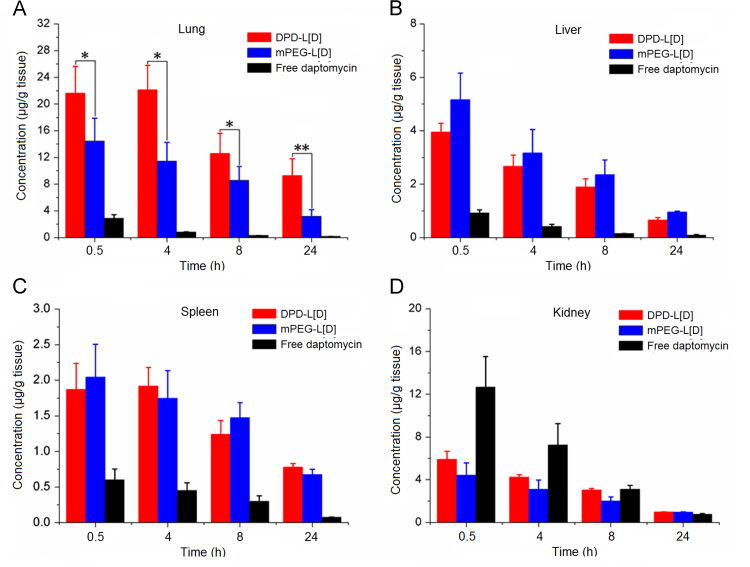
Both the pneumonia model and normal mice were used in the in vivo imaging assays. As shown in Figure 4, Figure 5, daptomycin-modified liposomes markedly increased the distribution of encapsulated fluorophore in infected lung compared with non-modified liposomes at each time point. However, in the treatment groups of normal mice there were no differences observed between those treated with daptomycin-modified liposomes and non-modified liposomes, indicating that specific interactions between daptomycin and MRSA were mainly responsible for the lung targeting of the liposomes. Furthermore, a quantitative analysis of drug distribution in multiple organs was performed after a single dose of 25 mg/kg of encapsulated daptomycin in bacterially infected mice (Fig. 6). It is clearly shown that the daptomycin concentration of lung tissues in the DPD-L[D] group were markedly higher than those in the mPEG-L[D] group and the free daptomycin injection group. Previously we have reported that conventional daptomycin-loaded liposomes increased level of daptomycin in the lung tissue of infected mice22, and here the further modification of liposomes by daptomycin itself has made a further improvement in the targeting efficiency.
Figure 4.
In vivo whole-body fluorescence imaging in mice administered DiR-loaded formulations via tail vein at 2 h (A), 4 h (B), 8 h (C) and 12 h (D): normal mice treated by DPD-L[DiR] (a), infected mice by DPD-L[DiR] (b), infected mice by mPEG-L[DiR] (c), and normal mice by mPEG-L[DiR] (d).
Figure 5.
Ex vivo fluorescent images of tissues dissected from mice (A) and the semi-quantitative analysis (B): normal mice treated by DPD-L[DiR] (a), infected mice by DPD-L[DiR] (b), infected mice by mPEG-L[DiR] (c) and normal mice by mPEG-L[DiR] (d).
Figure 6.
Biodistribution of daptomycin in lungs (A), livers (B), spleens (C) and kidneys (D) following tail vein injection of a dose (25 mg/kg) of DPD-L[D], mPEG-L[D] or free daptomycin in infected mice. All values are expressed as mean±SD (n = 6). *P<0.05, **P<0.01.
3.5. In vivo anti-infective efficacy
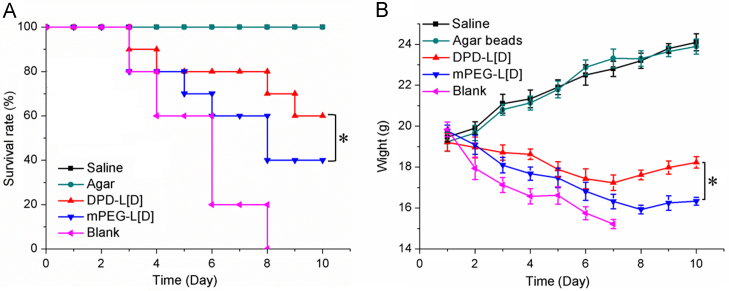
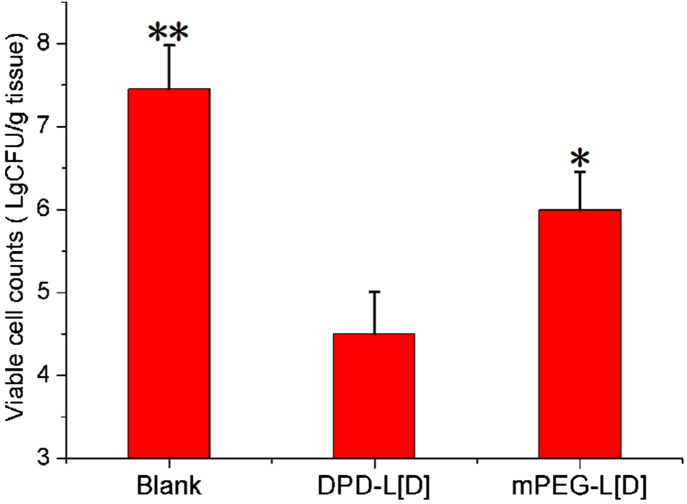
In comparison, the survival rates (Fig. 7A) of animals treated with DPD-L[D] or mPEG-L[D] were 60% and 40% at the 10th day, respectively. Obviously, the survival period of the DPD-L[D] group was significantly prolonged (P<0.05) compared to that of mPEG-L[D] group. Meanwhile, the mortality rate of the untreated group approached 100% by the 8th day post-administration. Normal mice given 0.04% agar beads or saline served as controls. As Fig. 7B shows, the body weight of the DPD-L[D], mPEG-L[D] and untreated groups showed a downward trend within the first week, especially the latter group, mainly due to the low dosage of daptomycin. However, the body weight of the DPD-L[D] group decreased more slowly than the other infected groups and increased gradually from the 8th day. An overall increase in body weight was observed in the two groups of normal mice. The bacteria present in the lung tissues of each group were counted on the third day post-treatment. As shown in Fig. 8, the concentration of bacteria in the DPD-L[D] treated group was almost three orders of magnitude lower than that of untreated group and more than one order of magnitude lower than that of the mPEG-L[D] treated group.
Figure 7.
Antibacterial effect of drug treatments on pneumonia-infected mice. Survival rate (A) and body weight (B) changes. DPD-L[D]: infected mice treated with DPD-L[D]; mPEG-L[D]: infected mice treated with mPEG-L[D]; blank: infected mice without treatment; agar: normal mice treated with agar bead suspension; saline: normal mice treated with physiological saline. The DPD-L[D] and mPEG-L[D] groups received a single dose of daptomycin (25 mg/kg). Data represented as mean ±SD (n = 10). *P < 0.05.
Figure 8.
Bacterial loads in lung tissues 3 days after intravenous tail injection of a dose (25 mg/kg) of DPD-L[D] or mPEG-L[D] formulations. Saline was used for the untreated group. Data represented as mean±SD (n = 6). *P<0.05, **P<0.01.
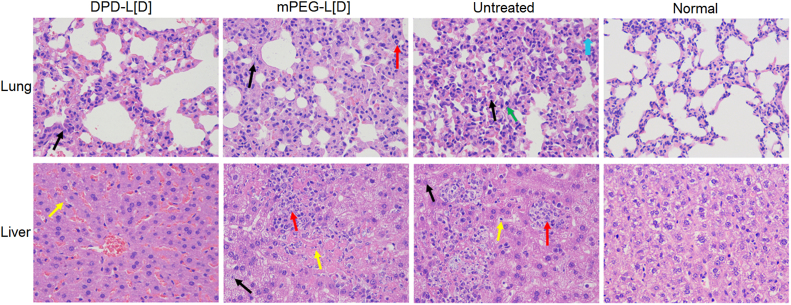
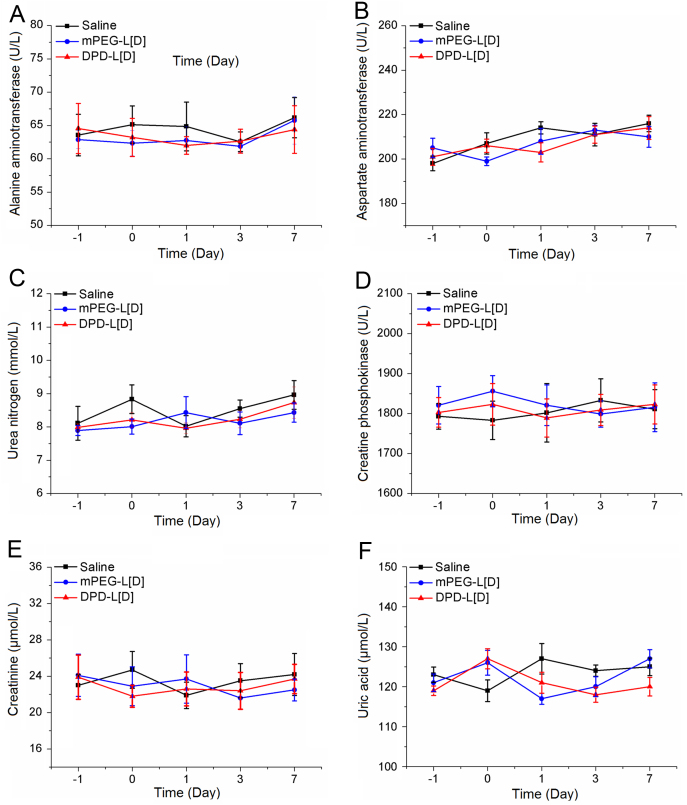
Histopathological assessments were performed on the 3rd day post-administration, as shown in Fig. 9. Lesions in liver and lung in the DPD-L[D] group were lighter than that in mPEG-L[D] group and untreated group. No significant difference was observed in serum biochemical parameters of all treatment groups compared with those of the saline group (Fig. 10). These results further demonstrate that DPD-L[D] had a much better antibacterial effect than the conventional liposomal formulation when given the same low dose of daptomycin.
Figure 9.
The histopathological examination of the major organs (lung and liver) 3 days post-treatment. Both DPD-L[D] and mPEG-L[D] groups received a single dose of daptomycin (25 mg/kg). Lung (H&E, 400×): ( ) indicates alveolar atrophy or not expansion and alveolar wall thickening; (
) indicates alveolar atrophy or not expansion and alveolar wall thickening; ( ) indicates neutrophils; (
) indicates neutrophils; ( ) indicates eosinophils; (
) indicates eosinophils; ( ) indicates dust cells. Liver (H&E 400×): (
) indicates dust cells. Liver (H&E 400×): ( ) indicates cell granular degeneration and edema; (
) indicates cell granular degeneration and edema; ( ) indicates Kupffer cells; (
) indicates Kupffer cells; ( ) indicates infiltration of inflammatory cells including lymphocytes and neutrophils.
) indicates infiltration of inflammatory cells including lymphocytes and neutrophils.
Figure 10.
Daily serum biochemical parameters detected during safety evaluation of daptomycin-modified daptomycin-loaded liposomes. Alanine aminotransferase (A), aspartate aminotransferase (B), urea nitrogen (C), creatinine phosphokinase (D), creatinine (E) and uric acid (F) levels from normal mice were detected on days −1, 0, 1, 3 and 7. Mice from saline group, DPD-L[D] group and mPEG-L[D] group were administrated a dose of saline, daptomycin-modified daptomycin-loaded liposomes (mPEG-L[D] 25 mg/kg in a single i.v. dose), and PEGylated liposomal daptomycin (mPEG-L[D], 25 mg/kg in a single i.v. dose) on day 0, respectively. Each data point represents the mean±SD, n=6.
4. Conclusions
In summary, we have demonstrated the self-enhanced targeted delivery of daptomycin against staphylococcal pneumonia, in which daptomycin has dual function of both an MRSA-targeting ligand and an antibiotic agent. This novel daptomycin-liposomal formulation exhibited highly specific targeting and promising therapeutic efficacy against MRSA, both in vitro and in vivo. In future research this self-enhanced targeting strategy can be extended to other cell wall–/membrane-active antimicrobial agents, creating innovative targeted therapeutic approaches against dangerous pathogens.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81102404), the Fundamental Research Funds for the Central Universities (Nos. XDJK2015A012 and XDJK2013A015), and the Program for Innovative Research Team in University of Chongqing.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Lagord C., Kearins O., Lawrence G., Bishop H., Monypenny I., Bates A.T. Paul Ehrlich: from magic bullets to chemotherapy. Colomb Méd. 2008;39:291–295. [Google Scholar]
- 2.Zhang L., Pornpattananangku D.C., Hu C.M., Huang C.M. Development of nanoparticles for antimicrobial drug delivery. Curr Med Chem. 2010;17:585–594. doi: 10.2174/092986710790416290. [DOI] [PubMed] [Google Scholar]
- 3.Gao H.F., Yang S.J., Han J., Xiong J., Kong W.J., Li C. Double-site recognition of pathogenic bacterium whole cells based on antibiotic-affinity strategy. Chem Commun. 2015;51:12497–12500. doi: 10.1039/c5cc02814k. [DOI] [PubMed] [Google Scholar]
- 4.Chen H.Y., Liu C.C., Chen D., Madrid K., Peng S.W., Dong X.Y. Bacteria-targeting conjugates based on antimicrobial peptide for bacteria diagnosis and therapy. Mol Pharm. 2015;12:2505–2516. doi: 10.1021/acs.molpharmaceut.5b00053. [DOI] [PubMed] [Google Scholar]
- 5.Beiras-Fernandez A., Vogt F., Sodian R., Weis F. Daptomycin: a novel lipopeptide antibiotic against Gram-positive pathogens. Infect Drug Resist. 2010;3:95–101. doi: 10.2147/IDR.S6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steenbergen J.N., Jeff A., Thorne G.M., Tally F.P. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother. 2005;55:283–288. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X.L., Huang X.L., Deng L., Li C., Liao G.J., Chen Z.B. Preparation and quality analysis of daptomycin transdermal delivery system. Chin J Antibiot. 2013;38:130–134. [Google Scholar]
- 8.Costa J.R., Silva N.C., Sarmento B., Pintado M. Potential chitosan-coated alginate nanoparticles for ocular delivery of daptomycin. Eur J Clin Microbiol Infect Dis. 2015;34:1255–1262. doi: 10.1007/s10096-015-2344-7. [DOI] [PubMed] [Google Scholar]
- 9.Boswell G.W., Buell D., Bekersky I. AmBisome (liposomal amphotericin B): a comparative review. J Clin Pharmacol. 1998;38:583–592. doi: 10.1002/j.1552-4604.1998.tb04464.x. [DOI] [PubMed] [Google Scholar]
- 10.Kim H.J., Jones M.N. The delivery of benzyl penicillin to Staphylococcus aureus biofilms by use of liposomes. J Liposome Res. 2004;14:123–139. doi: 10.1081/lpr-200029887. [DOI] [PubMed] [Google Scholar]
- 11.Onyeji C.O., Nightingale C.H., Marangos M.N. Enhanced killing of methicillin-resistant Staphylococcus aureus in human macrophages by liposome-entrapped vancomycin and teicoplanin. Infection. 1994;22:338–342. doi: 10.1007/BF01715542. [DOI] [PubMed] [Google Scholar]
- 12.Drulis-Kawa Z., Dorotkiewicz-Jach A. Liposomes as delivery systems for antibiotics. Int J Pharm. 2010;387:187–198. doi: 10.1016/j.ijpharm.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z.H., Yu Y., Dai W.B., Lu J.K., Cui J.R., Wu H.N. The use of a tumor metastasis targeting peptide to deliver doxorubicin-containing liposomes to highly metastatic cancer. Biomaterials. 2012;33:8451–8460. doi: 10.1016/j.biomaterials.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 14.European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect. 2003;9:ix–xv. doi: 10.1046/j.1469-0691.2000.00142.x. [DOI] [PubMed] [Google Scholar]
- 15.Lai C.C., Sheng W.H., Wang J.T., Liao C.H., Ho M.W., Chen C.J. Safety and efficacy of daptomycin for the treatment of hospitalized adult patients in Taiwan with severe staphylococcal infections. J Microbiol Immunol Infect. 2012;45:52–57. doi: 10.1016/j.jmii.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Sawai T., Tomono K., Yanagihara K., Yamamoto Y., Kaku M., Hirakata Y. Role of coagulase in a murine model of hematogenous pulmonary infection by intravenous injection of Staphylococcus aureus enmeshed in agar beads. Infect Immun. 1997;65:466–471. doi: 10.1128/iai.65.2.466-471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kihara R., Yanagihara K., Morinaga Y., Araki N., Nakamura S., Seki M. Potency of SMP-601, a novel carbapenem, in hematogenous murine bronchopneumonia caused by methicillin-resistant and vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:2163–2168. doi: 10.1128/AAC.01532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada Y., Yanagihara K., Yamada K., Migiyama Y., Nagaoka K., Morinaga Y. In vivo efficacy of daptomycin against methicillin-resistant Staphylococcus aureus in a mouse model of hematogenous pulmonary infection. Antimicrob Agents Chemother. 2013;57:2841–2844. doi: 10.1128/AAC.02331-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung H.J., Reiner T., Budin G., Min C., Liong M., Issadore D. Ubiquitous detection of Gram-positive bacteria with bioorthogonal magnetofluorescent nanoparticles. ACS Nano. 2011;5:8834–8841. doi: 10.1021/nn2029692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorian V., Zak O., Suter J., Bruecher C. Staphylococci, in vitro and in vivo. Diagn Microbiol Infect Dis. 1985;3:433–444. doi: 10.1016/0732-8893(85)90082-3. [DOI] [PubMed] [Google Scholar]
- 21.Orson F.M., Kinsey B.M., Hua P.J., Bhogal B.S., Densmore C.L., Barry M.A. Genetic immunization with lung-targeting macroaggregated polyethyleneimine-albumin conjugates elicits combined systemic and mucosal immune responses. J Immunol. 2000;164:6313–6321. doi: 10.4049/jimmunol.164.12.6313. [DOI] [PubMed] [Google Scholar]
- 22.Huang X.L., Wu J., Su T.T., Li Y.H., Chen Z.B., Li C. Long-circulating liposomal daptomycin enhances protection against systemic methicillin-resistant Staphylococcus aureus infection with improved therapeutic potential. Acta Pharm Sin. 2014;49:701–710. [PubMed] [Google Scholar]