Abstract
Background
Tumour necrosis factor (TNF) superfamily cytokines and their receptors regulate diverse immune system functions through a common set of signalling pathways. Genetic variants in and expression of individual TNF superfamily cytokines, receptors and signalling proteins have been associated with autoimmune and inflammatory diseases, but their interconnected biology has been largely unexplored.
Methods
We took a hypothesis-driven approach using available genome-wide datasets to identify genetic variants regulating gene expression in the TNF superfamily cytokine signalling network and the association of these variants with autoimmune and autoinflammatory disease. Using paired gene expression and genetic data, we identified genetic variants associated with gene expression, expression quantitative trait loci (eQTLs), in four peripheral blood cell subsets. We then examined whether eQTLs were dependent on gene expression level or the presence of active enhancer chromatin marks. Using these eQTLs as genetic markers of the TNF superfamily signalling network, we performed targeted gene set association analysis in eight autoimmune and autoinflammatory disease genome-wide association studies.
Results
Comparison of TNF superfamily network gene expression and regulatory variants across four leucocyte subsets revealed patterns that differed between cell types. eQTLs for genes in this network were not dependent on absolute gene expression levels and were not enriched for chromatin marks of active enhancers. By examining autoimmune disease risk variants among our eQTLs, we found that risk alleles can be associated with either increased or decreased expression of co-stimulatory TNF superfamily cytokines, receptors or downstream signalling molecules. Gene set disease association analysis revealed that eQTLs for genes in the TNF superfamily pathway were associated with six of the eight autoimmune and autoinflammatory diseases examined, demonstrating associations beyond single genome-wide significant hits.
Conclusions
This systematic analysis of the influence of regulatory genetic variants in the TNF superfamily network reveals widespread and diverse roles for these cytokines in susceptibility to a number of immune-mediated diseases.
Electronic supplementary material
The online version of this article (doi:10.1186/s13073-016-0329-5) contains supplementary material, which is available to authorized users.
Keywords: TNF superfamily, Autoimmunity, Autoinflammation, Genomics, eQTL, Gene set analysis, GWAS, Genetics
Background
The tumour necrosis factor (TNF) cytokine and receptor superfamilies are composed of 18 ligands (TNFSF) and 29 receptors (TNFRSF), respectively, that share both structural and signalling characteristics [1–3]. Members of these superfamilies modulate immunological responses via co-stimulation, maturation and cell death signalling pathways. In addition, they play important roles in bone homeostasis, eccrine gland development and the nervous system. The expression of TNFSF and TNFRSF molecules is often limited to particular cell types and modulated by the maturation or activation status of these cells [4]. TNFRSF ligation has three broad consequences: cellular activation via TRAF family proteins leading to NF-κB and MAP kinase activity; caspase-dependent death via FADD and caspase-8; and caspase-independent necroptosis mediated by RIP1 and RIP3 kinases. Certain TNFRSF members recruit the adaptor protein TRADD, which subsequently signals through either TRAFs or FADD, depending on cellular context. In addition, decoy receptors within the superfamily can inhibit TNFSF signalling by binding specific ligands without initiating downstream signalling. Recent research has begun to shed light on additional complexities of these signalling pathways. For example, the molecules cFLIP, RIP3 and caspase-8 co-ordinately drive TNF signalling through TRADD to initiate survival, apoptotic or necroptotic pathways depending on their relative concentrations and/or activities [5].
Genetic studies have implicated TNFSF and TNFRSF members in immune-mediated diseases. Mendelian syndromes such as autoimmune lymphoproliferative syndrome (ALPS) and TNF receptor associated periodic syndrome (TRAPS) are caused by mutations in FAS (or other members of the FAS signalling pathway) and TNFRSF1A, respectively [6–8]. The mechanisms by which missense mutations drive these two syndromes differ: heterozygous dominant negative FAS mutations lead to defective signalling in ALPS patients [6], while heterozygous TNFRSF1A mutations in TRAPS patients result in endoplasmic reticulum retention of mutant proteins and exacerbated inflammatory signalling [9]. Mutations in TNFRSF members can also lead to common variable immunodeficiency (CVID): approximately 9 % of patients carry one or two variant alleles of TNFRSF13B (encoding TACI) [10] and a few patients carry biallelic mutations of TNFRSF13C (encoding BAFF-R) [11]. Although CVID is by definition an immunodeficiency, many CVID patients suffer from autoimmune diseases [12]. For example, heterozygous carriers of TNFRSF13B mutations are susceptible to autoimmunity via the failure of central tolerance to select against autoreactive B cells [13]. Genome-wide association studies (GWASs) of common autoimmune and autoinflammatory diseases have identified associations with single nucleotide polymorphisms (SNPs) near a quarter of the 88 autosomal genes encoding TNFSF cytokines, their receptors and downstream signalling molecules [14] (Additional files 1, 2 and 3). Many genetic variants in the TNFSF network are associated with multiple diseases and many diseases are associated with multiple variants in TNFSF network genes. Whether the same genetic variant truly underlies different diseases is likely to remain ambiguous until the causal variants are fine-mapped [15–17].
Increased expression of TNFSF and TNFRSF members has been observed in the serum and/or at the site of inflammation in patients with immune-mediated disease, including rheumatoid arthritis (RA) [18–20], inflammatory bowel disease (IBD) [21–25] and systemic lupus erythematosus (SLE) [26–28]. In addition, mouse models of both autoimmune disease and allergic asthma can be ameliorated by genetic or therapeutic blockade of numerous TNFRSF signalling pathways [29]. TNFSF pathogenicity in these diseases is further corroborated by the success of therapeutically targeting TNF [30] and TNFSF13B (BAFF) [31], as well as on-going development of therapeutics against additional family members [32]. Given that the majority of disease-associated genetic variants in TNFSF-related genes are non-coding and that expression of many of these genes is dysregulated in the same diseases, the question arises as to whether genetic variants directly drive pathogenic expression changes. Recent genome-wide expression quantitative trait loci (eQTL) studies have uncovered disease-associated SNPs that may regulate expression of nearby TNFSF and TNFRSF members in several primary leucocyte subsets [33–40]. In-depth studies of specific polymorphisms have revealed direct consequences on gene expression and occasionally downstream phenotype for disease-associated variants located near TNFSF4 [41, 42], TNFRSF1A [43], TNFSF14 [44], CD40 [45], TNFRSF6B [46] and TNFSF15 [47–50]. However, most of these studies focus on a single leucocyte subset or whole blood measurements.
Here we took a hypothesis-driven approach to investigate how genetic variants that regulate genes encoding TNFSF and TNFRSF members, as well as key downstream signalling molecules, influence disease susceptibility. Our workflow is depicted in Fig. 1. We examined regulation of these genes across peripheral blood leucocyte subsets by mapping eQTLs. Using these eQTL SNPs as genetic markers of TNFSF-related genes, we performed gene set association analysis with autoimmune and autoinflammatory diseases. This revealed widespread association with the TNFSF gene network.
Fig. 1.
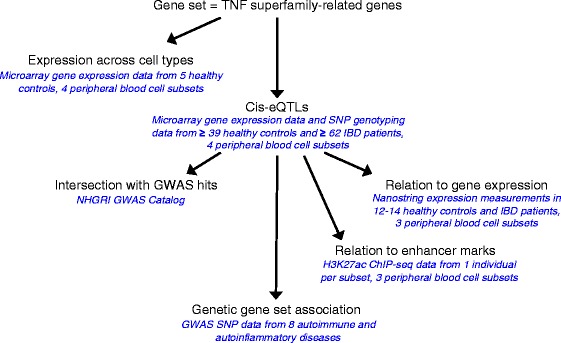
Flow chart of analyses. Flow chart demonstrates how results from each analysis feed into the next. Datasets analysed are listed in blue italics
Methods
GWAS Catalog search for TNFSF-related genes and intersection with eQTLs
Processing and analysis of data from the NHGRI GWAS Catalog [14] is described in Additional file 4: Supplemental Methods.
Sorting peripheral blood subsets from individuals
Whole blood collection for this study was approved by the Cambridgeshire 3 Research Ethics Committee (08/H0306/21). Written informed consent was obtained from all participants. Whole blood from healthy controls and individuals with newly diagnosed, flaring Crohn’s disease (CD) or ulcerative colitis (UC) was separated into peripheral blood leucocyte subsets by magnetic bead-based positive selection [51, 52] as described in Additional file 4: Supplemental Methods. CD4+ T cells, CD8+ T cells, CD14+ monocytes and CD16+ neutrophils were used in this study, a subset of the samples described in [53].
Gene expression measurements and data processing
RNA (200 ng) from each sample was prepared for Human Gene 1.1 ST 96-Array (Affymetrix) using the Ambion WT Expression Kit and GeneChip WT Terminal Labeling and Controls Kit (Affymetrix). These samples were run in batches of 96 on a Gene Titan Multi-Channel (MC) Instrument (Affymetrix). Gene expression data are available through ArrayExpress, accession numbers E-MTAB-3554 ([53] eQTL analysis) and E-MTAB-4887 (comparison across leucocyte subsets in healthy controls). Processing of gene expression data is described in Additional file 4: Supplemental Methods.
Genotyping for eQTL analysis
DNA samples were extracted with the Qiagen All-Prep DNA/RNA Mini kit from peripheral blood cells. DNA was genotyped on the Illumina Beadchip HumanOmniExpress-12v1 platform at the Wellcome Trust Sanger Institute in two batches. These data have been deposited in the European Genome-phenome Archive (EGA; accession number EGAS00001001251 [53]) and are available on request. Genotype calls were made using GenoSNP software. Processing of genotype data is described in Additional file 4: Supplemental Methods.
eQTL analysis
Details of samples used in eQTL analyses are tabulated in Additional file 5. Cis-eQTL mapping to autosomal TNFSF-related genes was carried out in each cell type separately using the All.cis function of the GGtools Bioconductor package [54]. This method fits a generalised linear model with expression as the dependent variable and then performs score tests (one degree of freedom asymptotic chi-squared tests) for the addition of genotype to the model. P values were calculated in a one-tailed test from the chi-squared scores. Probe set location annotation was based on Ensembl release 71 and SNPs were annotated with SNPlocs.Hsapiens.dbSNP.20120608 [55]. SNPs were filtered for minor allele frequency above 5 %. False discovery rate (FDR) was estimated by sample label permutation [56] with a threshold of 10 % applied for significance. Because our ultimate goal in this re-analysis study was to perform gene set disease association analysis, we wished to uncover additional eQTLs to tag our TNFSF-related genes. We felt that a 10 % FDR threshold provided a reasonable compromise between maximising eQTL discovery and minimising false positives. We found 320 permutations to be more than sufficient to achieve FDR stability (Additional file 4: Figure S1a). Regulatory elements have been shown to primarily reside within 50 kbp of the transcription start and end sites of a gene [57]. By varying the radius from 50 kbp to 1 Mbp while controlling the FDR at 10 %, we found that the number of eQTL discoveries increased from 50 to 100 kbp and then declined at larger radii tested (Additional file 4: Figure S1b). Thus, for each gene, SNPs within the region from 100 kbp upstream to 100 kbp downstream of the gene were designated as cis. For each cis-eQTL in the combined IBD and healthy control cohort, we also performed linear regression of expression on SNP genotype in patients and controls separately. We then plotted the coefficients of the genotype term for each eQTL in IBD patients versus healthy controls to compare effect sizes and directions. See Additional file 4: Supplemental Methods for variable selection at loci with multiple significant cis-eQTL SNPs.
Nanostring nCounter measurements and data processing
RNA was previously measured by the nCounter Analysis System (Nanostring Technologies) [58]. See Additional file 4: Supplemental Methods for sample, measurement, and normalisation details.
Intersection of H3K27ac chromatin immunoprecipitation sequencing data with eQTLs
H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) and ChIP input DNA sequencing .bed files for CD4+ T cells, CD8+ T cells and CD14+ monocytes were obtained from the NIH Roadmap Epigenomics Project datasets [59] available through the Gene Expression Omnibus (GEO) database [60]: H3K27ac CD4+ CD25− primary cells, GSM997239; input CD4+ CD25− primary cells, GSM1112781; H3K27ac CD8 primary cells, GSM1102781; input CD8 primary cells, GSM1102806; H3K27ac CD14 primary cells, GSM1102782; input CD14 primary cells, GSM1102807. The available H3K27ac immunoprecipitated and input DNA sequencing data from CD4+ CD25− T cells were not from the same sample. Processing and analysis of H3K27ac ChIP-seq data are described in Additional file 4: Supplemental Methods.
Hypothesis-driven genetic gene set analyses in previous GWAS datasets
Genetic studies typically examine association of individual SNPs with disease. This approach fails to exploit functional relationships between SNPs affecting the same gene or biological pathway. To address this, we performed gene set association analysis using data from previous GWASs. Previous GWAS datasets are detailed in Additional file 6. Processing of these genetic data is described in Additional file 4: Supplemental Methods. SNPs to represent the TNFSF-related gene set were chosen as follows. In each cell subset, we selected SNPs in linkage disequilibrium (LD) r2 ≥ 0.8 with the strongest significant cis-eQTL SNP for each TNFSF-related gene. This was performed using 1000 Genomes Phase 1 EUR population vcf files [61] in PLINK [62, 63]. In each GWAS dataset, we then extracted all of these SNPs that were present on the SNP chip used. Next, we filtered the SNPs for relative independence (multiple correlation coefficient ≤ 0.33) to make our SNP set representative of TNFSF-related genes, referred to hereafter as the TNFSF eQTL SNP set. In each GWAS dataset, hypothesis-driven gene set association analysis was based on that of Sun et al. [64] as follows. We calculated chi-squared allelic case-control association statistics and inflation factor (λ) for the TNFSF eQTL SNP set. The same independence filtering and association testing was then performed genome-wide. Qq-plots were compared between TNFSF eQTL SNPs and SNPs genome-wide. λ1000 values were calculated by rescaling λ for 1000 cases and 1000 controls:
To calculate a self-contained gene set association statistic [65, 66], phenotypes were permuted 10,000 times and chi-squared disease association statistics were calculated in each permuted dataset for the TNFSF eQTL SNP set. We then summed chi-squared scores across SNPs in the original data and in each permuted dataset. Empirical p values were calculated as the fraction of summed scores from permuted datasets that were greater than that from the original data. A similar procedure was followed to estimate gene-level disease association. For each gene, the sum of chi-squared statistics was compared with the sum of chi-squared statistics in the permuted data to obtain an empirical p value. Gene-level p values were then adjusted for the multiple genes tested using the Benjamini–Hochberg method. Comparison of gene length with disease association p values revealed negligible impact of gene length on association statistics (Additional file 4: Figure S2).
Results
TNF superfamily-related genes are differentially regulated among leucocyte subsets
We curated genes that encode members of the TNFSF and TNFRSF and their downstream signalling molecules from the literature (Additional file 2) and examined their expression in CD4+ T cells, CD8+ T cells, CD14+ monocytes and CD16+ neutrophils sorted from peripheral blood from five healthy individuals (Fig. 2). The signalling molecules downstream of superfamily receptors were generally expressed more broadly across cell subsets than TNFSF ligands or TNFRSF receptors. Hierarchical clustering of gene expression levels across cell types revealed cell type-specific expression, separating the lymphoid (CD4+ and CD8+ T cells) and myeloid (monocytes and neutrophils) lineages and clearly distinguishing monocytes from neutrophils. Such cell type clustering occurred even when only TNFSF, TNFRSF or signalling molecules were considered (Additional file 4: Figure S3).
Fig. 2.
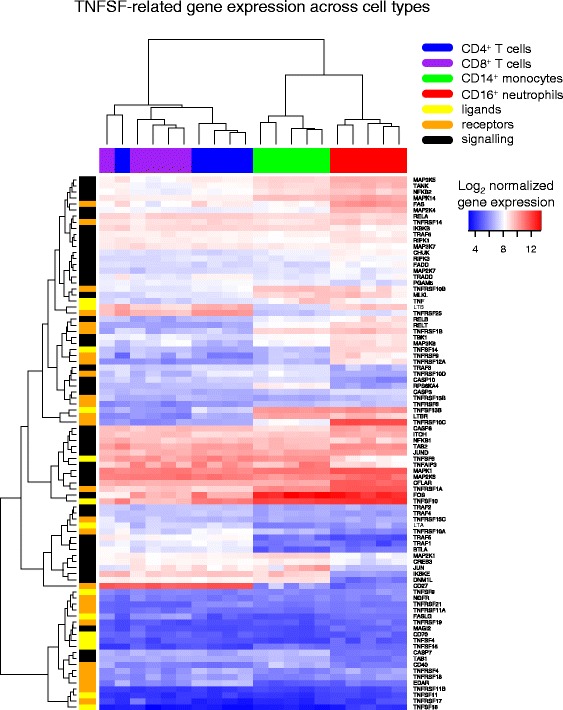
Expression of TNFSF-related genes differs across leukocyte subsets. Expression of TNFSF-related genes was measured across four cell subsets from five healthy controls by microarray. Expression values are hierarchically clustered. Cell types are coloured blue (CD4+ T cells), purple (CD8+ T cells), green (CD14+ monocytes) and red (CD16+ neutrophils). Genes are grouped by function and coloured yellow (TNFSF member ligands), orange (TNFRSF member receptors) and black (adaptors and signalling molecules in TNFSF signalling network)
To examine the relationship between genetic variation and expression of these TNFSF-related genes, we performed targeted cis-eQTL mapping in a previously analysed cohort of combined healthy controls and individuals with newly diagnosed IBD [53]. Although eQTLs have been mapped for TNFSF-related genes in genome-wide studies, our targeted approach reduced the multiple testing burden and thereby found significant associations for additional regulatory variants. We mapped eQTLs in CD4+ T cells, CD8+ T cells, CD14+ monocytes and CD16+ neutrophils, accounting for potential confounders including disease status, gender and age (Additional file 4: Supplemental Methods). At a 10 % FDR threshold, we identified 51 genes with a cis-eQTL in at least one cell type (Additional file 7). eQTLs have been mapped in CD4+ T cells, monocytes and neutrophils in other cohorts [34, 35, 37–40]. Of the genes we identified with eQTLs in these subsets, 56 % were previously reported in at least one of these other cohorts (Additional file 7). eQTL effects on gene expression were found to be generally concordant between IBD patients and healthy controls (Additional file 4: Figure S4). Only six genes, such as the apoptosis inducer FAS (also known as CD95; Fig. 3a), had detectable eQTLs across all cell types examined (Additional file 4: Figure S5). Four of these six genes function in the apoptotic pathway by which FAS signals through FADD to activate caspase-induced death and one (MLKL) is integral to the necroptotic pathway [67]. TNFSF and TNFRSF genes often appear in clusters throughout the genome due to their origin via gene duplication [68] and therefore many share cis elements. Intriguingly, we identified one SNP for which the minor allele was associated with increased neutrophil expression of the TRAIL receptor TNFRSF10B (DR5) and decreased monocyte expression of the decoy TRAIL receptor TNFRSF10C (DcR1) approximately 34 kbp away (Fig. 3b).
Fig. 3.
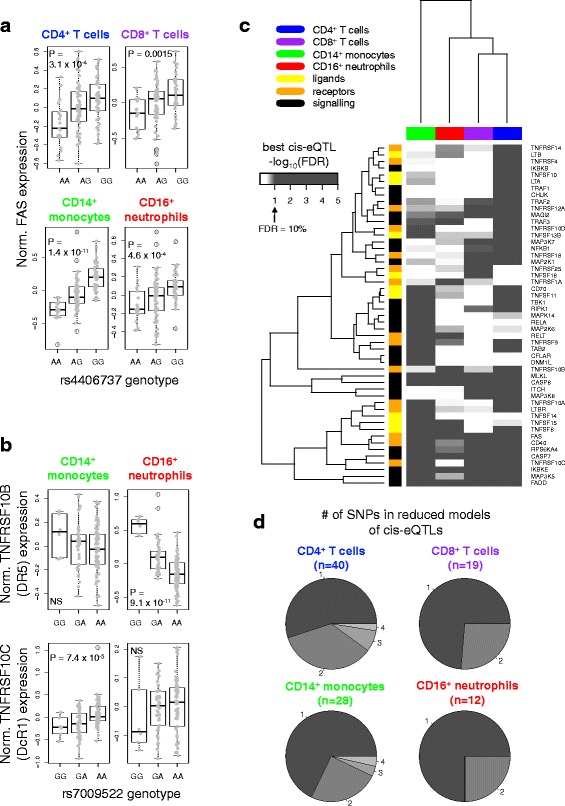
TNFSF-related genes are under extensive genetic regulation. a Normalised FAS expression in each cell subset is plotted against rs4406737 genotype. Association p values are indicated for eQTLs with FDR < 0.1. b Normalised TNFRSF10B and TNFRSF10C expression in monocytes and neutrophils is plotted against rs7009522 genotype. NS not significant. c TNFSF-related genes with a significant cis-eQTL (FDR < 0.1) in any cell type were extracted. For each gene, the SNP most significantly associated with expression in each cell type was extracted (best cis-eQTL) and the FDR corresponding to its p value in that dataset calculated. The plot depicts hierarchical clustering of the negative logarithm of these FDRs. Colours are as in Fig. 2. d The number of SNPs in cis-eQTL models after variable selection. Numbers around the periphery and grey shades indicate the number of eQTL SNPs remaining as predictors in the model for each gene. “n” indicates the number of genes with cis-eQTLs represented by each pie chart
Use of a fixed FDR threshold can lead to underestimation of the extent of eQTL sharing across tissues due to varying power from different sample sizes and effect sizes, as well as random sampling error. A heatmap of the strongest eQTL SNP for each gene in each cell type revealed a greater level of common regulation than evidenced by a rigid FDR threshold (Fig. 3c; Additional file 4: Figure S5). Some genes, such as TNFSF14 (LIGHT), exhibited strong subset-specific regulation, while others, such as its receptor TNFRSF14 (HVEM), met our significance threshold in only one subset but showed a trend toward association in other cell types. Clustering these test statistics revealed that the greatest similarity between cell subsets was among T cells, similar to observations at the expression level.
To better understand the complexity of cis genetic control over gene expression in TNFSF-related genes, we examined each gene in each cell type with more than one significant cis-eQTL SNP. By fitting a linear model with all significant eQTL SNPs for the gene as predictors, we performed exhaustive variable selection to find the most informative set of genetic predictors (Fig. 3d; Additional file 8). Most cis-eQTLs could be attributed to a single SNP, while some could be explained by up to four contributing SNPs. Genetic fine-mapping at these loci could clarify whether these multiple contributing SNPs were truly independent or participating in mutual tagging of an un-typed causal variant. Cell types with a greater number of genes with cis-eQTLs also exhibited greater tendency to have complex, multi-SNP cis-eQTLs.
eQTLs for TNFSF-related genes are not associated with average gene expression level or enhancer marks
In microarray measurements, probe effects can hinder comparisons of expression levels between genes and saturation and noise can impact measurements at the extremes [69, 70]. To understand how average gene expression related to eQTL detection, we utilised expression measurements of TNFSF and TNFRSF members acquired by the Nanostring nCounter technology, which provides a count-based measurement without nucleotide amplification steps. These data encompassed three of the four cell types investigated in a similar cohort of healthy controls and IBD patients [58] (Fig. 4a). Discovery of an eQTL for a gene in a given cell type was not related to its average expression in relation to other genes or other cell types. Presence or absence of eQTLs may instead be regulated by other factors such as transcription factor expression or chromatin state.
Fig. 4.
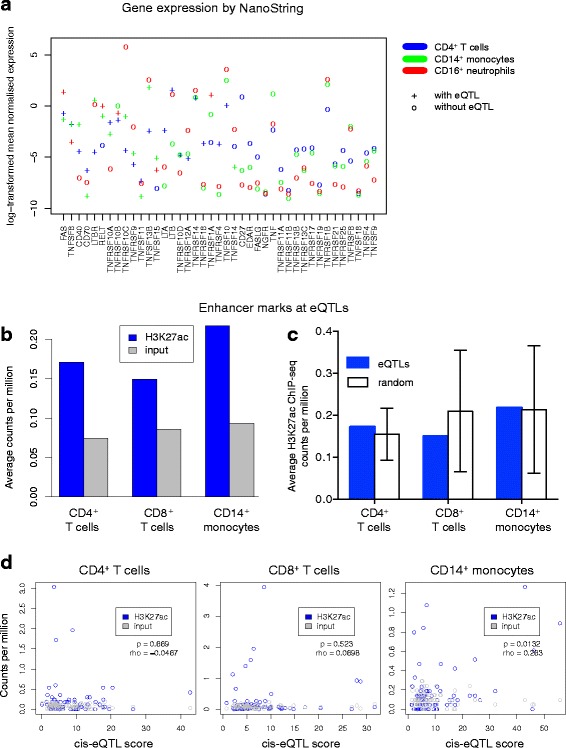
eQTLs are independent of total magnitude of gene expression and not preferentially associated with active enhancer marks. a Expression of TNFSF and TNFRSF members was measured by the NanoString nCounter analysis system. Each point represents average expression over eight or more individuals, including healthy controls and individuals with IBD. Genes are listed left to right in order of decreasing number of cell types in which an eQTL was detected. b Average H3K27ac ChIP-seq or input DNA sequencing counts per million intersecting TNFSF-related eQTL SNPs in the same cell type are plotted. c Average H3K27ac ChIP-seq counts per million intersecting eQTL SNPs in the same cell type are compared with a random distribution of intersections created by selecting the same number of SNPs from the cis genomic regions used for eQTL search. Error bars represent the standard deviation of 10,000 iterations of random selection. d eQTL chi-squared scores from the strongest eQTL SNP for each TNFSF-related gene (regardless of whether the association passed our eQTL significance threshold) are compared with H3K27ac ChIP-seq or input DNA sequencing counts per million at the same SNPs. Spearman correlation coefficient (rho) and correlation p values (p) are indicated for H3K27ac ChIP-seq counts per million versus eQTL score
To investigate the possibility that eQTLs were associated with chromatin marks of enhancers, we used primary leucocyte data from the NIH Roadmap Epigenomics Project. Acetylation of histone 3, lysine 27 (H3K27ac) has been shown to delineate active enhancers [71]. Extracting the most significantly associated eQTL SNP for each gene in each cell type with a cis-eQTL (FDR < 0.1), we examined these loci in H3K27ac ChIP-seq data from primary CD4+ T cells, CD8+ T cells and CD14+ monocytes. In all cell types, eQTLs were, on average, enriched by H3K27ac immunoprecipitation compared with input control DNA (Fig. 4b; Additional file 4: Figure S6a). However, randomly sampled SNPs from the same cis regions around TNFSF-related genes showed acetylation similar to that of eQTL SNPs (Fig. 4c), suggesting that H3K27ac marks are not specific for eQTLs but rather are characteristic of genic regions. To control for the fact that we did not have fine-mapped eQTLs, we repeated this comparison to include all SNPs tagged (LD r2 ≥ 0.8) by our eQTL SNPs (Additional file 4: Figure S6b) and again found no difference in acetylation compared with a random selection of SNPs from the same genic regions. To examine whether eQTL strength correlated with acetylation level, the most significant eQTL SNP for each gene (regardless of whether the eQTL passed our FDR threshold) was extracted. The eQTL chi-squared association statistics were then plotted against acetylation at the same loci (Fig. 4d). In monocytes, but not in other cell types, we found correlation between eQTL strength and H3K27ac enrichment. Indeed, relatively few significant eQTLs were strongly acetylated in the cell type of their discovery, though many monocyte eQTLs did exhibit greater acetylation in monocytes than in other cell types (Additional file 4: Figure S6c, d). Together, these data demonstrate that, within our gene set, eQTLs are not enriched for active enhancer marks over other SNPs within the genic regions.
TNFSF-related eQTLs are associated with a variety of autoimmune and autoinflammatory diseases
Loci near 24 % of autosomal TNFSF-related genes have been associated with autoimmune and inflammatory diseases by GWAS (Additional file 3, “Mapped Genes” column), resulting in highly significant enrichment of these gene loci with autoimmune diseases (Fisher’s exact test p = 1.4 × 10−10). However, these SNPs are attributed to genes by physical proximity, not by evidence of functional relationship. To understand the role of SNPs that affect gene expression in immune-mediated disease susceptibility, we searched for TNFSF-related gene eQTLs among autoimmune and autoinflammatory disease-associated SNPs in a comprehensive database of previous GWASs (the NHGRI GWAS Catalog; Additional file 9). We found that approximately equal numbers of disease risk alleles were associated with increased and decreased gene expression (Fig. 5a). For example, the multiple sclerosis (MS) protective allele near the co-stimulatory ligand LIGHT (encoded by TNFSF14) was associated with increased expression of this molecule in monocytes (Fig. 5b). Examining the cell types in which these GWAS SNPs were eQTLs revealed a variety of effects across diseases and cell types, potentially suggesting protective effects of TNFSF-related genes in myeloid cells but disease risk effects of members of this signalling network in T cells (Fig. 5c). Such cellular diversity emphasises the distinct influences of TNFSF-related gene variants in autoimmune and autoinflammatory disease onset. A caveat to this analysis is that because many of the GWASs in the GWAS Catalog are not fine-mapped, we cannot confirm whether eQTL and disease association signals at the same locus are due to the same causal variant.
Fig. 5.
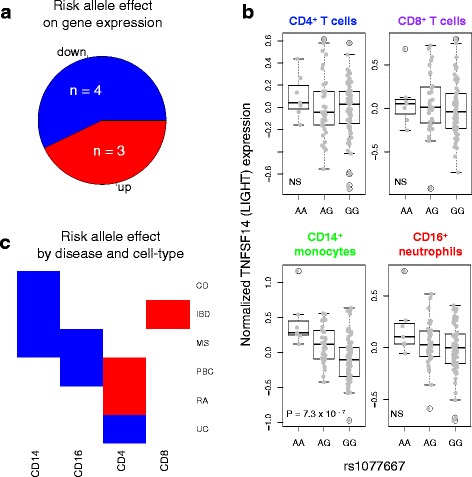
Immune-mediated disease risk alleles can either increase or decrease TNFSF-related gene expression. a Autoimmune and autoinflammatory disease GWAS hits tagged by TNFSF-related cis-eQTLs were identified and the directions of effect of the risk alleles on expression are plotted. Disease-associated SNPs that are eQTL SNPs in multiple cell types or are associated with multiple diseases are counted only once. “n” indicates the number of SNPs in each slice of the pie. b TNFSF14 expression is plotted against MS-associated SNP rs1077667 genotype. Allele A is protective. P values are provided for eQTLs with FDR < 0.1; NS indicates not significant. c Disease-associated eQTL SNPs depicted in a are plotted by eQTL cell type and disease association. Effect directions are coloured as in a. SNPs associated with more than one disease are plotted once per disease
Given the immunological roles and interconnected nature of the TNFSF-related gene network, we wished to examine genetic association of the whole gene set with autoimmune and autoinflammatory disease. Assignment of eQTL SNPs to genes for gene set analysis has previously been proposed to ensure functional relevance of variants used in gene set testing of genetic data [72]. Using this strategy, we re-analysed available GWAS data from eight autoimmune and autoinflammatory diseases for association with TNFSF-related genes. A TNFSF eQTL SNP set was created by combining the strongest significant eQTL SNP for each TNFSF-related gene in each cell type and then filtering these SNPs for relative independence. LTA, TNF and LTB are located within the major histocompatibility complex (MHC) and SNPs near these genes might, therefore, appear disease-associated due to LD with MHC variants that are strongly associated with disease. TNF and LTB lacked eQTLs and were therefore not included in the TNFSF eQTL SNP set. Disease association with the LTA eQTL SNP could not be proven independent of the strong MHC association with many diseases and thus the LTA eQTL was excluded from the set. The inflation factor (λ) for disease association with the TNFSF eQTL SNP set was computed and compared with that from genome-wide SNPs thinned for LD (Table 1; Additional file 4: Figure S7). This analysis revealed inflated λ values for TNFSF eQTL SNP set association with six of the eight diseases, in particular CD and Behçet’s disease (BEH), but not type 1 diabetes (T1D) or anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV).
Table 1.
Autoimmune and autoinflammatory diseases show widespread association with functional variants of the TNFSF network
| Disease | λ whole genomea | λ eQTL SNP setb | eQTL SNP set association p valuec |
|---|---|---|---|
| Behçet’s disease (BEH) | 1.05 | 2.48 | 0.0002 |
| Crohn’s disease (CD) | 1.04 | 2.39 | 0.0003 |
| Multiple sclerosis (MS) | 1.06 | 1.59 | <0.0001 |
| Primary biliary cirrhosis (PBC) | 1.06 | 1.49 | <0.0001 |
| Rheumatoid arthritis (RA) | 1.02 | 1.48 | 0.003 |
| Ulcerative colitis (UC) | 1.04 | 1.44 | 0.0021 |
| ANCA-associated vasculitis (AAV) | 1.11 | 1.25 | 0.17 |
| Type 1 diabetes (T1D) | 1.04 | 1.00 | 0.77 |
a λ values were calculated for the whole genome filtered for relative SNP independence
b λ values were calculated for the TNFSF eQTL SNP set defined as follows: for each TNFSF-related gene in each cell subset with a significant cis-eQTL (FDR < 0.1), the strongest eQTL SNP was identified; SNPs with LD r2 ≥ 0.8 with these eQTL SNPs were then extracted from the GWAS dataset and filtered for relative independence
c Permutation-based p values for TNFSF eQTL SNP set association with disease were calculated from the same set of SNPs as in the TNFSF eQTL SNP setb
To test total association of TNFSF-related genes with disease, we employed a self-contained method for gene set disease association analysis. Self-contained analyses compare gene set association with a simulated null distribution of no association. This answers the question of whether the gene set is associated with disease, without requiring competitive comparisons with other gene sets that we cannot a priori assume are not associated with disease. To this end, we combined disease association test statistics across the TNFSF eQTL SNP set and used phenotype permutation to simulate a null distribution, modelling our method after that of Sun et al. [64]. Total disease association of a SNP set can be driven by either a single, highly associated variant or multiple, less associated variants. This analysis found significant gene set association with the same six diseases as the inflation factor comparison, particularly with primary biliary cirrhosis (PBC), MS, CD and BEH (column 3 in Table 1; Additional file 4: Figure S8).
To reveal whether gene set association results were driven by eQTLs for particular genes or a more general inflation of test statistics for these variants, we examined disease association of eQTL SNPs at the gene level by the same method (Fig. 6). After correcting for the number of genes tested, most TNFSF-related genes were not found to be significantly associated with disease (Fig. 6; blue shades indicate Benjamini–Hochberg FDR > 0.1). Most disease-associated genes were unique to individual diseases but associations with eQTL SNPs for TNFRSF1A, TNFSF15 and RPS6KA4 were shared across two or three diseases, similar to results observed by Parkes et al. [73] in a comparison of genome-wide significant loci across diseases. A striking cluster of genes was associated with PBC, including LTBR (TNFR3), TNFRSF1A (TNFR1), NFKB1 (the p50 subunit of the classical NF-κB transcription factor complex), CHUK (NF-κB inhibitor kinase α) and IKBKB (NF-κB inhibitor kinase β), that all play roles in TNF and lymphotoxin alpha (LTA) signalling.
Fig. 6.

Genes regulated by disease-associated eQTL SNPs differ across diseases. Within each disease, permutation-based p values for gene-level disease association were calculated by combining the strongest significant cis-eQTL SNPs in each cell type. As in Table 1, proxy eQTL SNPs in each genetic dataset were filtered for relative independence before computation of a permutation-based disease association p value. P values were corrected within each disease by the Benjamini–Hochberg FDR method. The heatmap represents the negative logarithm of these corrected values such that genes marked in white-to-red shades show disease association, FDR < 0.1. Grey indicates no data available because the GWAS dataset did not include SNPs that tagged eQTLs for this gene with LD r2 ≥ 0.8. Gene colours on the left side correspond to TNFSF ligands (yellow), TNFRSF receptors (orange) and adaptors/signalling molecules (black) as in Fig. 2. Genes are presented in order of decreasing association with any disease
Discussion
We have investigated the contribution of genetic variation in TNFSF-related genes to gene expression and susceptibility to autoimmune and autoinflammatory disease. Previous studies have examined the intersection of a similar variety of genomic data [16] but our approach is unique in its hypothesis-driven investigation of a biological pathway. Targeted eQTL analysis revealed extensive and variable genetic regulation. Shared eQTLs across cell types in the FAS-mediated apoptosis and necroptosis pathways suggest universality of genetic regulation of programmed cell death responses. In the case of TRAIL receptor expression, opposing regulation of competing signalling and decoy receptors by the same eQTL implies enhanced upregulation of this TRAIL-induced death pathway in individuals carrying the minor allele. eQTL detection was not dependent on gene expression levels and eQTL SNPs were not preferentially marked by H3K27ac chromatin modifications. It is possible that, under conditions of acute cellular activation, a different relationship between eQTLs and enhancer loci might emerge.
We studied genes in the TNFSF, their receptors and associated signalling proteins based on the strong involvement of genes in this cytokine superfamily with inflammatory processes and their sharing of downstream signalling pathways. The downstream molecules in TNFRSF signal transduction can play roles in additional pathways, such as pattern recognition receptor signalling [74], and thus association of these genes with autoimmune disease may also reflect involvement of additional pathways not addressed in this study. By using eQTL SNPs as previously suggested by Zhong et al. [72], we ensured that the variants defining our gene set were functionally relevant. Through empirical examination of disease association inflation factors and application of a phenotype-permutation-based test for significance, we were able to demonstrate association of TNFSF-related genes with CD, BEH, PBC, MS, UC and RA, often beyond that of known GWAS loci, but we did not find association with AAV or T1D.
The fact that the TNFSF eQTL SNP set exhibited elevated λ in diseases associated by GWASs with only one or two of these variants (Additional file 8) suggests that additional TNFSF-related eQTLs may have subtle effects on disease susceptibility and lead to the observed cumulative disease association. None of the TNFSF eQTL SNPs have been found to be associated with BEH by GWASs, but their cumulative association is highly significant compared with our permuted null dataset. This suggests that insufficient power may have prevented individual variants from reaching genome-wide significance in the original study that we re-analysed. Such associations might reach genome-wide significance if larger cohorts become available. Association of the TNFSF eQTL SNP set with BEH predicts that expression of TNFSF cytokines, receptors and signalling molecules contributes to the pathogenesis of this disease more than has been previously appreciated. Indeed, rare variants in the NF-κB inhibitor TNFAIP3 (A20) have recently been associated with a BEH-like familial disease [75] and variants contributing to dysregulation of NF-κB signalling may thus also contribute to the more common form of BEH.
GWASs have identified associations between PBC and genomic loci near LTBR/TNFRSF1A and NFKB1 (Additional file 3) [76] but the genetic associations with the NF-κB inhibitor kinase subunits CHUK and IKBKB that we describe here have not been previously established. Interestingly, a variant near CHUK has been associated with plasma liver enzyme levels [77] and these two kinases have been found to co-ordinately protect liver bile ducts from inflammatory destruction in mice [78]. Thus, our targeted analysis indicates that such associations may reach genome-wide significance with larger PBC GWASs and further implicates the TNF/LTA pathway in PBC pathogenesis. In fact, evidence of the power of a targeted gene set analysis approach to identify sub-threshold associations is already apparent within this study: eQTL SNPs for TNFSF15 and NFKB1 did not meet genome-wide significance in the CD [79] and UC [80] GWAS datasets used for re-analysis, respectively, but did contribute to the SNP set association (Fig. 6) and were later found to be genome-wide significant in larger studies (Additional file 9). These data provide support for the use of such a targeted approach to highlight potential disease-associated genes and pathways.
Finally, we found that while the TNFSF-related SNP set was associated with six of the eight diseases examined, the genes contributing to association with these six diseases often differed. This variation suggests that while the pathway is relevant in many immune-mediated syndromes, particular branches are more influential in specific diseases, shedding some light on their independent aetiologies.
Conclusions
We performed a targeted analysis of TNFSF cytokines, their receptors and signalling molecules to better understand their regulation and association with autoimmune and autoinflammatory diseases. By mapping eQTLs and using these regulatory variants in GWAS gene set analysis, we demonstrated association of TNFSF-related genes with six of the eight immune-mediated diseases examined. Through this hypothesis-driven approach, we have suggested disease association of this gene set beyond individual variants identified in genome-wide SNP association testing.
Abbreviations
AAV, ANCA-associated vasculitis; ALPS, autoimmune lymphoproliferative syndrome; ANCA, anti-neutrophil cytoplasmic antibody; BEH, Behçet’s disease; CD, Crohn’s disease; ChIP, chromatin immunoprecipitation; CVID, common variable immunodeficiency; EGA, European Genome-phenome Archive; eQTL, expression quantitative trait locus; FDR, false discovery rate; GEO, Gene Expression Omnibus; GWAS, genome-wide association study; H3K27ac, histone 3 lysine 27 acetylation; IBD, inflammatory bowel disease; LD, linkage disequilibrium; LTA, lymphotoxin alpha; MHC, major histocompatibility complex; MS, multiple sclerosis; PBC, primary biliary cirrhosis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; SNP, single nucleotide polymorphism; T1D, type 1 diabetes; TNF, tumour necrosis factor; TNFRSF, TNF receptor superfamily; TNFSF, TNF superfamily; TRAPS, TNF receptor associated periodic syndrome; UC, ulcerative colitis
Acknowledgements
We thank all individuals who contributed samples to this study. We thank Alexander Hatton and Huzefa Ratlamwala for peripheral blood sample processing and RNA extraction and John Ferdinand for critical reading of the manuscript. The nCounter Analysis System was available through the Translational Immunology Section of the Office of Science and Technology in NIAMS. We thank Ivan Ovcharenko at the National Center for Biotechnology Information for assistance with our IRB application to access data in dbGaP.
Funding
This research was supported in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) and the National Library of Medicine (NLM) of the NIH, the Wellcome Trust (080327/Z/06/Z, 087007/Z/08/Z and 094227/Z/10/Z), the Medical Research Council (G0400929) and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. KGCS is an NIHR Senior Investigator. ACR was funded by the NIH-Oxford-Cambridge Scholars Program. JEP and JCL were funded by the Wellcome Trust Clinical PhD Programme. The Cambridge Institute for Medical Research is in receipt of a Wellcome Trust Strategic Award (079895).
H3K27ac ChIP-seq data are available from the NIH Roadmap Epigenomics Project (http://nihroadmap.nih.gov/epigenomics/). The data used for analysis of Behçet’s disease were obtained from dbGaP, Genome-Wide Association Study of Behçet’s Disease in the Turkish Population (dbGaP accession number phs000272) provided by Daniel L. Kastner and Elaine F. Remmers. Funding support for the original study was provided by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the US National Institutes of Health, by the Istanbul University Research Fund, by the UK Behcet’s Syndrome Society and by other sources as detailed in [81]. Institutional review board approval was obtained as part of NIH human subjects protocol 10-HG-N045. AAV GWAS data were provided by the European Vasculitis Genetics Consortium. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk/. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475.
Availability of data and material
Gene expression microarray data for the five healthy control samples used in Fig. 2 are available through ArrayExpress, accession number E-MTAB-4887. Gene expression microarray data for the eQTL study are available through ArrayExpress, accession number E-MTAB-3554. Genotyping data for the eQTL analysis are available on request from EGA, accession number EGAS00001001251. H3K27ac ChIP-seq data are available from the NIH Roadmap Epigenomics Project (http://nihroadmap.nih.gov/epigenomics/) through GEO at the accession numbers listed in Methods. GWAS datasets used in re-analysis are available from dbGaP and EGA as described in “Ethics approval and consent to participate” and tabulated in Additional file 6.
Authors’ contributions
ACR carried out analyses and wrote the manuscript. JEP assisted with eQTL analyses, provided statistical advice and edited the manuscript. JCL recruited and processed patient samples. GV designed and advised enhancer analysis. AAS assisted in dataset acquisition and usage, provided advice for analyses and helped write the manuscript. RMS, PAL and KGCS conceived and supervised the project and edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Whole blood collection for this study was approved by the Cambridgeshire 3 Research Ethics Committee (08/H0306/21). Written informed consent was obtained from all participants. Datasets used in re-analysis of previous GWAS studies are detailed in Additional file 6. Database of Genotypes and Phenotypes (dbGaP) datasets were obtained through the controlled access portal of dbGaP [82], with approval of the National Center for Biotechnology Information (NCBI) Institutional Review Board (IRB). IRB approval was obtained as part of National Institutes of Health (NIH) human subjects protocol 10-HG-N045. EGA datasets were obtained through the controlled access portal of EGA [83], with approval of the appropriate data access committees. The datasets used were as follows: CD (EGA EGAD00000000005), RA (EGA EGAD00000000007) and T1D (EGA EGAD00000000008) [79], UC (EGA EGAD00000000025) [80], MS (EGA EGAD00000000120, UK individuals only) [84], PBC (EGA EGAD00000000056) [85], AAV [86], BEH (dbGaP phs000272.v1.p1) [81] and healthy controls (EGA EGAD00000000001, EGAD00000000002, EGAD00000000021, EGAD00000000022, EGAD00000000023, EGAD00000000024).
Additional files
Autoimmune and autoinflammatory disease terms were curated from the list of disease terms available in the NHGRI GWAS Catalog. (XLSX 51 kb)
Autosomal TNFSF-related genes were identified and probesets extracted from the Hugene1.1St Array. Gene names are from Ensembl release 71. (XLSX 31 kb)
The NHGRI GWAS Catalog was filtered for autoimmune and autoinflammatory disease-associated variants near TNFSF or TNFRSF members or downstream signalling molecules (both “Mapped” and “Reported Gene” categories). (XLSX 16 kb)
This file includes Supplemental Methods and Figures S1–S8 with legends. (PDF 954 kb)
eQTL samples are broken down by cell type and diagnosis. (XLSX 36 kb)
GWAS datasets were used for hypothesis-driven gene set analysis. (XLSX 40 kb)
The best-associated SNP is listed for each cis-eQTL (FDR < 0.1). Studies that have previously identified eQTLs for these genes in the same cell types are tabulated. (XLSX 56 kb)
Exhaustive variable selection to minimise the Bayesian information criterion (BIC) was used to determine the best model of SNPs for each cis-eQTL (FDR < 0.1). (XLSX 33 kb)
GWAS hits tagged by TNFSF-related gene eQTLs (Additional file 7) were examined for risk allele effects. SNPs associated with gene expression in multiple cell types are repeated, one line per cell type. Duplicate associations from different studies were removed in plotting Fig. 5 but are retained here due to different references, p values and odds ratios. (XLSX 55 kb)
Contributor Information
Arianne C. Richard, Email: acr62@cam.ac.uk
James E. Peters, Email: jp549@cam.ac.uk
James C. Lee, Email: jcl65@cam.ac.uk
Golnaz Vahedi, Email: vahedi@mail.med.upenn.edu.
Alejandro A. Schäffer, Email: aschaffe@helix.nih.gov
Richard M. Siegel, Email: SiegelR@mail.nih.gov
Paul A. Lyons, Email: pal34@cam.ac.uk
Kenneth G. C. Smith, Email: kgcs2@cam.ac.uk
References
- 1.Grivennikov SI, Kuprash DV, Liu ZG, Nedospasov SA. Intracellular signals and events activated by cytokines of the tumor necrosis factor superfamily: from simple paradigms to complex mechanisms. Int Rev Cytol. 2006;252:129–61. doi: 10.1016/S0074-7696(06)52002-9. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Yin Q, Wu H. Structural basis of signal transduction in the TNF receptor superfamily. Adv Immunol. 2013;119:135–53. doi: 10.1016/B978-0-12-407707-2.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119(3):651–65. doi: 10.1182/blood-2011-04-325225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol. 2009;9(4):271–85. doi: 10.1038/nri2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberst A, Green DR. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nat Rev Mol Cell Biol. 2011;12(11):757–63. doi: 10.1038/nrm3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, et al. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81(6):935–46. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 7.McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97(1):133–44. doi: 10.1016/S0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- 8.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IA, Debatin KM, Fischer A, et al. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268(5215):1347–9. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 9.Rigante D, Lopalco G, Vitale A, Lucherini OM, De Clemente C, Caso F, et al. Key facts and hot spots on tumor necrosis factor receptor-associated periodic syndrome. Clin Rheumatol. 2014;33(9):1197–207. doi: 10.1007/s10067-014-2722-z. [DOI] [PubMed] [Google Scholar]
- 10.Salzer U, Bacchelli C, Buckridge S, Pan-Hammarstrom Q, Jennings S, Lougaris V, et al. Relevance of biallelic versus monoallelic TNFRSF13B mutations in distinguishing disease-causing from risk-increasing TNFRSF13B variants in antibody deficiency syndromes. Blood. 2009;113(9):1967–76. doi: 10.1182/blood-2008-02-141937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnatz K, Salzer U, Rizzi M, Fischer B, Gutenberger S, Bohm J, et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc Natl Acad Sci U S A. 2009;106(33):13945–50. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwal S, Cunningham-Rundles C. Autoimmunity in common variable immunodeficiency. Curr Allergy Asthma Rep. 2009;9(5):347–52. doi: 10.1007/s11882-009-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romberg N, Chamberlain N, Saadoun D, Gentile M, Kinnunen T, Ng YS, et al. CVID-associated TACI mutations affect autoreactive B cell selection and activation. J Clin Invest. 2013;123(10):4283–93. doi: 10.1172/JCI69854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindorff LA, MacArthur JEBI, Morales JEBI, Junkins HA, Hall PN, Klemm AK, et al. A Catalog of Published Genome-Wide Association Studies. https://www.genome.gov/gwastudies/. Accessed 8 March 2015.
- 15.Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7(8):e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace C. Statistical testing of shared genetic control for potentially related traits. Genet Epidemiol. 2013;37(8):802–13. doi: 10.1002/gepi.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bamias G, Siakavellas SI, Stamatelopoulos KS, Chryssochoou E, Papamichael C, Sfikakis PP. Circulating levels of TNF-like cytokine 1A (TL1A) and its decoy receptor 3 (DcR3) in rheumatoid arthritis. Clin Immunol. 2008;129(2):249–55. doi: 10.1016/j.clim.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Petrovic-Rackov L, Pejnovic N. Clinical significance of IL-18, IL-15, IL-12 and TNF-alpha measurement in rheumatoid arthritis. Clin Rheumatol. 2006;25(4):448–52. doi: 10.1007/s10067-005-0106-0. [DOI] [PubMed] [Google Scholar]
- 20.Vinay DS, Kwon BS. Targeting TNF superfamily members for therapeutic intervention in rheumatoid arthritis. Cytokine. 2012;57(3):305–12. doi: 10.1016/j.cyto.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Bamias G, Kaltsa G, Siakavellas SI, Gizis M, Margantinis G, Zampeli E, et al. Differential expression of the TL1A/DcR3 system of TNF/TNFR-like proteins in large vs. small intestinal Crohn’s disease. Dig Liver Dis. 2012;44(1):30–6. doi: 10.1016/j.dld.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu M, Kobayashi D, Saito K, Furuya D, Yagihashi A, Araake H, et al. Tumor necrosis factor-alpha in serum of patients with inflammatory bowel disease as measured by a highly sensitive immuno-PCR. Clin Chem. 2001;47(7):1297–301. [PubMed] [Google Scholar]
- 23.Wang J, Anders RA, Wang Y, Turner JR, Abraham C, Pfeffer K, et al. The critical role of LIGHT in promoting intestinal inflammation and Crohn’s disease. J Immunol. 2005;174(12):8173–82. doi: 10.4049/jimmunol.174.12.8173. [DOI] [PubMed] [Google Scholar]
- 24.Bamias G, Kaltsa G, Siakavellas SI, Papaxoinis K, Zampeli E, Michopoulos S, et al. High intestinal and systemic levels of decoy receptor 3 (DcR3) and its ligand TL1A in active ulcerative colitis. Clin Immunol. 2010;137(2):242–9. doi: 10.1016/j.clim.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Slebioda TJ, Kmiec Z. Tumour necrosis factor superfamily members in the pathogenesis of inflammatory bowel disease. Mediators Inflamm. 2014;2014:325129. doi: 10.1155/2014/325129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boghdadi G, Elewa EA. Increased serum APRIL differentially correlates with distinct cytokine profiles and disease activity in systemic lupus erythematosus patients. Rheumatol Int. 2014;34(9):1217–23. doi: 10.1007/s00296-014-3020-4. [DOI] [PubMed] [Google Scholar]
- 27.Stohl W, Metyas S, Tan SM, Cheema GS, Oamar B, Xu D, et al. B lymphocyte stimulator overexpression in patients with systemic lupus erythematosus: longitudinal observations. Arthritis Rheum. 2003;48(12):3475–86. doi: 10.1002/art.11354. [DOI] [PubMed] [Google Scholar]
- 28.Rajabi P, Alaee M, Mousavizadeh K, Samadikuchaksaraei A. Altered expression of TNFSF4 and TRAF2 mRNAs in peripheral blood mononuclear cells in patients with systemic lupus erythematosus: association with atherosclerotic symptoms and lupus nephritis. Inflamm Res. 2012;61(12):1347–54. doi: 10.1007/s00011-012-0535-6. [DOI] [PubMed] [Google Scholar]
- 29.Croft M, Duan W, Choi H, Eun SY, Madireddi S, Mehta A. TNF superfamily in inflammatory disease: translating basic insights. Trends Immunol. 2012;33(3):144–52. doi: 10.1016/j.it.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sfikakis PP. The first decade of biologic TNF antagonists in clinical practice: lessons learned, unresolved issues and future directions. Curr Dir Autoimmun. 2010;11:180–210. doi: 10.1159/000289205. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Davidson A. BAFF inhibition: a new class of drugs for the treatment of autoimmunity. Exp Cell Res. 2011;317(9):1270–7. doi: 10.1016/j.yexcr.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croft M, Benedict CA, Ware CF. Clinical targeting of the TNF and TNFR superfamilies. Nat Rev Drug Discov. 2013;12(2):147–68. doi: 10.1038/nrd3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325(5945):1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343(6175):1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44(5):502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343(6175):1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344(6183):519–23. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5(5):e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naranbhai V, Fairfax BP, Makino S, Humburg P, Wong D, Ng E, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. doi: 10.1038/ncomms8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S, Becker J, Bechheim M, Kaiser V, Noursadeghi M, Fricker N, et al. Characterizing the genetic basis of innate immune response in TLR4-activated human monocytes. Nat Commun. 2014;5:5236. doi: 10.1038/ncomms6236. [DOI] [PubMed] [Google Scholar]
- 41.Cunninghame Graham DS, Graham RR, Manku H, Wong AK, Whittaker JC, Gaffney PM, et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat Genet. 2008;40(1):83–9. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manku H, Langefeld CD, Guerra SG, Malik TH, Alarcon-Riquelme M, Anaya JM, et al. Trans-ancestral studies fine map the SLE-susceptibility locus TNFSF4. PLoS Genet. 2013;9(7):e1003554. doi: 10.1371/journal.pgen.1003554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory AP, Dendrou CA, Attfield KE, Haghikia A, Xifara DK, Butter F, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488(7412):508–11. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malmestrom C, Gillett A, Jernas M, Khademi M, Axelsson M, Kockum I, et al. Serum levels of LIGHT in MS. Mult Scler. 2013;19(7):871–6. doi: 10.1177/1352458512463766. [DOI] [PubMed] [Google Scholar]
- 45.Li G, Diogo D, Wu D, Spoonamore J, Dancik V, Franke L, et al. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 2013;9(5):e1003487. doi: 10.1371/journal.pgen.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kugathasan S, Baldassano RN, Bradfield JP, Sleiman PM, Imielinski M, Guthery SL, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40(10):1211–5. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hedl M, Abraham C. A TNFSF15 disease-risk polymorphism increases pattern-recognition receptor-induced signaling through caspase-8-induced IL-1. Proc Natl Acad Sci U S A. 2014;111(37):13451–6. doi: 10.1073/pnas.1404178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kakuta Y, Ueki N, Kinouchi Y, Negoro K, Endo K, Nomura E, et al. TNFSF15 transcripts from risk haplotype for Crohn’s disease are overexpressed in stimulated T cells. Hum Mol Genet. 2009;18(6):1089–98. doi: 10.1093/hmg/ddp005. [DOI] [PubMed] [Google Scholar]
- 49.Michelsen KS, Thomas LS, Taylor KD, Yu QT, Mei L, Landers CJ, et al. IBD-associated TL1A gene (TNFSF15) haplotypes determine increased expression of TL1A protein. PLoS One. 2009;4(3):e4719. doi: 10.1371/journal.pone.0004719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zucchelli M, Camilleri M, Andreasson AN, Bresso F, Dlugosz A, Halfvarson J, et al. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut. 2011;60(12):1671–7. doi: 10.1136/gut.2011.241877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyons PA, Koukoulaki M, Hatton A, Doggett K, Woffendin HB, Chaudhry AN, et al. Microarray analysis of human leucocyte subsets: the advantages of positive selection and rapid purification. BMC Genomics. 2007;8:64. doi: 10.1186/1471-2164-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JC, Lyons PA, McKinney EF, Sowerby JM, Carr EJ, Bredin F, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest. 2011;121(10):4170–9. doi: 10.1172/JCI59255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters JE, Lyons PA, Lee JC, Richard AC, Fortune MD, Newcombe PJ, et al. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet. 2016;12(3):e1005908. doi: 10.1371/journal.pgen.1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carey VJ, Davis AR, Lawrence MF, Gentleman R, Raby BA. Data structures and algorithms for analysis of genetics of gene expression with Bioconductor: GGtools 3.x. Bioinformatics. 2009;25(11):1447–8. doi: 10.1093/bioinformatics/btp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pages H. SNPlocs.Hsapiens.dbSNP.20120608: SNP locations for Homo sapiens (dbSNP build 137). R package version 0.99.9. 2012. http://bioconductor.org/packages/release/data/annotation/html/SNPlocs.Hsapiens.dbSNP.20120608.html.
- 56.Hastie T, Tibshirani R, Friedman J. The false discovery rate. In: The elements of statistical learning: data mining, inference, and prediction. 2nd ed. New York: Springer-Verlag; 2009.
- 57.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4(10):e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richard AC, Lyons PA, Peters JE, Biasci D, Flint SM, Lee JC, et al. Comparison of gene expression microarray data with count-based RNA measurements informs microarray interpretation. BMC Genomics. 2014;15:649. doi: 10.1186/1471-2164-15-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Center for Statistical Genetics, University of Michigan. 1000G Phase I Integrated Release Version 3 Haplotypes. 2012. ftp://share.sph.umich.edu/1000genomes/fullProject/2012.03.14/phase1_release_v3.20101123.snps_indels_svs.genotypes.refpanel.EUR.vcf.gz.tgz. Accessed 3 Dec 2012.
- 62.Purcell S. PLINK v 1.07. http://pngu.mgh.harvard.edu/purcell/plink/.
- 63.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun L, Rommens JM, Corvol H, Li W, Li X, Chiang TA, et al. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat Genet. 2012;44(5):562–9. doi: 10.1038/ng.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fridley BL, Biernacka JM. Gene set analysis of SNP data: benefits, challenges, and future directions. Eur J Hum Genet. 2011;19(8):837–43. doi: 10.1038/ejhg.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98(1):1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39(3):443–53. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 68.Wiens GD, Glenney GW. Origin and evolution of TNF and TNF receptor superfamilies. Dev Comp Immunol. 2011;35(12):1324–35. doi: 10.1016/j.dci.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 69.Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nat Methods. 2007;4(11):911–3. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Irizarry RA, Wu Z, Jaffee HA. Comparison of Affymetrix GeneChip expression measures. Bioinformatics. 2006;22(7):789–94. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
- 71.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6(5):e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14(9):661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 74.Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8(1):7. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Q, Wang H, Schwartz DM, Stoffels M, Park YH, Zhang Y, et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat Genet. 2016;48(1):67–73. doi: 10.1038/ng.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu JZ, Almarri MA, Gaffney DJ, Mells GF, Jostins L, Cordell HJ, et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2012;44(10):1137–41. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83(4):520–8. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luedde T, Heinrichsdorff J, de Lorenzi R, De Vos R, Roskams T, Pasparakis M. IKK1 and IKK2 cooperate to maintain bile duct integrity in the liver. Proc Natl Acad Sci U S A. 2008;105(28):9733–8. doi: 10.1073/pnas.0800198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41(12):1330–4. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat Genet. 2010;42(8):698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Database of Genotypes and Phenotypes (dbGaP). Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine. 2009. http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap.
- 83.European Genome-phenome Archive (EGA). Wellcome Trust Genome Campus, Hinxton, Cambridgeshire, UK: EMBL-EBI. http://www.ebi.ac.uk/ega/home.
- 84.International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium. Sawcer S, Hellenthal G, Pirinen M, Spencer CC, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43(4):329–32. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lyons PA, Rayner TF, Trivedi S, Holle JU, Watts RA, Jayne DR, et al. Genetically distinct subsets within ANCA-associated vasculitis. N Engl J Med. 2012;367(3):214–23. doi: 10.1056/NEJMoa1108735. [DOI] [PMC free article] [PubMed] [Google Scholar]


