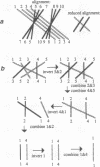
Abstract
Detailed knowledge of gene maps or even complete nucleotide sequences for small genomes leads to the feasibility of evolutionary inference based on the macrostructure of entire genomes, rather than on the traditional comparison of homologous versions of a single gene in different organisms. The mathematical modeling of evolution at the genomic level, however, and the associated inferential apparatus are qualitatively different from the usual sequence comparison theory developed to study evolution at the level of individual gene sequences. We describe the construction of a database of 16 mitochondrial gene orders from fungi and other eukaryotes by using complete or nearly complete genomic sequences; propose a measure of gene order rearrangement based on the minimal set of chromosomal inversions, transpositions, insertions, and deletions necessary to convert the order in one genome to that of the other; report on algorithm design and the development of the DERANGE software for the calculation of this measure; and present the results of analyzing the mitochondrial data with the aid of this tool.
Full text
PDF

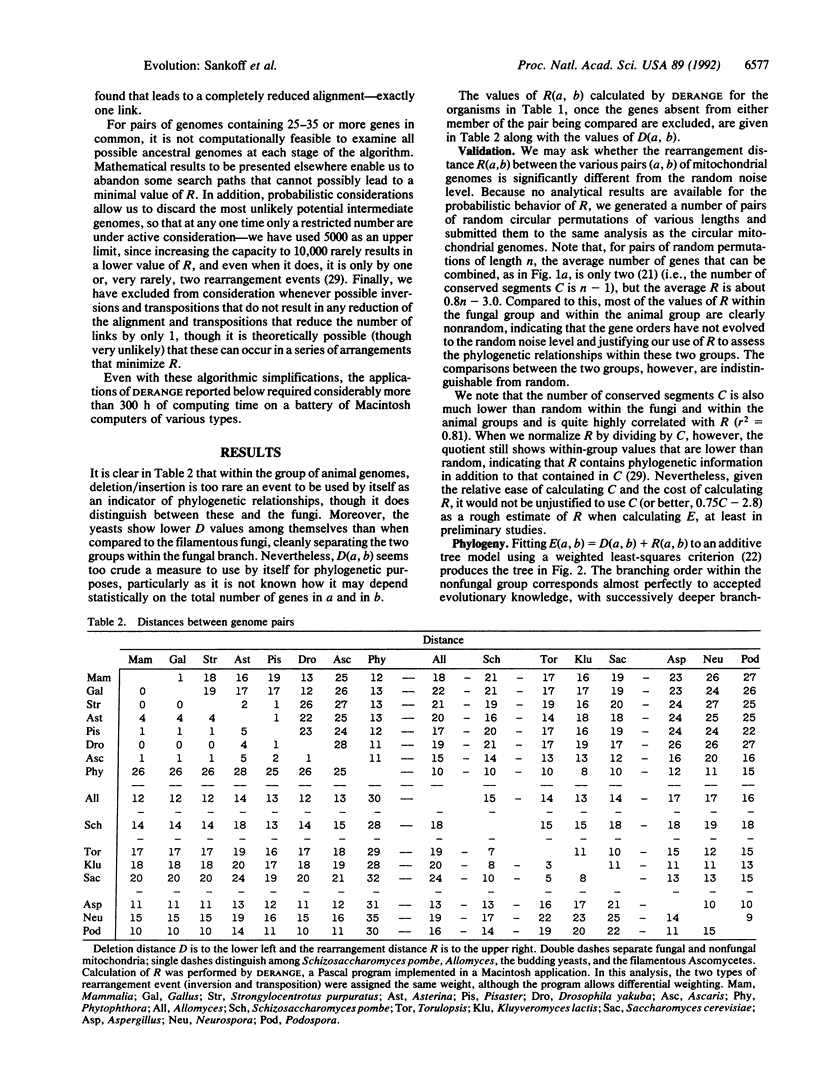
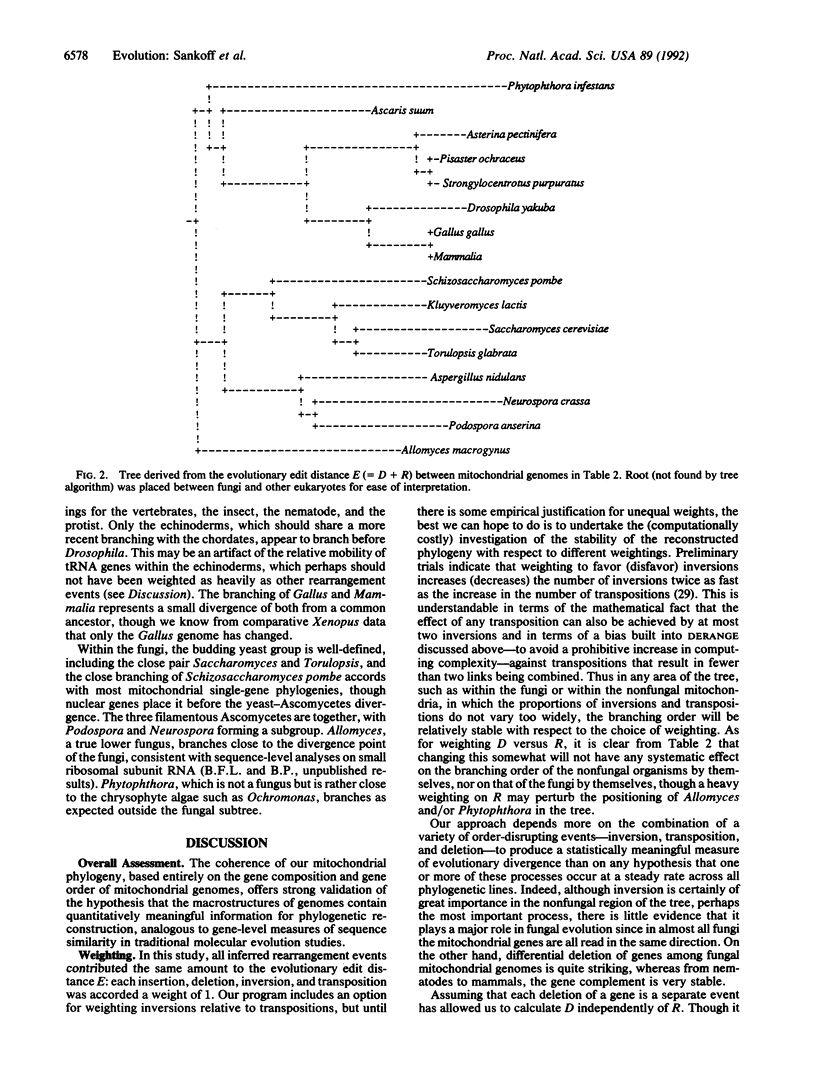

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Asakawa S., Kumazawa Y., Araki T., Himeno H., Miura K., Watanabe K. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J Mol Evol. 1991 Jun;32(6):511–520. doi: 10.1007/BF02102653. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore P., Gadaleta M. N., Roberti M., Saccone C., Wilson A. C. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. 1987 Oct 29-Nov 4Nature. 329(6142):853–855. doi: 10.1038/329853a0. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985 Feb;4(2):465–473. doi: 10.1002/j.1460-2075.1985.tb03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., McNally K. L., Domenico J. M., Matsuura E. T. The complete DNA sequence of the mitochondrial genome of Podospora anserina. Curr Genet. 1990 May;17(5):375–402. doi: 10.1007/BF00334517. [DOI] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Jacobs H. T., Asakawa S., Araki T., Miura K., Smith M. J., Watanabe K. Conserved tRNA gene cluster in starfish mitochondrial DNA. Curr Genet. 1989 Mar;15(3):193–206. doi: 10.1007/BF00435506. [DOI] [PubMed] [Google Scholar]
- Jacobs H. T., Elliott D. J., Math V. B., Farquharson A. Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol. 1988 Jul 20;202(2):185–217. doi: 10.1016/0022-2836(88)90452-4. [DOI] [PubMed] [Google Scholar]
- Lang B. F. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe: highly homologous introns are inserted at the same position of the otherwise less conserved cox1 genes in Schizosaccharomyces pombe and Aspergillus nidulans. EMBO J. 1984 Sep;3(9):2129–2136. doi: 10.1002/j.1460-2075.1984.tb02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H., Taylor B. A. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci U S A. 1984 Feb;81(3):814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Päbo S., Thomas W. K., Whitfield K. M., Kumazawa Y., Wilson A. C. Rearrangements of mitochondrial transfer RNA genes in marsupials. J Mol Evol. 1991 Nov;33(5):426–430. doi: 10.1007/BF02103134. [DOI] [PubMed] [Google Scholar]
- Sankoff D., Cedergren R., Abel Y. Genomic divergence through gene rearrangement. Methods Enzymol. 1990;183:428–438. doi: 10.1016/0076-6879(90)83028-8. [DOI] [PubMed] [Google Scholar]
- Sankoff D., Goldstein M. Probabilistic models of genome shuffling. Bull Math Biol. 1989;51(1):117–124. doi: 10.1007/BF02458839. [DOI] [PubMed] [Google Scholar]
- Smith M. J., Banfield D. K., Doteval K., Gorski S., Kowbel D. J. Nucleotide sequence of nine protein-coding genes and 22 tRNAs in the mitochondrial DNA of the sea star Pisaster ochraceus. J Mol Evol. 1990 Sep;31(3):195–204. doi: 10.1007/BF02109496. [DOI] [PubMed] [Google Scholar]
- Wilson C., Ragnini A., Fukuhara H. Analysis of the regions coding for transfer RNAs in Kluyveromyces lactis mitochondrial DNA. Nucleic Acids Res. 1989 Jun 26;17(12):4485–4491. doi: 10.1093/nar/17.12.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme D. R., Macfarlane J. L., Okimoto R., Clary D. O., Wahleithner J. A. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1324–1328. doi: 10.1073/pnas.84.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]