Ectrodactyly-ectodermal dysplasia-clefting (EEC) syndrome is a rare autosomal dominant disease caused by mutations in the p63 gene. This report describes homozygous-heterozygous mosaicism occurring in a young woman with EEC syndrome and corneal dysfunction, and suggests the heterozygous mutant R311K-p63 stem cells may be isolated, regenerated, and used to correct the patient’s corneal defects.
Keywords: Ectrodactyly-ectodermal dysplasia-clefting syndrome, Cell therapy, p63, Mosaicism, Gene conversion
Abstract
Ectrodactyly-ectodermal dysplasia-clefting (EEC) syndrome is a rare autosomal dominant disease caused by mutations in the p63 gene. To date, approximately 40 different p63 mutations have been identified, all heterozygous. No definitive treatments are available to counteract and resolve the progressive corneal degeneration due to a premature aging of limbal epithelial stem cells. Here, we describe a unique case of a young female patient, aged 18 years, with EEC and corneal dysfunction, who was, surprisingly, homozygous for a novel and de novo R311K missense mutation in the p63 gene. A detailed analysis of the degree of somatic mosaicism in leukocytes from peripheral blood and oral mucosal epithelial stem cells (OMESCs) from biopsies of buccal mucosa showed that approximately 80% were homozygous mutant cells and 20% were heterozygous. Cytogenetic and molecular analyses excluded genomic alterations, thus suggesting a de novo mutation followed by an allelic gene conversion of the wild-type allele by de novo mutant allele as a possible mechanism to explain the homozygous condition. R311K-p63 OMESCs were expanded in vitro and heterozygous holoclones selected following clonal analysis. These R311K-p63 OMESCs were able to generate well-organized and stratified epithelia in vitro, resembling the features of healthy tissues. This study supports the rationale for the development of cultured autologous oral mucosal epithelial stem cell sheets obtained by selected heterozygous R311K-p63 stem cells, as an effective and personalized therapy for reconstructing the ocular surface of this unique case of EEC syndrome, thus bypassing gene therapy approaches.
Significance
This case demonstrates that in a somatic mosaicism context, a novel homozygous mutation in the p63 gene can arise as a consequence of an allelic gene conversion event, subsequent to a de novo mutation. The heterozygous mutant R311K-p63 stem cells can be isolated by means of clonal analysis and given their good regenerative capacity, they may be used to successfully correct the corneal defects present in this unique case of ectrodactyly-ectodermal dysplasia-clefting syndrome.
Introduction
Ectrodactyly-ectodermal dysplasia-clefting (EEC) syndrome (MIM#604292) is an autosomal dominant disease, clinically characterized by limb defects, orofacial clefting, ectodermal dysplasia, and ocular defects. It is related to mutations of the transcription factor p63, a master regulator of gene expression for squamous epithelial proliferation, differentiation, and maintenance. Mutations in p63 account for 98% of patients with typical EEC features and the majority are heterozygous missense mutations located in the DNA-binding domain [1]. To date, approximately 40 different pathogenic p63 mutations have been identified in EEC syndrome [2]. Nearly 90% of these mutations involve 5 arginine residues in the DNA-binding domain: p.R204, p.R227, p.R279, p.R280, and p.R304 [3, 4]. The arginine codons at 304 and 279 are mutational hotspots [4, 5]. Most of cases are sporadic (approximately 70%), related to de novo mutations frequently arising during early-stage development. Moreover, germline mosaicism cases were also described [6]. Familial cases show an autosomal dominant inheritance with variable penetrance. The incidence in the population is 1:900,000, according to the Italian Ministry of Health. We recently found that the major cause of visual morbidity in EEC syndrome is due to a premature ageing and progressive deficiency of limbal stem cells, causing corneal blindness [4–6]. No definitive cures are currently available to treat or at least arrest the progression of these corneal disorders.
Limbal stem cell deficiency (LSCD) is characterized by conjunctival epithelial ingrowth, neovascularization, chronic inflammation, opacification, recurrent corneal erosion, persistent ulcers, destruction of basement membrane components, and fibrous tissue ingrowth [4]. Many causes can lead to severe LSCD, including physical-chemical traumas, infections, irradiations, chronic therapies, inflammatory, autoimmune, and congenital diseases such as aniridia and EEC syndrome [7]. Transplantation of cultured, autologous limbal epithelial cell sheets (CALECSs) has shown to be a valid and successful treatment for acquired unilateral LSCD but not applicable in cases with bilateral LSCD, which are characterized by complete stem-cell deficiency in both eyes [8, 9]. Transplantation of allogenic limbus is feasible but requires immunosuppressive treatments and has a success rate that tends to decrease gradually over time (graft survival rate of 40% at 1 year and 33% at 2 years) [10]. For patients with total and bilateral LSCD, transplantation of cultured autologous oral mucosal epithelial stem cell sheets (CAOMESCs) represents the only effective alternative for reconstructing the ocular surface [11–13]. Nevertheless, for patients with EEC who have a total bilateral LSCD resulting from a genetic alteration, the ocular pathology should be treated by making use of gene therapy-based approaches able to disrupt the deleterious effects of the p63 mutation
Materials and Methods
Cell Culture and Clonal Analysis
Primary human keratinocytes were isolated from fresh oral mucosal biopsies of healthy subjects or patients with EEC syndrome after an informed consent form was signed. The research study and the informed consent forms were approved by the Venetian Ethical Committee for Clinical Research Studies (Prot. 2009/77661, dated November 19, 2009). Once isolated, cells were cultivated onto a feeder layer of lethally irradiated murine 3T3-J2 fibroblasts, as previously described [9]. Subconfluent primary cultures were passaged, serially diluted to obtain 0.5 cells per well, and plated into 96-well plates. The obtained clones were passaged and plated in 24-well plates (three-fourths of the well) for the extraction of DNA and RNA, and in 100-mm dishes (one-fourth of the well) for a colony-forming efficiency (CFE) assay. Human embryonic kidney cells stably transduced to express the simian virus 40 T antigen (293T; provided by D. Baltimore, Rockefeller University, New York, NY) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, San Diego, CA, https://www.thermofisher.com) supplemented with 10% (volume per volume) heat-inactivated fetal bovine serum (Thermo Fisher Scientific), 100 U/ml penicillin, and 100 mg/ml streptomycin (Sigma-Aldrich, St Louis, MO, http://www.sigmaaldrich.com).
Luciferase Reporter Assay
For the luciferase assay, HEK293T cells were plated at density of 2 × 105 cells per well in a 24-well plate and transfected 24 hours later. Lipofectamine 2000 (Thermo Fisher Scientific) was used as transfecting agent. A plasmid containing the luciferase gene under the control of the K14 promoter and expression vectors encoding for WT-, R311K-, R311G-, R304Q-, and R279H-DNp63a were used. When needed, the pcDNA empty vector (Thermo Fisher Scientific) was added to reach the total amount of DNA (500 ng) used in each transfection. In all cases, 10 ng of Renilla luciferase vector (pRL-CMV; Promega, Madison, WI, http://www.promega.com) was cotransfected as a control of the transfection efficiency. Cells were transfected at 80%–90% confluence and incubated at 37°C for 6 hours before medium change. Luciferase activities of cellular extracts were measured by using a dual luciferase reporter assay system (Promega) and light emission was measured over 1 and 5 seconds using a Modulus Microplate Reader luminometer (Promega). All experiments were performed in triplicate.
Human Keratoplasty Lenticules and Hemicornea Preparation In Vitro
Human keratoplasty lenticules (HKLs) were prepared as previously described [14]. Briefly, corneoscleral rims were preserved at 4°C and used within 1–2 days from excision. HKLs were obtained by microkeratome resection of epithelium-free corneas. Epithelial stem cells were plated onto HKLs at a concentration of 5 × 104 cells per cm2. Hemicorneas were cultured under submerged conditions for 7 days and air-lifted for 14 more days.
Histology, Immunofluorescence, and Imaging
Hemicorneas were fixed in 4% paraformaldehyde (PFA) overnight at 4°C, embedded in optimal cutting-temperature compound, frozen, and sectioned. Sections (5–7 μm) were analyzed by indirect immunofluorescence. For immunofluorescence, after 4% PFA fixation (10 minutes at room temperature), samples were blocked in PBS containing 1% bovine serum albumin and the assays were performed by using antibodies against ΔNp63α (rabbit polyclonal, 1:200; Primmbiotech, Milan, Italy, http://www.primmbiotech.com), keratin 3 (AE5 clone, mouse monoclonal, 1:100; MP Biomedicals, Solon, OH, http://www.mpbio.com), involucrin (goat polyclonal, 1:100; Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com), and laminin-β3 (goat polyclonal, 1:200; Santa Cruz Biotechnology) for 1 hour at 37°C. Fluoroscein-isothiocyanate and rhodamine-conjugated secondary antibodies (1:100; Santa Cruz Biotechnology) were incubated for 1 hour at room temperature. Nuclei were stained with far-red fluorescent DNA dye (DRAQ5, 1:1,000; Cell Signaling Technology, Boston, MA, http://www.cellsignal.com) for 10 minutes at room temperature. For Z-stack acquisitions and 3D analysis, cells were directly plated and cultured on coverslips. Specimens were analyzed with a Nikon confocal laser scanning microscope (Nikon, Tokyo, Japan, https://www.nikoninstruments.com) and image analysis was performed using the NIS-Elements Advanced Research software (Nikon).
Polymerase Chain Reaction of the Exon 8 of the Tp63 Gene
DNA was extracted directly from oral mucosal biopsy or from the primary culture by using the DNeasy Blood & Tissue Kit DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany, https://www.qiagen.com), following the manufacturer’s instructions and procedures. Polymerase chain reaction (PCR) was performed on the genomic DNA extracted from tissue or cells, using forward and reverse primers designed to amplify exons 8 (61.6°C; amplicon length: 351 base pairs).
Sanger Sequencing
PCR products (6 ml) were submitted to a phase of the BigDye Terminator kit (Thermo Fisher Scientific). BigDye mix contained 2 ml of buffer, 2 ml of terminators, and 5 ml of primer mix (1 mM). The sequencing program was as follows: a first step of 96°C for 3′, continued with 30 cycles of 96°C for 10′, 50°C for 5′, and 60°C for 4′. After the BigDye treatment, sequencing was performed according to the Sanger method.
cDNA Synthesis and Quantitative Real-Time PCR
Total RNA was extracted from cells and concentration determined using a NanoDropND-1000 spectrophotometer (Thermo Fisher Scientific). Total RNA (0.2–2 μg) was reverse transcribed to first-strand cDNA, using the MoMULV reverse transcription kit (Thermo Fisher Scientific) with random hexamer primers. To calculate the copy number of ΔNp63α transcript molecules, normalized for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in each OMESC clone (n = 24) and autologous limbal stem cell grafts (n = 21), the single-stranded cDNAs were subjected to quantitative real-time PCR (qPCR). A standard curve method was used and reactions carried out in an Applied Biosystems 7800 Real-Time PCR System (Thermo Fisher Scientific). The sequences of the primers and probes are as follows:
p63-Exon 8 FW: 5′-GGTAGATCTTCAGGGGACTTTC-3′
p63-Exon 8 Rev: 5′-TTCTCACTGGCTCTGAGGG-3′
GAPDH FW: 5′-CCACTCCTCCACCTTTGACG-3′
GAPDH Rev: 5′-CATGAGGTCCACCACCCTGT-3′
GAPDH probe: 5′-TET-TTGCCCTCAACGACCACTTT-TAMRA-3′
Seq FW: 5′-GTAGCAACCCTCTATTGTGTG-3′
Seq Rev: 5′-GCCGTTGGATGGGAAGTTC-3′
ΔNp63α FW: 5′-GCATTGTCAGTTTCTTAGCGAG-3′
ΔNp63α Rev: 5′-CCATGGAGTAATGCTCAATCTG-3′
ΔNp63α probe: 5′-FAM-GGACTATTTCACGACCCAGG-BHQ1-3′
Molecular Cytogenetics
Bacterial artificial chromosome (BAC) genomic clones were obtained from Source BioScience (Berlin, Germany, http://www.us.sourcebioscience.com) and labeled by random priming (BioPrime DNA labeling system; Thermo Fisher Scientific) with Biotin-14-dCTP (Thermo Fisher Scientific) or digoxigenin-11-dUTP. Molecular combing analysis was performed as previously described [15]. Briefly, R311K keratinocytes were harvested and immobilized in agarose plugs. High-molecular-weight DNA obtained after β-agarase I digestion (3 U per 1–2 plugs; EuroClone, Milan, Italy) in 0.1 M MES, pH 6.5, was combed on silanized surfaces, according to a standard procedure [16]. A range of 250–300 ng of each labeled probe was hybridized per each slide in the presence of 13× Cot-1 DNA (Thermo Fisher Scientific) and 10 μg of salmon sperm DNA (Thermo Fisher Scientific). To detect the biotin-labeled probes, 594 Alexa Fluor-conjugated streptavidin (1:50; Thermo Fisher Scientific) and biotinilated anti-streptavidin antibodies (1:50; Rockland, Limerick, PA, http://www.rockland-inc.com) were used. The digoxigenin-labeled probe was detected by a monoclonal anti-digoxigenin antibody (1:25; Roche, South San Francisco, CA, https://www.gene.com) and a 488 Alexa Fluor-conjugated anti-mouse IgG (1:50; Thermo Fisher Scientific).
Combed DNA was scored under a ×40 oil immersion objective (numerical aperture [NA]: 1.30) by using a motorized fluorescence microscope (Zeiss Axio Imager.M1; Zeiss International, Oberkochen, Germany, http://www.zeiss.com) equipped with a charge-coupled device (CCD) camera (Coolsnap HQ2; Photometrics, Tucson, AZ, http://www.photometrics.com). Single wavelength images were merged and adjacent fields were aligned using Adobe Photoshop CS2 (Adobe Systems, San Jose, CA, https://www.adobe.com). Measurements of fluorescent signals were done using Metavue software (Molecular Devices, Sunnyvale, CA, https://www.moleculardevices.com) according to the molecular combing calibration factor (1 μm = 2 kb) and the magnification features of the objective and the CCD camera (1 pixel = 0.16125 μm = 0.3225 kb).
For interphase fluorescence in situ hybridization (FISH), cells were layered onto microscope slides by using the Cytospin III (Thermo Fisher Scientific). Chromosome spreads were obtained by standard procedures. FISH was carried out with a cocktail of three BAC probes directly labeled with Cy5-AP3-dUTP (GE Healthcare, Little Chalfont, UK, http://www3.gehealthcare.com), or indirectly labeled with Biotin-16-dUTP or digoxigenin-11-dUTP (Roche Biochemicals). Biotin- and digoxigenin-labeled probes were acquired in red and green fluorescences, respectively. Slides were analyzed under a ×40 or a ×100 oil immersion objectives with an NA of 1.30. Images were captured and merged as previously in this article [16].
Results and Discussion
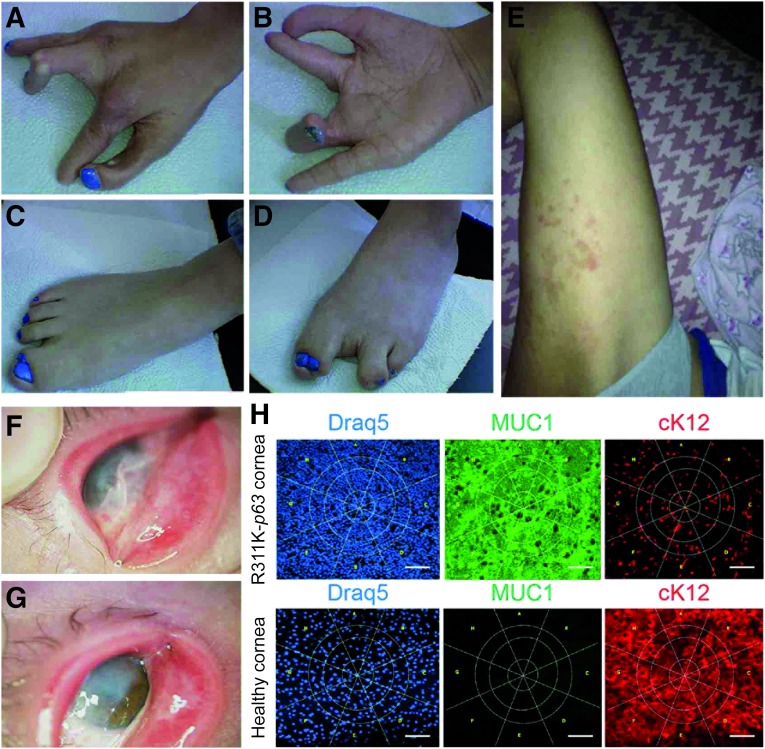
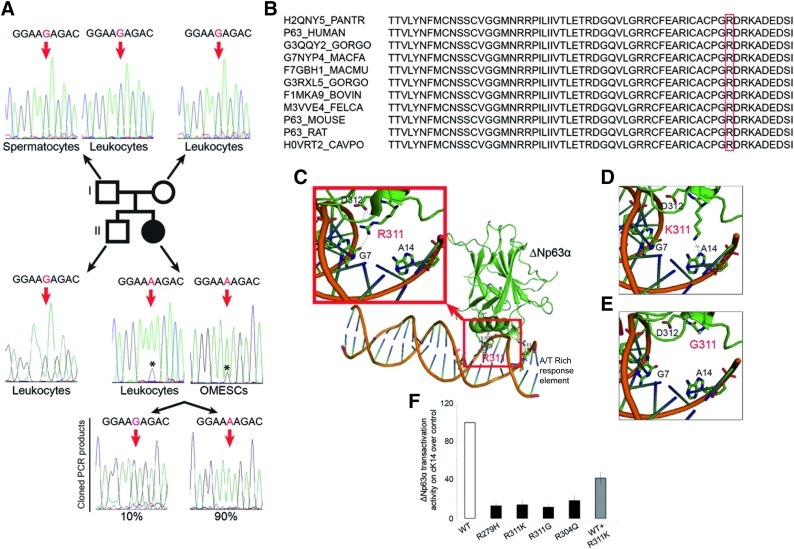
Here we report a singular and rare case of EEC syndrome (Fig. 1) associated with severe conjunctivalization of the cornea and symblepharon (Fig. 1F, 1G). The clinical setting was confirmed through impression cytology analysis using antibodies against cytokeratin (cK) 12 and mucin 1 (MUC1) (Fig. 1H) [17]. The genetic analysis showed that the patient was unexpectedly homozygous for a novel and de novo missense mutation in the p63 gene, R311K. The homozygous mutation detected in this patient occurred in codon 311 at nucleotide 1049 and caused the conversion from arginine to lysine in exon 8 (R311K; AGA > AAA; c.1049G > A) (Fig. 2A). Sequencing of the corresponding region in the DNAs obtained from the parents, brother, and paternal sperm (Fig. 2A) confirmed that the mutation was de novo. The assessment of paternity through a 16-microsatellite marker analysis (D8S1179, D21S11, D7S820, CSF1P0, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, amelogenin, D5S818, FGA), elaborated by PatCan2 software (Cantabria, Spain, https://patcan.es), showed a 99,999989515% compatibility (data not shown). The involved amino acid has a high degree of phylogenetic conservation (Fig. 2B), and the mutation is located in the same position as another EEC causative mutation, p.R311G, which was previously described [18]. The position of the mutation is given according to the original published TA-p63α sequence (GenBank accession no. AF075430), which does not enclose the 39 additional amino acids at the N-terminus, as reported by GenBank (accession no. AF091627; gi:3695081) [19].
Figure 1.
Phenotypic features of R311K-p63 mutation. (A–G): Photographs showing the phenotypic tracts of ectrodactyly-ectodermal dysplasia-clefting syndrome in the patient. Ectrodactyly and sindactily are visible in hands and feet (A–D). Dermatitis often appears on the skin (E). The pannus, due to the conjunctivalization of the ocular surface, and the symblepharon are present in both eyes (F, G). (H): Analysis of impression cytology specimens from the patient and from a healthy subject. Specimens were stained with antibodies against the K12/MUC1 couple of markers and DRAQ5 (for DNA staining). Panel staining: DRAQ5 (blue); MUC1 (green), and cK12 (red). Immunostaining with MUC1 and cK12 shows a severe conjunctivalized cornea. Scale bar = 100 μm. Abbreviations: cK12, cytokeratin 12; MUC1, mucin 1.
Figure 2.
Genotype and functional characterization of R311K-p63 mutation. (A): Schematic representation of R311K-p63 mutation in the patient’s family. The chromatograms of exon 8 of the p63 gene in the father’s spermatocytes and leukocytes and in the mother’s leukocytes are shown at the top. The wild-type sequence is shown; the red arrows indicate the base involved in the point mutation. The chromatograms of exon 8 of the p63 gene in the brother’s leukocytes and in the patient’s leukocytes and OMESCs are shown in the lower part of this panel. The sequence from the brother’s cells indicates the wild-type pattern; those of the patient with ectrodactyly-ectodermal dysplasia-clefting (EEC) show the G-to-A transition at nucleotide position 1049 (in red), resulting in the R311K codon. The low wild-type G peak (*) below the mutant A peak is indicative of a mosaicism condition. The sequencing of single molecules of exon 8 of the patient, amplified from leukocytes and OMESCs by PCR and cloned into a TOPO vector, shows the presence of 90% of mutated and 10% of wild-type PCR fragments. (B): Alignment of p63 protein sequence from different species. (C–E): Three-dimensional model of ΔNp63α protein. The arginine in the wild-type form of the p63 protein can bind DNA in G7 through 2 hydrogen bonds and the amino acid in D312 through a hydrogen bond (C). The glycine in the same position (311), clinically responsible for a severe EEC syndrome, leads to a complete loss of the ability to bind the DNA and flanking amino acid (E). Meanwhile, the lysine loses the 3 hydrogen bonds of the wild-type protein but still binds DNA, acquiring a hydrogen bond in A14 on the consensus DNA-binding site (D). (F): Transactivation potential of ΔNp63α protein and of its EEC mutants determined by transient transfection into HEK293T cells [34]. Keratin-14 promoter, cloned in the luciferase reporter vector, was cotransfected along with an empty expression vector (pcDNA 3.1) or the indicated ΔNp63α expressing plasmid: wild-type (WT), DNA-binding domain mutants (R279H; R311K; R311G; R304Q), and a 1:1 combination of WT and R311K mutant. Transfection efficiency was normalized with a Renilla reporter vector. The result is the mean of four independent experiments. Abbreviations: OMESCs, oral mucosal epithelial stem cells; PCR, polymerase chain reaction.
An in-depth analysis of the sequence chromatogram revealed a small G peak, corresponding to the wild-type sequence, below the A peak, corresponding to the mutant sequence (Fig. 2A). The faint G peak was detectable in all the sequence chromatograms, probably because of a low level of mosaicism, both in leukocytes from the peripheral blood and in OMECs from biopsy specimens of buccal mucosa.
To better explore and obtain an estimate of the degree of mosaicism, we cloned the PCR products from both leukocytes and OMECs. Sequence analysis of 100 clones showed that approximately 90% of the clones were derived from the mutant allele, whereas 10% of clones represented the normal allele (Fig. 2A). This result suggested a somatic mosaicism in which 80% of the patient cells carry 2 mutant alleles and, thus, are homozygous, and approximately 20% are heterozygous for the wild-type and mutant alleles. Homozygous mutations in dominant disorders are frequently lethal in embryos. Consistent with this, p63-null mice die soon after birth [20, 21], and no other mutation in the p63 gene has ever been found in a homozygous condition. We envisage that when the mutation arises in heterozygosity, as all the well-characterized EEC causative mutations (R279H, R304Q, and R311G), a milder form of EEC syndrome is generated. In silico analysis (Fig. 2C-2E) supported this hypothesis and the R311K mutation was still able to bind the p63 site on the genomic DNA. Meanwhile, the severe heterozygous EEC-causing mutation, such as R311G, lost this capacity.
To determine the transactivation capacity of the mutated sequences (and, therefore, the severity of a specific p63 mutation), a luciferase assay was developed by transfecting cell lines with either wild-type or mutant p63 sequences together with a reporter construct containing the luciferase gene under the control of the K14 promoter. The luciferase assay showed a reduced R311K-p63 transcriptional activity on the p63 specific promoter, thus confirming its pathogenicity, but a milder effect in the heterozygous state (Fig. 2F).
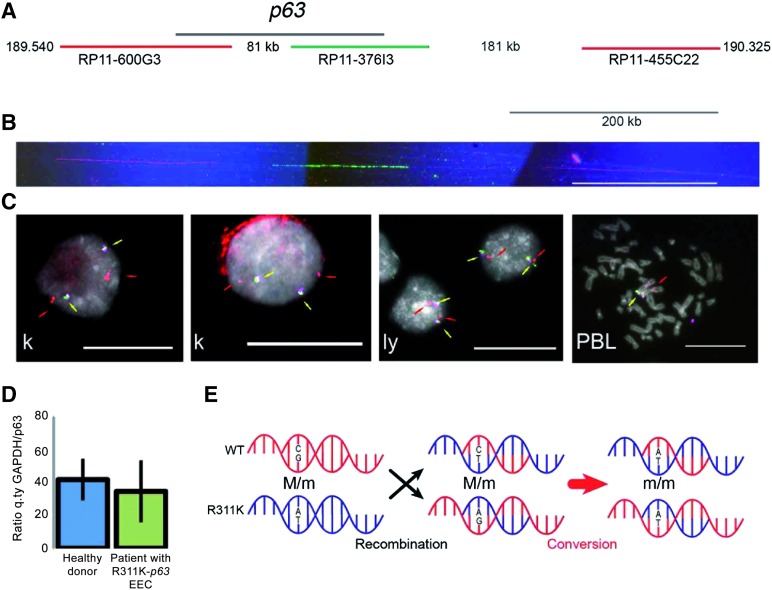
To find a possible explanation, cytogenetic and molecular analyses were performed. Neither parent carried the R311K mutation. By molecular cytogenetics (Fig. 3A-3C), we excluded the presence of large genomic rearrangements, deletions, or duplications in R311K-p63 OMESCs [15, 16]. The resolution of the analysis on DNA extended fibers (molecular combing; Fig. 3A, 3B) is 5 kb; interphase FISH with 2 BAC clones closely mapping at p63 and a distal probe mapping in the short arm of chromosome 3 had a resolution of 100 kb (Fig. 3A, 3C). The presence of genomic rearrangements, deletions, or duplications within the p63 gene was excluded by quantitative p63-specific real-time PCR analysis [22] (Fig. 3D). Our overall hypothesis is, therefore, that a likely mechanism explaining the homozygous status of the patient could be tracked back to a de novo mutation followed by an allelic gene conversion of the wild-type allele by the de novo mutant allele in the p63 gene (Fig. 3E). Forty-two single nucleotide polymorphisms analyzed in the exon 8 flanking the mutation were monomorphic in both parents (data not shown) and, therefore, we were unable to definitively confirm allelic gene conversion, likely because of the small size of the gene conversion event [23, 24]. To our knowledge, this is the first report describing these pathogenetic events in the p63 gene.
Figure 3.
Cytogenetic and molecular analyses suggest an allelic-conversion hypothesis. (A): Schematic representation of the p63 genomic region at 3q27, indicating the distance from the centromere (Mb). The black line identifies the p63 gene; the red and green lines represent the genomic clones (bacterial artificial chromosome [BAC]) used as probes in molecular combing analysis. Probes are differentially spaced and allowed to orientate the molecule in the centromere-telomere direction. Their identity is revealed through red and green fluorescence. (B): An example of fluorescent in situ hybridization (FISH) on DNA extended molecules of R311K keratinocytes prepared through the molecular combing assay. In red and green are visible three probes that hybridize as the expected pattern represented in (A). To verify the integrity of the molecules, DNA was counterstained with an anti-ssDNA antibody (blue fluorescence). Only the expected pattern was observed in mutated keratinocytes (a normal human cell line was used as positive control). Calibration bar = 200 kb. (C): Representative images from interphase FISH analysis of R311K keratinocytes. A human normal lymphoblastoid cell line and human PBLs were used as controls. Yellow arrows indicate the overlapping hybridization signals from two BAC clones closely mapping at p63 (RP11-373I6: magenta fluorescence; RP11-600G3: green fluorescence; see [A]). Because of the proximity of the probes, the hybridization signals are merging in a yellow fluorescence. Red arrows indicate the position of a third probe (RP11-468L11: red fluorescence) mapping in the short arm of chromosome 3 at 3p14.2, and used to monitor the chromosome integrity. In all cell types, the hybridization signals are consistent with a normal chromosome set. The location of the same probes is also shown in a metaphase spread from human PBLs. Calibration bars = 20 μm. (D): Comparative real-time quantitative PCR analysis of genomic DNA extracted from healthy donors (n = 5) and from the patient with R311K-p63 mutation EEC (three different sample preparations) excluded large and small rearrangements, deletions, or duplications in the p63 gene. (E): Schematic representation of the allelic gene conversion event that occurred after the de novo R311K-p63 mutation, causing the generation of the homozygous mutation in the patient with EEC. Abbreviations: EEC, ectrodactyly-ectodermal dysplasia-clefting; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ly, lymphoblastoid cell line; k, keratinocyte; PBL, peripheral blood lymphocyte; q.ty, quantity.
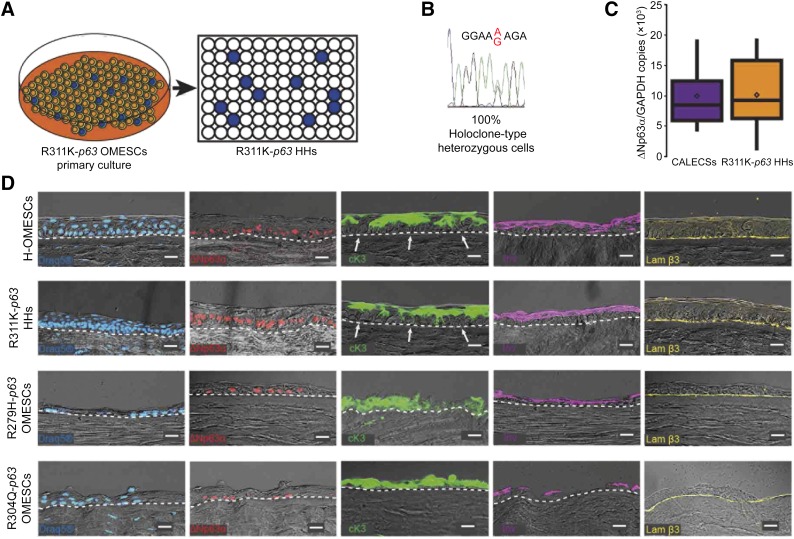
R311K-p63 OMESCs were characterized, ex vivo expanded, and analyzed by clonal analysis (Fig. 4A). Of approximately 400 clones, a CFE assay showed that only 24 had a high clonogenic ability and proliferative potential in vitro; in addition, at least 6 of these resulted as holoclones (data not shown) [25]. Results from the sequencing analysis demonstrated that all the 24 clones were heterozygous (Fig. 4B), thus suggesting a selective growth advantage of the heterozygous stem cells compared with the homozygous ones. We were unable to obtain homozygous clonogenic cells. Stem cell content evaluated by quantifying ΔNp63α expression through real-time qPCR [9, 26, 27] was comparable to that observed in CALECSs successfully transplanted in patients with LSCD caused by traumatic events (Fig. 4C).
Figure 4.
Isolation of R311K-p63 heterozygous holoclones and expression of epithelial cell markers in reconstructed hemicorneas. (A): Cultivation of primary mosaic R311K-p63 OMESCs and isolation of 400 heterozygous R311K-p63 clones through clonal analyses. (B): A representative chromatogram of the sequence around the R311K mutation site of the p63 gene obtained from all the 24 holoclones, previously selected through colony-forming efficiency assay after single-cell clonal amplification. (C): Real-time quantitative analysis of ΔNp63α expression found in CALECSs (n = 19) successfully transplanted in patients with limbal stem cell deficiency, and in R311K-p63 HHs (n = 24). A comparable stem cell content is observed. (D): DRAQ5 staining and expression of epithelial cell markers in reconstructed hemicorneas generated by growing (a) healthy OMESCs, (b) R311K-p63 HHs, (c) R279H-p63 OMESCs, and (d) R304Q-p63 OMESCs onto human keratoplasty lenticules. Cryosections were analyzed through immunofluorescence using ΔNp63α (red), cK3 (green), Inv (violet), and Lam β3 (yellow) antibodies (n = 3) Scale bars = 20 μm. Note that the R311K-p63 HHs’ resulting epithelium was well organized and stratified into four to five cell layers, with basal cuboidal cells differentiating upward to winged cells. The strong expression of the stem cell marker ΔNp63α confirms the maintenance of basal and undifferentiated progenitor cells, which are also negative for cK3 (white arrows). Abbreviations: CALECS, cultured, autologous limbal epithelial cell sheet; cK, cytokeratin; HHs, heterozygous holoclones; Inv, involucrin; Lam β3, laminin β3; OMESCs, oral mucosal epithelial stem cell sheet.
Finally, to test the regenerative ability of the selected R311K-p63 OMESCs carrying the hypomorphic allele, we set up organotypic cultures derived by pooling all R311K-p63 heterozygous holoclones (HHs) grown on a HKL model [14] (Fig. 4D). Compared with OMESCs from healthy (H-OMESCs; n = 3) and patients with EEC (EEC-OMESCs; n = 2, with severe heterozygous p.R279H and p.R304Q p63 mutations), epithelia generated by p.R311K-p63 HHs were more similar to H-OMESCs and well organized into (a) basal column-shaped cells expressing ΔNp63α; (b) suprabasal cuboid wing cells, expressing cK3; and (c) flat, squamous, superficial terminally differentiated cells, as indicated by the expression of involucrin. The basal cuboidal cell layer was firmly attached to the underlying extracellular matrix and to the basement membrane through laminin β3. In sharp contrast, tissues generated from R279H and R304Q-p63 OMESCs showed defects in both stratification and differentiation, thus resulting in severe tissue hypoplasia and lack of proper tissue polarity (Fig. 4D), consistent with previous reports [28–35]. Collectively, these results strongly support the potential clinical use of p.R311K-p63 HHs for the development of CAOMESCs, to reconstruct the ocular surface of this unique case of EEC syndrome.
In this study, we report the first and only case, to our knowledge, of an 18-year-old patient with EEC with a homozygous mutation in the p63 gene. To our knowledge, this patient shows the most severe ocular phenotype. The fact that the patient survived despite having a homozygous mutation in the p63 gene is intriguing, thus predicting a milder severity of this mutation when in the heterozygous state. Arginine and lysine are both positively charged and both are polar amino acids, thus suggesting that, in some cases, their exchange could be well tolerated. Our hypothesis is that the somatic mosaicism combined with (a) the homozygosity for a hypomorphic allele rather than a loss-of-function allele and (b) the heterozygosity consisting of at least 20% of cells, contributed to the survival of the patient, despite what would otherwise represent a lethal condition.
Conclusion
The novelty and the importance of this study are related to the ability to isolate and recover stem cells heterozygous for p63, which appear to have an extraordinary regenerative capacity comparable to that of healthy cells, thus representing a valuable source for starting a customized clinical trial for this unique case of EEC syndrome based solely on epithelial stem cell manipulation.
Patients with LSCD have become quite optimistic after the approval of Holoclar (Holostem Terapie Avanzate, Modena, Italy, http://www.holostem.com), the first advanced-therapy medicinal product containing stem cells to be used for the treatment of moderate to severe LSCD, by the European Medicines Agency. Holoclar consists of cells taken from the patient’s limbus (at the edge of the cornea) and then grown in a laboratory so that they can be used to repair the damaged corneal surface in cases of LSCD caused by alkali burns and similar traumatic events. Likewise, OMESCs therapy is becoming an advanced and valid clinical practice, used in severe bilateral cases of ocular surface reconstruction. This study (a) highlights and emphasizes the potential of regenerative and personalized medicine, obtained through the development of CAOMESCs, to reconstruct the ocular surface of this unique case of EEC syndrome, and (b) demonstrates a proof-of-principle approach, based only on stem cell strategy, that bypassing gene therapy approaches provides evidence of the feasibility of an innovative strategy to correct a severe corneal pathology.
Acknowledgments
We thank Dr. Pio D’Adamo for single nucleotide polymorphism analysis. The work was supported through the Italian Ministry of Health (GR-2009-1555694, CUP n. H31J11000260001; GR-2010-2316138, CUP n. H41J12000100001). The study was supported by COST action BM1302, “Joining Forces in Corneal Regeneration Research.”
Author Contributions
V.B. and E.D.I.: conception and design, data analysis and interpretation, manuscript writing; A.A.N., P.R., A.M., P.N., E.P., M.B., C.B., and F.M.: collection and assembly of data, data analysis and interpretation; S.F., A.R., L.C., and D.P.: data analysis and interpretation; G.P. and C.P.: supervision of the project and final approval of the manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.van Bokhoven H, Hamel BC, Bamshad M, et al. p63 Gene mutations in EEC syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet. 2001;69:481–492. doi: 10.1086/323123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clements SE, Techanukul T, Coman D, et al. Molecular basis of EEC (ectrodactyly, ectodermal dysplasia, clefting) syndrome: Five new mutations in the DNA-binding domain of the TP63 gene and genotype-phenotype correlation. Br J Dermatol. 2010;162:201–207. doi: 10.1111/j.1365-2133.2009.09496.x. [DOI] [PubMed] [Google Scholar]
- 3.van Bokhoven H, Brunner HG. Splitting p63. Am J Hum Genet. 2002;71:1–13. doi: 10.1086/341450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Iorio E, Kaye SB, Ponzin D, et al. Limbal stem cell deficiency and ocular phenotype in ectrodactyly-ectodermal dysplasia-clefting syndrome caused by p63 mutations. Ophthalmology. 2012;119:74–83. doi: 10.1016/j.ophtha.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 5.Hamada T, Chan I, Willoughby CE, et al. Common mutations in Arg304 of the p63 gene in ectrodactyly, ectodermal dysplasia, clefting syndrome: Lack of genotype-phenotype correlation and implications for mutation detection strategies. J Invest Dermatol. 2002;119:1202–1203. doi: 10.1046/j.1523-1747.2002.19526.x. [DOI] [PubMed] [Google Scholar]
- 6.Barbaro V, Nardiello P, Castaldo G, et al. A novel de novo missense mutation in TP63 underlying germline mosaicism in AEC syndrome: Implications for recurrence risk and prenatal diagnosis. Am J Med Genet A. 2012;158A:1957–1961. doi: 10.1002/ajmg.a.35414. [DOI] [PubMed] [Google Scholar]
- 7.Di Iorio E, Barbaro V, Del Vecchio C, et al. Gene therapy approaches for corneal diseases. In: Caenazzo L, editor. New Insights on Biobanks. 1st ed. Padua, Italy: CLEUP; 2013. pp. 87–112. [Google Scholar]
- 8.Daya SM, Ilari FA. Living related conjunctival limbal allograft for the treatment of stem cell deficiency. Ophthalmology. 2001;108:126–133; discussion 133–134. doi: 10.1016/s0161-6420(00)00475-9. [DOI] [PubMed] [Google Scholar]
- 9.Di Iorio E, Ferrari S, Fasolo A, et al. Techniques for culture and assessment of limbal stem cell grafts. Ocul Surf. 2010;8:146–153. doi: 10.1016/s1542-0124(12)70225-2. [DOI] [PubMed] [Google Scholar]
- 10.Santos MS, Gomes JA, Hofling-Lima AL, et al. Survival analysis of conjunctival limbal grafts and amniotic membrane transplantation in eyes with total limbal stem cell deficiency. Am J Ophthalmol. 2005;140:223–230. doi: 10.1016/j.ajo.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Nishida K, Yamato M, Hayashida Y, et al. Corneal reconstruction with tissue-engineered cell sheets composed of autologous oral mucosal epithelium. N Engl J Med. 2004;351:1187–1196. doi: 10.1056/NEJMoa040455. [DOI] [PubMed] [Google Scholar]
- 12.Chen HC, Chen HL, Lai JY, et al. Persistence of transplanted oral mucosal epithelial cells in human cornea. Invest Ophthalmol Vis Sci. 2009;50:4660–4668. doi: 10.1167/iovs.09-3377. [DOI] [PubMed] [Google Scholar]
- 13.Burillon C, Huot L, Justin V, et al. Cultured autologous oral mucosal epithelial cell sheet (CAOMECS) transplantation for the treatment of corneal limbal epithelial stem cell deficiency. Invest Ophthalmol Vis Sci. 2012;53:1325–1331. doi: 10.1167/iovs.11-7744. [DOI] [PubMed] [Google Scholar]
- 14.Barbaro V, Ferrari S, Fasolo A, et al. Reconstruction of a human hemicornea through natural scaffolds compatible with the growth of corneal epithelial stem cells and stromal keratocytes. Mol Vis. 2009;15:2084–2093. [PMC free article] [PubMed] [Google Scholar]
- 15.Palumbo E, Tosoni E, Matricardi L, et al. Genetic instability of the tumor suppressor gene FHIT in normal human cells. Genes Chromosomes Cancer. 2013;52:832–844. doi: 10.1002/gcc.22079. [DOI] [PubMed] [Google Scholar]
- 16.Palumbo E, Matricardi L, Tosoni E, et al. Replication dynamics at common fragile site FRA6E. Chromosoma. 2010;119:575–587. doi: 10.1007/s00412-010-0279-4. [DOI] [PubMed] [Google Scholar]
- 17.Barbaro V, Ferrari S, Fasolo A, et al. Evaluation of ocular surface disorders: A new diagnostic tool based on impression cytology and confocal laser scanning microscopy. Br J Ophthalmol. 2010;94:926–932. doi: 10.1136/bjo.2009.164152. [DOI] [PubMed] [Google Scholar]
- 18.Willoughby CE, Chen I, Ellis I, et al. Mutations in p63 gene in ectrodactyly-ectodermal dysplasia-clefting (EEC) syndrome and their relevance to the ocular phenotype. Invest Ophthalmol Vis Sci. 2005;46:1833a. [Google Scholar]
- 19.McGrath JA, Duijf PH, Doetsch V, et al. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- 20.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 21.Amelio I, Grespi F, Annicchiarico-Petruzzelli M, et al. p63 the guardian of human reproduction. Cell Cycle. 2012;11:4545–4551. doi: 10.4161/cc.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbaro V, Confalonieri L, Vallini I, et al. Development of an allele-specific real-time PCR assay for discrimination and quantification of p63 R279H mutation in EEC syndrome. J Mol Diagn. 2012;14:38–45. doi: 10.1016/j.jmoldx.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Yang S, Yuan Y, Wang L, et al. Great majority of recombination events in Arabidopsis are gene conversion events. Proc Natl Acad Sci USA. 2012;109:20992–20997. doi: 10.1073/pnas.1211827110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benovoy D, Drouin G. Ectopic gene conversions in the human genome. Genomics. 2009;93:27–32. doi: 10.1016/j.ygeno.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Iorio E, Barbaro V, Ruzza A, et al. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc Natl Acad Sci USA. 2005;102:9523–9528. doi: 10.1073/pnas.0503437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Iorio E, Barbaro V, Ferrari S, et al. Q-FIHC: Quantification of fluorescence immunohistochemistry to analyse p63 isoforms and cell cycle phases in human limbal stem cells. Microsc Res Tech. 2006;69:983–991. doi: 10.1002/jemt.20375. [DOI] [PubMed] [Google Scholar]
- 28.Koster MI, Kim S, Mills AA, et al. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll DK, Carroll JS, Leong CO, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 30.Senoo M, Pinto F, Crum CP, et al. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 31.Candi E, Rufini A, Terrinoni A, et al. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci USA. 2007;104:11999–12004. doi: 10.1073/pnas.0703458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melino G, Memmi EM, Pelicci PG, et al. Maintaining epithelial stemness with p63. Sci Signal. 2015;8:re9. doi: 10.1126/scisignal.aaa1033. [DOI] [PubMed] [Google Scholar]
- 33.Memmi EM, Sanarico AG, Giacobbe A, et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2015;112:3499–3504. doi: 10.1073/pnas.1500762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Browne G, Cipollone R, Lena AM, et al. Differential altered stability and transcriptional activity of ΔNp63 mutants in distinct ectodermal dysplasias. J Cell Sci. 2011;124:2200–2207. doi: 10.1242/jcs.079327. [DOI] [PubMed] [Google Scholar]
- 35.Barbaro V, Nasti AA, Del Vecchio C, et al. Correction of mutant p63 in EEC syndrome using siRNA mediated allele specific silencing restores defective stem cell function. Stem Cells. 2016 doi: 10.1002/stem.2343. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]