During their synthesis at the ribosome, many proteins have to be either translocated across or inserted into the endoplasmic reticulum (ER) membrane by the translocon,1 a multi-subunit complex located in the ER membrane. The universally conserved protein-conducting channel Sec61 forms the functional core of the translocon. Accessory translocon components, most notably the stoichiometric translocon associated protein complex (TRAP) and the near-stoichiometric oligosaccharyl-transferase (OST) complex,2 complement Sec61 and assist in protein transport and membrane protein integration or facilitate maturation of nascent chains by covalent modifications. Early characterization of Sec61 by conductance measurements indicated that it adopts at least 2 distinct conformational states to enable protein translocation and membrane insertion while preventing extensive ion flux:3 a more-conductive state when bound to the ribosome and a less-conductive state upon ribosome release.
Sec61 is a hetero-trimeric complex, consisting of the central Sec61α subunit and 2 much smaller peripheral subunits, Sec61β and Sec61γ. X-ray crystallographic analyses of prokaryotic Sec61 homologs revealed that Sec61α consists of 2 pseudo-symmetrical N- and C-terminal halves, each comprising 5 transmembrane helices, which form the translocation channel.4 The two domains are connected by a short ‘hinge’ helix allowing a jaw-like motion of the N- and C-terminal halves with respect to each other. Consistent with the early characterization of the protein-conducting channel,3 Sec61 was found to adopt 2 functionally different conformations: a state with a lateral opening between the 2 Sec61α halves, which allows hydrophobic helices to partition into the lipid bilayer (termed the lateral gate), as well as a laterally closed state (Fig. 1).
Figure 1.
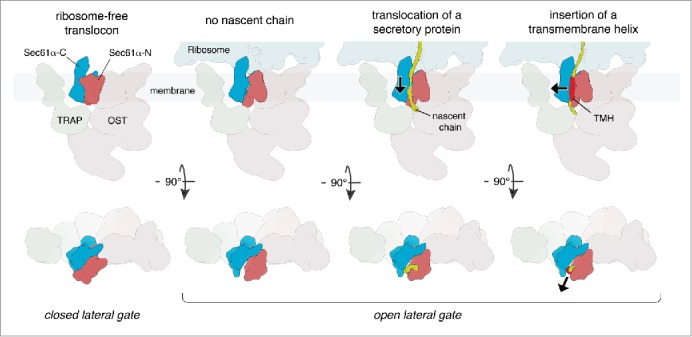
Mechanistic model for protein transport and membrane insertion by Sec61. Ribosome-free Sec61 likely adopts a laterally closed conformation. Upon ribosome binding, the lateral gate of Sec61α opens. Nascent proteins slide along the interior surface of the lateral gate and are either translocated into the lumen or inserted into the membrane as a transmembrane helix (TMH). For each state, views perpendicular to the membrane (upper row) and views from the cytoplasm (lower row) are shown.
Recent mechanistic models for the interplay of the ribosome and Sec61 were derived from single particle cryo-EM structures of ribosome-bound, detergent-solubilized Sec61 in distinct functional states.5,6 They suggested that ribosome-bound Sec61 is mostly present in a closed state and opens only transiently for integration of a nascent transmembrane helix into the membrane. However, these models were inconsistent with the earlier conductance measurements, which indicated that ribosome binding alone induces conformational changes of the native protein-conducting channel toward a more conductive state.3 This discrepancy illustrates the need for visualizing the conformation of ribosome-bound Sec61 in a lipid environment and in presence of all other translocon components.
Cryo-electron tomography (CET) in combination with subtomogram analysis is an excellent method for studying the structures of large macromolecules in their natural environment. It is particularly attractive for studying membrane-embedded and –associated complexes, because detergent solubilization is not required, avoiding destabilization of the complex during purification. Developments in direct detector technology, automated tomography data acquisition, and image processing software recently allowed us to obtain a subtomogram average of mammalian ribosomes bound to the native translocon in canine rough ER-vesicles at subnanometer resolution.7 In the density, secondary structure elements can be distinguished for the ribosome and the Sec61 protein-conducting channel, visualizing the conformation of Sec61 in a lipid environment and in presence of all other translocon components. In the native translocon, Sec61 was clearly found in a laterally open conformation, the highest resolution structure of eukaryotic Sec61 in this state to date.
In order to be able to mechanistically interpret the laterally open conformation of Sec61, we characterized the functional states of the imaged ribosome-translocon complexes. Subtomogram classification focused on the ribosomal tRNA binding sites revealed defined density for peptidyl tRNAs on only 29% of ribosomes, suggesting that the majority of imaged ribosome translocon complexes were idle. Sequencing of ribosome-protected mRNA segments (ribosome profiling) demonstrated that the subset of actively translating ribosomes is almost exclusively engaged in the synthesis of soluble secretory proteins, not containing a transmembrane helix. Consequently, the vast majority of depicted translocon complexes are not in the process of inserting a nascent transmembrane helix into the lipid bilayer. In agreement with the early conductance measurements, our study of the native ribosome-Sec61 complex therefore suggests that the laterally open conformation of Sec61 does not only occur transiently during transmembrane helix insertion, but may be the only major conformation present in the fully assembled ribosome-bound translocon complex irrespective of its functional state (Fig. 1). The discrepancy between these findings and the recent mechanistic models might be explained by the effects of detergent solubilization: the absence of a native membrane environment and separation of Sec61 from physiological interaction partners, in particular TRAP and OST, may destabilize the laterally open conformation.
The observation of a constitutively open lateral gate of ribosome-bound Sec61 significantly changes our understanding, of how membrane proteins are inserted into the lipid bilayer. While recent mechanistic models suggested an active switching of Sec61, significantly influencing the energy landscape of transmembrane helix insertion, a largely invariant conformation of ribosome-associated Sec61 predicts that membrane protein insertion is primarily driven by the thermodynamic behavior of the growing nascent chain.8 In such a model, nascent chains would be sliding along the interior surface of the constitutively opened lateral gate and – if hydrophobic enough - partition into the lipid bilayer (Fig. 1). Clearly, follow-up studies visualizing the partitioning of defined insertion intermediates in a native membrane environment will be required for a detailed mechanistic understanding of secretory protein transport and membrane protein insertion by Sec61.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Johnson AE, et al.. Annu Rev Cell Dev Biol 1999; 15:799-842; PMID:10611978; http://dx.doi.org/ 10.1146/annurev.cellbio.15.1.799 [DOI] [PubMed] [Google Scholar]
- [2].Pfeffer S, et al.. Nat Comm 2014; 5:3072; PMID: 24407213; http://dx.doi.org/ 10.1038/ncomms4072 [DOI] [PubMed] [Google Scholar]
- [3].Simon SM, et al.. Cell 1991; 65:371-80. PMID:1902142; http://dx.doi.org/ 10.1016/0092-8674(91)90455-8 [DOI] [PubMed] [Google Scholar]
- [4].Park E, et al.. Annu Rev Biophys 2012; 41:21-40; PMID:22224601; http://dx.doi.org/ 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- [5].Gogala M, et al.. Nature 2014; 506:107-110; PMID:24499919; http://dx.doi.org/ 10.1038/nature12950 [DOI] [PubMed] [Google Scholar]
- [6].Voorhees RM, et al.. Cell 2014; 157:1632-1643; PMID:24930395; http://dx.doi.org/ 10.1016/j.cell.2014.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pfeffer S, et al.. Nat Comm 2015; 6:8403; PMID:26411746; http://dx.doi.org/ 10.1038/ncomms9403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cymer F, et al.. J Mol Biol 2015; 427:999-1022; PMID:25277655; http://dx.doi.org/ 10.1016/j.jmb.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]


