ABSTRACT
Intracellular Fibroblast Growth Factor 14 (iFGF14) and the other intracellular FGFs (iFGF11-13) regulate the properties and densities of voltage-gated neuronal and cardiac Na+ (Nav) channels. Recent studies have demonstrated that the iFGFs can also regulate native voltage-gated Ca2+ (Cav) channels. In the present study, a mass spectrometry (MS)-based proteomic approach was used to identify the components of native cerebellar iFGF14 complexes. Using an anti-iFGF14 antibody, native iFGF14 complexes were immunoprecipitated from wild type adult mouse cerebellum. Parallel control experiments were performed on cerebellar proteins isolated from mice (Fgf14−/−) harboring a targeted disruption of the Fgf14 locus. MS analyses of immunoprecipitated proteins demonstrated that the vast majority of proteins identified in native cerebellar iFGF14 complexes are Nav channel pore-forming (α) subunits or proteins previously reported to interact with Nav α subunits. In contrast, no Cav channel α or accessory subunits were revealed in cerebellar iFGF14 immunoprecipitates. Additional experiments were completed using an anti-PanNav antibody to immunoprecipitate Nav channel complexes from wild type and Fgf14−/− mouse cerebellum. Western blot and MS analyses revealed that the loss of iFGF14 does not measurably affect the protein composition or the relative abundance of Nav channel interacting proteins in native adult mouse cerebellar Nav channel complexes.
KEYWORDS: cerebellum, intracellular fibroblast growth factors, native interactomes, proteomics, voltage-gated Na+ channels
Introduction
Members of the intracellular Fibroblast Growth Factor (iFGF) family of proteins, which include iFGFs 11-14, share significant sequence and structural homology with canonical FGFs, but are functionally unrelated as they are not secreted and do not activate FGF receptors.1,2 Mutations in iFGF14 in humans have been linked to spinocerebellar ataxia 27 (SCA27), an autosomal-dominant disorder characterized by gait and movement disorders, nystagmus, and cognitive impairment.3-7 Mice lacking iFGF14 (Fgf14−/−) exhibit an ataxia phenotype resembling SCA27,8,9 accompanied by marked changes in the excitability of cerebellar granule and Purkinje neurons.10 Numerous previous studies have linked iFGFs-mediated effects on neuronal excitability to the regulation of voltage-gated Na+ (Nav) channels.10-14 Through binding to the C-terminal domain of Nav channel pore-forming (α) subunits, iFGFs have most consistently been reported to increase the availability of Nav channels by shifting the voltage-dependence of steady-state inactivation toward depolarized potentials.11,14-17 It was recently also reported, however, that the iFGFs also regulate voltage-gated Ca2+ (Cav) channels. In cardiac myocytes, for example, iFGF13 was shown to modulate Cav current densities and the cell surface expression of Cav channel α subunits.18 In addition, iFGF14 was shown to regulate Cav current densities in (presynaptic) cerebellar granule neurons and synaptic transmission at granule to Purkinje neuron synapses.19 Taken together, these observations suggest that iFGF14 (and the other iFGFs) may regulate multiple types of ion channels and subserve multiple physiological functions.
The goals of the studies reported here were to identify the proteins that co-immunoprecipitate with iFGF14 from adult wild-type (WT) mouse cerebellum and to determine if the loss of iFGF14 alters the protein components of native Nav channel complexes in adult (Fgf14−/−) mouse cerebellum. Native iFGF14 and Nav channel protein complexes were immunoprecipitated from adult WT (and Fgf14−/−) mouse cerebella and the protein components of these complexes were identified using a mass spectrometry (MS)-based proteomic approach and Western blotting.
Results
Immunoprecipitation of cerebellar iFGF14 complexes
Initial experiments were focused on optimizing the experimental conditions for the IP of iFGF14 complexes from adult WT cerebella using an anti-iFGF14 mouse monoclonal antibody (mαFGF14) from the UC Davis/NIH NeuroMab Facility. Brains from animals (Fgf14−/−) harboring a targeted disruption of the Fgf14 locus (which encodes iFGF14)8 were used as a control. As illustrated in Figure 1A, Western blots probed with the rabbit polyclonal anti-FGF14 (RbαFGF14) antiserum revealed robust expression of iFGF14 in lysates from WT animals and efficient IP of iFGF14 from WT mouse cerebellum with mαFGF14. Western blot analysis of the IP supernatant revealed ∼60% depletion of iFGF14 compared with the starting cerebellar lysate. No iFGF14 protein was detected in Western blots of total protein lysates or mαFGF14-IPs from Fgf14−/− cerebella (Fig. 1A, bottom).
Figure 1.
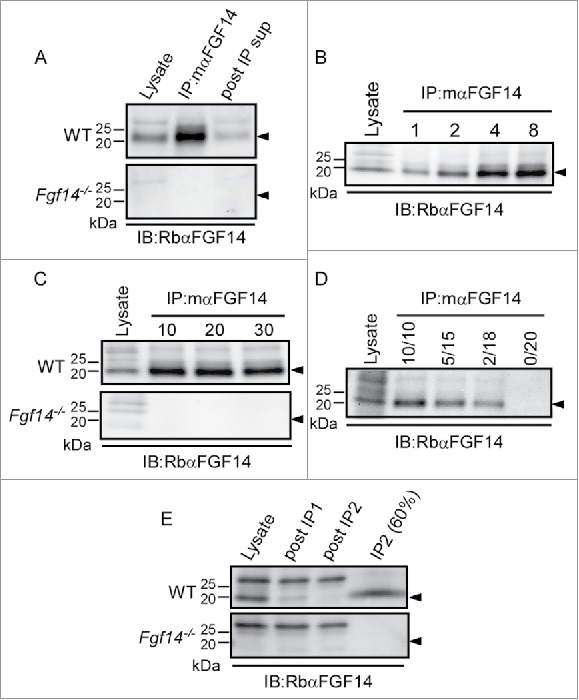
Optimization of mαFGF14 Immunoprecipitations. All blots were probed (IB) with a RbαFGF14 polyclonal antiserum as described in Materials and Methods. A. Representative Western blots of WT (top) and Fgf14−/− (bottom) cerebellar lysates, proteins immunoprecipitated with the mouse monoclonal anti-iFGF14 (mαFGF14) antibody, and the corresponding post immunoprecipitation supernatants (post IP sup). The ∼20 kDa iFGF14 protein is clearly evident in the WT lanes and absent in the Fgf14−/− lanes. Analyses of these blots revealed approximately 60% depletion of iFGF14 from WT mouse cerebellar lysates following IP with the mαFGF14 antibody. B. Western blots of WT cerebellar lysates before and after IP using variable amounts (1, 2, 4, or 8 mg) of protein lysates and a constant amount (20 μl) of mαFGF14-coupled sepharose beads. Analysis of IP samples revealed that increasing amounts of iFGF14 were immunoprecipitated from increasing amounts of cerebellar lysate, suggesting that the binding capacity of the mαFGF14 antibody-conjugated beads was not saturated. C. Western blots of WT (upper) and Fgf14−/− (lower) cerebellar lysates before and after IPs using 8 mg of cerebellar proteins with variable volumes (10, 20, or 30 μl) of mαFGF14 antibody-conjugated beads. Analysis of the IP samples revealed no significant increase in the amount of iFGF14 immunoprecipitated when the bead volume was increased. D. Western blot of WT cerebellar lysates and proteins immunoprecipitated from 8 mg of cerebellar proteins with decreasing volumes of mαFGF14 antibody-conjugated beads. Non-conjugated control sepharose beads were used to maintain the bead amount constant; the numbers above the lanes refer to the mαFGF14 antibody-conjugated bead volumes (left) and the control bead volumes (right). Decreasing amounts of iFGF14 were immunoprecipitated as the mαFGF14-antibody-conjugated bead volume was decreased. E. Western blots of WT and Fgf14−/− cerebellar lysates, post IP supernatants following sequential mαFGF14-IPs (post IP1 and post IP2), and proteins immunoprecipitated after IP2; 60% of the IP2 fraction was loaded onto the gel. Analysis of these blots revealed that approximately 85% depletion of iFGF14 was achieved with the first IP and a 90% depletion was achieved with the second IP.
To determine the optimal amount of protein for IP of cerebellar iFGF14 complexes, a constant amount (20 μl) of pre-conjugated anti-iFGF14 sepharose beads was used to IP proteins from varying amounts of WT cerebellar lysates (Fig. 1B). Western blots of cerebellar lysates and immunoprecipitated proteins revealed an increase in the amount of immunoprecipitated iFGF14 with the higher lysate inputs, suggesting that the binding capacity of the mαFGF14- beads was not saturated, even when the largest amount (8 mg) of lysate was used. To optimize the amount of mαFGF14-bead volume for IP of cerebellar iFGF14 complexes, variable amounts of mαFGF14-beads were used to IP proteins from a constant amount (8 mg) of WT or Fgf14−/− cerebellar lysates. As illustrated in Figure 1C, Western blot analyses of WT cerebellar lysates and immunoprecipitated proteins revealed that increasing the amount of mαFGF14-beads did not result in detectable increases in the amount of iFGF14 immunoprecipitated. The maximum amount of iFGF14 was precipitated with 10 μl of mαFGF14-beads. No iFGF14 was detected in the total protein lysates or in the mαFGF14-IPs from Fgf14−/− cerebella (Fig. 1C, bottom).
To optimize the relative amounts of mαFGF14-beads and cerebellar lysates, IPs were also performed using 8 mg WT cerebellar lysates and decreasing amounts of the mαFGF14-conjugated beads (Fig. 1D). Antibody-conjugated beads were mixed with non-conjugated control sepharose beads to maintain equal bead volumes. As illustrated in Figure 1D, Western blots of WT cerebellar lysates and immunoprecipitated proteins revealed that decreasing the amount of mαFGF14-conjugated beads resulted in reduced IP of iFGF14. In an effort to improve the yield of iFGF14, sequential IPs were performed using the post IP supernatant from the first IP as the input for the second IP (Fig. 1E). Western blot analyses of the WT lysates and the supernatants from the 2 sequential IPs revealed ∼85% depletion of iFGF14 in the first IP and ∼90% depletion of FGF14 in the second IP. Together, these observations suggest that 8 mg of cerebellar protein and 10 μl of mαFGF14-conjugated beads are optimal for robust IP of native iFGF14 complexes, and these relative amounts were scaled for subsequent proteomic analyses.
Identification of proteins immunoprecipitating with iFGF14
To identify the protein components of native iFGF14 protein complexes in WT mouse cerebellum, the proteins immunoprecipitating with the mαFGF14 antibody-conjugated beads were digested with trypsin and the resulting tryptic peptides were analyzed using 2D-LC-MS/MS. This in-solution analysis yielded a total of 12 iFGF14 peptides and an amino acid sequence coverage of 59% (51% in average, Fig. 2 and Table 1). A representative fragmentation spectrum of an iFGF14 tryptic peptide, as well as the amino acid sequence derived from this spectrum, is illustrated in Figure 2A. An amino acid sequence alignment of iFGF14B, the major iFGF14 splice variant in the adult mouse brain,20,21 with iFGF14A and the other iFGFs, iFGF11A, iFGF12A and iFGF13A, is depicted in Figure 2B. Although iFGF14 and the other iFGFs are highly homologous, most (10) of the peptides identified are unique to FGF14. Two (of the 12) peptides, however, are also present in iFGF12 (Fig. 2B). One peptide specific to the unique iFGF14B N-terminus was identified, whereas the other iFGF14 peptides could reflect the presence of either iFGF14A or iFGF14B.
Figure 2.
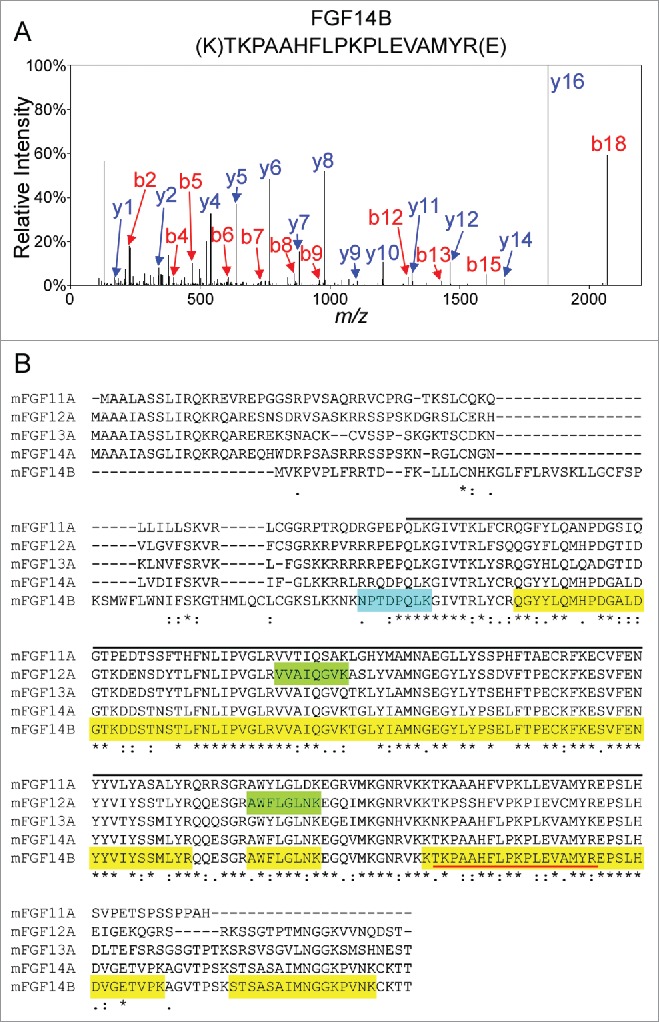
Mass Spectrometric Identification of iFGF14 Using In-Solution 2D-LC-MS/MS. A. Representative MS2 fragmentation spectrum of one of the identified iFGF14 tryptic peptides, NH2-(K)TKPAAHFLPKPLEVAMYR(E), with the y- (in blue) and b- (in red) ions highlighted. B. Sequence alignment of mouse iFGF11A, iFGF12A, iFGF13A, iFGF14A and iFGF14B with the amino acid sequence coverage for iFGF14 obtained following high resolution LC-MS/MS proteomic analysis of immunoprecipitated cerebellar iFGF14 complexes shown. All detected peptides are highlighted in yellow. The identified peptides that are also present in other iFGFs are also highlighted in green and the identified peptide that is unique to the iFGF14B isoform is highlighted in blue. The peptide identified by the fragmentation spectrum in A is underlined in red. The iFGF core homology domain is underlined in black. Asterisks (*) indicate fully conserved residues, colons (:) indicate residues with strongly similar properties, and periods (.) indicate residues with weakly similar properties.
Table 1.
Proteins identified in immunoprecipitated cerebellar iFGF14 complexes.
| Identified Protein | Gene | Average number of unique peptides (total spectra) | Average % amino acid sequence coverage | |
|---|---|---|---|---|
| WT | Fgf14−/− | |||
| Voltage-gated Na+ channel α subunit Nav1.2 | Scn2a | 58 (158) | 0 | 27 |
| Voltage-gated Na+ channel α subunit Nav1.1 | Scn1a | 35 (73) | 0 | 24 |
| Voltage-gated Na+ channel α subunit Nav1.6 | Scn8a | 23 (45) | 0 | 17 |
| FGF14 | Fgf14 | 10 (26) | 0 | 51 |
| Calmodulin | Calm | 9 (33) | 2 (3) | 55 |
| Voltage-gated Na+ channel β2 subunit | Scn2b | 9 (21) | 0 | 35 |
| Voltage-gated Na+ channel α subunit Nav1.4 | Scn4a | 2 (12) | 0 | 3 |
| Synaptotagmin-1 | Syt1 | 2 (3) | 0 | 16 |
| Synaptotagmin-2 | Syt2 | 15 (32) | 2 (2) | 57 |
| Voltage-gated Na+ channel β1 subunit | Scn1b | 8 (17) | 0 | 28 |
| Ankyrin R | Ank1 | 4 (4) | 0 | 3 |
| Ankyrin G | Ank3 | 9 (10) | 0 | 6 |
| Inositol 1,4,5-trisphosphate receptor type 1 | Itpr1 | 4 (4) | 0 | 2 |
| Voltage-dependent anion-selective channel protein 1 | Vdac1 | 2 (2) | 0 | 7 |
| Vesicle-associated membrane protein-associated protein A | Vapa | 2 (4) | 0 | 13 |
| Voltage-gated Na+ channel β4 subunit | Scn4b | 2 (3) | 0 | 7 |
| Excitatory amino acid transporter 1 | Slc1a3 | 5 (7) | 2 (2) | 14 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | Atp2a2 | 2 (3) | 0 | 3 |
| Casein kinase II subunit β | Csnk2b | 1 (1) | 0 | 5 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β−1 | Gnb1 | 1 (1) | 0 | 4 |
| Solute carrier family 12 member 5 | Slc12a5 | 1 (1) | 0 | 1 |
| Protein-glutamine gamma-glutamyltransferase 2 | Tgm2 | 1 (1) | 0 | 3 |
| Neurofascin | Nfasc | 1 (1) | 0 | 1 |
| 14-3-3 protein theta | Ywhaq | 1 (1) | 0 | 14 |
| Synaptosomal-associated protein 25 | Snap25 | 1 (1) | 0 | 9 |
Note. The average number of exclusive unique peptides and total spectra as well as the average percent amino acid sequence coverage obtained for each protein (from a total of 4 runs in each condition) are presented. Rows in bold indicate voltage-gated Na+ (Nav) channel α subunits or previously identified Nav channel interacting proteins.
In addition to iFGF14, the Nav α subunits, Nav1.1, Nav1.2, Nav1.4, and Nav1.6, as well as several proteins previously shown to interact with Nav α subunits, including Navβ1, Navβ2, Navβ4, calmodulin, synaptotagmin-2, ankyrin G, casein kinase II and 14.3.3,22-27 were also identified (Fig. 3 and Table 1). Importantly, with the exceptions of calmodulin and synaptotagmin-2, none of these proteins were present in the Fgf14−/− control IPs. As illustrated in the pie chart in Figure 3, the vast majority of peptides identified were Nav α subunits or previously described Nav α subunit interacting proteins. The average numbers of exclusive unique peptides and total spectra, as well as the average percent amino acid sequence coverage obtained for each protein are provided in Table 1 (and Table S1).
Figure 3.
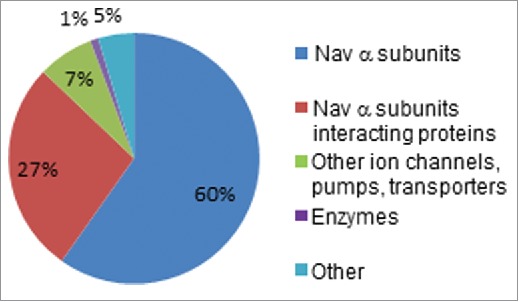
2D-LC-MS/MS Proteomic Analysis of mαFGF14-IPs. Pie chart representing the percentages of unique peptides identified in mαFGF14 antibody immunoprecipitated samples from WT cerebellum that are Nav α subunits, Nav channel accessory subunits, and other, non-Nav channel associated proteins. The vast majority (87%) of the identified peptides correspond to Nav α subunits or proteins known to associate with Nav α subunits. iFGF14 peptides are not included.
Proteins immunoprecipitated with the mαFGF14 antibody from WT and Fgf14−/− cerebellar lysates were also analyzed by Western blot (Fig. 4). As illustrated previously (Fig. 1), iFGF14 is readily detected in, and immunoprecipitated from, WT mouse cerebellar lysates, whereas no iFGF14 was present in lysates or mαFGF14 immunoprecipitated samples from Fgf14−/− mouse cerebellum (Fig. 4A). A high molecular weight (∼250 kDa) band was also detected in the WT IP with the mαFGF14 antibody. This high molecular weight band may reflect iFGF14 bound to Nav α subunits. Consistent with this suggestion, Western blots probed with antibodies against the Nav1.1, Nav1.2 or Nav1.6 revealed high molecular weight bands at ∼250 kDa (Fig. 4A). Immunoblots with the anti-Nav1.1, anti-Nav1.2 and anti-Nav1.6 specific antibodies revealed no apparent difference in the levels of these Nav α subunit proteins in WT and Fgf14−/− cerebellar lysates (Fig. 4A). As expected, however, the various Nav channel α subunits were immunoprecipitated with the mαFGF14 antibody only from WT, and not from Fgf14−/−, cerebellar lysates (Fig. 4A).
Figure 4.
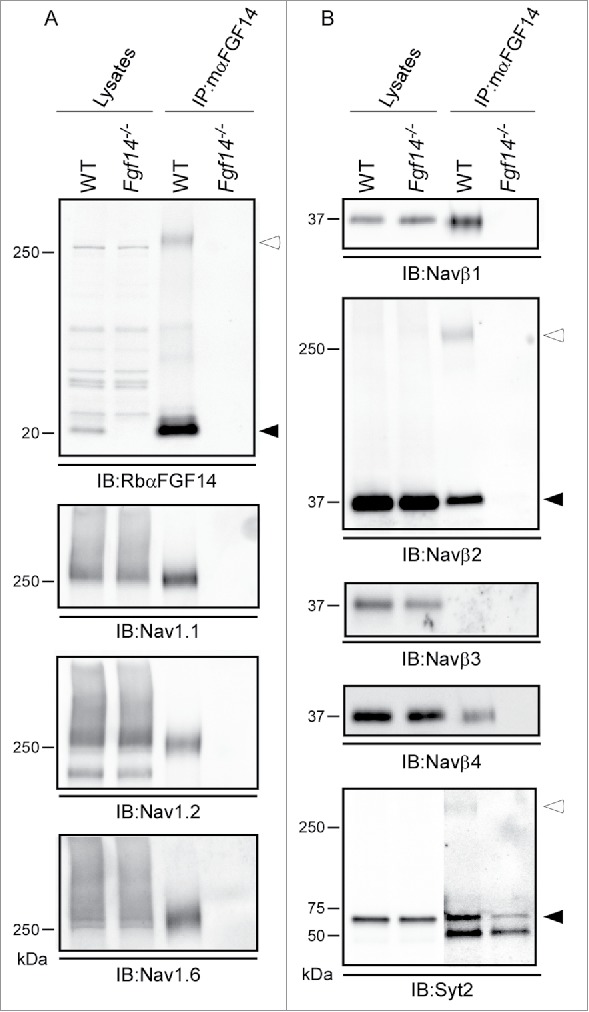
Western Blot Validation of Selected mαFGF14 Immunoprecipitated Proteins Identified by High Resolution 2D-LC-MS/MS. Representative Western blots of cerebellar lysates from WT and Fgf14−/− mice before and after IP with the mαFGF14 antibody. A. mαFGF14-IP of iFGF14 and Nav α subunits. Immunoblots (IB) with RbαFGF14 revealed a 20 kDa band (closed arrowhead) in the WT lysate and IP, but not the Fgf14−/− lysate or IP. An additional ∼250 kDa band (open arrowhead) is present in the WT IP lane, and likely represents iFGF14 bound to Nav α subunits (see text). IB with the anti-Nav1.1, anti-Nav1.2 and anti-Nav1.6 antibodies revealed that these Nav α subunits are present at comparable levels in WT and Fgf14−/− cerebellar lysates, but only in IP with the mαFGF14 antibody from WT cerebellum. B. IBs with the anti-Navβ1, anti-Navβ2, anti-Navβ3, anti-Navβ4 and anti-Syt2 antibodies revealed comparable levels of each of these proteins in the WT and Fgf14−/− cerebellar lysates. Navβ1, Navβ2 and Navβ4, but not Navβ3, were also identified in Western blots following IP with the mαFGF14 antibody from WT cerebellar lysates. Two bands for Navβ2 were evident in the WT-IPs, a ∼37 kDa band (closed arrowhead) representing Navβ2 and a ∼250 kDa band (open arrowhead), likely corresponding to Navβ2 bound to Nav α subunits. IB with the anti-Syt2 antibody revealed that the ∼60 kDa Syt2 protein (closed arrowhead) was immunoprecipitated with the mαFGF14 antibody from WT and Fgf14−/− cerebellar lysates, but was present at a higher level in the WT IP. The anti-Syt2 antibody also recognized an additional ∼250 kDa band (open arrowhead) in the WT IP, suggestive of Syt2 bound to Nav α subunits.
Western blots probed with anti-Nav β subunit (anti-Navβ1, anti-Navβ2, anti-Navβ3 and anti-Navβ4) specific antibodies revealed no apparent differences in the amounts of the Navβ subunit proteins present in WT and Fgf14−/− cerebellar lysates (Fig. 4B). Consistent with the proteomic data, Navβ1, Navβ2 and Navβ4 were readily identified in the observed mαFGF14-IPs from WT, but not from Fgf14−/− cerebellum (Fig. 4B). A high molecular weight (∼250 kDa) band was also identified with the anti-Navβ2 antibody, likely reflecting Navβ2 bound to Nav α subunits. Similar to the proteomic findings (Table 1), Navβ3 was not identified in the Western blot analyses of the proteins that immunoprecipitate with the mαFGF14 antibody-conjugated beads from WT or from Fgf14−/− cerebellar lysates (Fig. 4B). Immunoblots with the anti-synaptotagmin-2 (Syt2) specific antibody revealed no apparent difference in Syt2 protein expression levels in the WT and the Fgf14−/− cerebellar lysates (Fig. 4B). Interestingly, however, and consistent with the MS data (Table 1), the Syt2 protein was present at a much higher level in the blot of the proteins immunoprecipitated with the mαFGF14 antibody from WT, compared with Fgf14−/−, cerebellum (Fig. 4B) (see Discussion).
Western blots of Nav α subunit interacting proteins in WT and Fgf14−/− cerebella
Numerous previous studies have demonstrated that the iFGFs modulate the densities and the voltage-dependent properties of native and heterologously expressed Nav channels.10-14,28-31 In addition, it is clear that the modulatory effects of the iFGFs vary with the cellular environment in which the Nav channels and iFGFs are co-expressed,14 suggesting that the iFGFs may affect the interactions between Nav α and accessory subunits and/or with other Nav channel regulatory proteins in assembled Nav channel complexes. Additional experiments were conducted here to explore the impact of the loss of iFGF14 on the protein components of native cerebellar Nav channel complexes immunoprecipitated from WT and Fgf14−/− cerebellar lysates using an anti-PanNav α subunit-specific mouse monoclonal (mαPanNav) antibody (Fig. 5). WT and Fgf14−/− cerebellar lysates were fractionated and probed with the RbαFGF14, anti-Navβ1, anti-Navβ2 or anti-Pan-iFGF-A antibody (Fig. 5A). The intensities of the identified bands were measured and normalized to the intensities of the Transferrin Receptor (Transferrin-R) bands on the same blots. Analysis of the blots probed with the anti-Navβ1 and anti-Navβ2 antibodies revealed no significant difference in the expression of these proteins in the WT and Fgf14−/− cerebellar lysates (Fig. 5A). Although it appears that the intensity of the bands detected with the anti-pan-iFGF-A antibody, which is targeted against the common region of the iFGF proteins (Fig. 2B), is higher in the Fgf14−/−, than in the WT, cerebellar samples, quantitation (not illustrated) revealed that the difference was not statistically significant.
Figure 5.
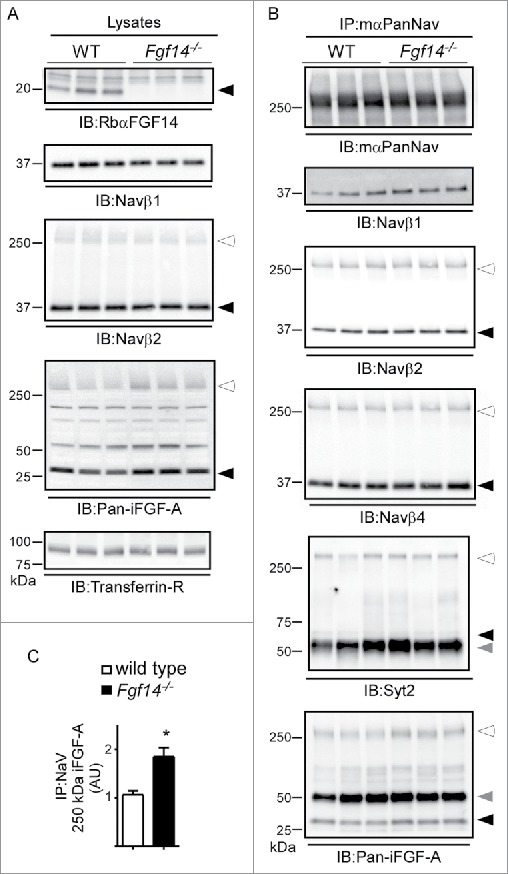
Western Blot Analysis of Nav Channel α Subunits and Nav Channel Interacting Proteins in WT and Fgf14−/− Cerebellar Lysates. A. Representative Western blots of cerebellar lysates from 3 WT and 3 Fgf14−/− animals. Immunoblots (IB) with the RbαFGF14 antibody identified iFGF14 in the 3 WT lanes but not the 3 Fgf14−/− lanes. IB with the anti-Navβ1 and anti-Navβ2 antibodies revealed no significant difference in the amounts of the Navβ1 and Navβ2 proteins in the WT and Fgf14−/− cerebellar lysates. IB with the anti-pan-iFGF-A antibody revealed no significant difference in A-type iFGF expression in WT and Fgf14−/− cerebellar lysates (see text). Closed arrowheads indicate dissociated protein bands and open arrowheads indicate proteins bound to Nav α subunits. B. Native Nav channel complexes were immunoprecipitated from the 3 WT and 3 Fgf14−/− cerebellar lysates depicted in using a monoclonal anti-PanNav α subunit-specific (mαPanNav) antibody and analyzed by Western blot. Similar amounts of Nav α subunit proteins immunoprecipitate from WT and Fgf14−/− cerebellar lysates with the mαPanNav antibody. IB with the anti-Navβ1, anti-Navβ2, anti-Navβ4 and anti-Syt2 antibodies revealed no significant differences in the amounts of the Navβ1, Navβ2, Navβ4 or Syt2 proteins that co-IP with the mαPanNav antibody from WT and Fgf14−/− cerebellar lysates. IB with the anti-Pan-iFGF-A antibody revealed that the amount of A-type iFGFs that co-IP with the mαPanNav antibody and that remain bound to Nav α subunits (˜250 kDa; open arrowhead) are greater in the WT, compared with the Fgf14−/−, cerebellar lysates. The gray arrowheads correspond to IgG heavy chain. C. Quantification of the intensities of the ˜250 kDa anti-Pan-iFGF-A bands, determined from blots such as those in (B), normalized to the amount of immunoprecipitated Nav α subunits in the same samples, revealed a significantly increased (*P<0.05) mean ± SEM band intensity in Fgf14−/−, compared to WT, IPs.
Immunoblots with the mαPanNav antibody revealed no significant difference in Nav α subunits immunoprecipitated from WT and Fgf14−/− cerebella (Fig. 5B). No significant differences in the amounts of Navβ1, Navβ2 or Navβ4 subunits immunoprecipitating from WT and Fgf14−/− with the mαPanNav antibody were observed. Consistent with previous reports that Syt2 directly interacts with Nav α subunits,25 there was no apparent difference in the amount of Syt2 that could be precipitated from WT versus Fgf14−/− cerebellar lysates with the mαPanNav antibody (Fig. 5B). Analyses of the amounts of iFGF-A immunoprecipitating with the mαPanNav antibody revealed that significantly (P < 0.05) more Nav-bound iFGF-A immunoprecipitates with the mαPanNav antibody in Fgf14−/− compared with WT cerebellar lysates (Fig. 5C), suggesting that A-type iFGFs association with Nav channels is increased with the loss of iFGF14 (see Discussion).
Immunoprecipitation of cerebellar Nav channel complexes
To explore the composition of cerebellar Nav channel protein complexes, IP conditions using the mαPanNav antibody and a non-specific mouse immunoglobulin G (mIgG) as a control were optimized. Initial experiments varying the amounts of mαPanNav antibody were used to immunoprecipitate proteins from a constant amount (0.5 mg) of WT cerebellar lysates. As illustrated in Figure 6A, Western blot analyses of cerebellar lysates and immunoprecipitated proteins probed with the mαPanNav antibody revealed robust IP of Nav α subunits from WT mouse cerebellar lysates. The IP of Nav α subunit proteins was specific, as evidenced by the absence of signals in the mIgG control IPs. Increasing the amount of the mαPanNav antibody used in the IP increased the amount of Nav α subunit proteins obtained until a plateau at 3 μg of the antibody was reached. Immunoblotting of the post IP supernatants revealed that about 70% depletion of the Nav α subunit proteins was achieved with 3 (and with 5) μg of the mαPanNav antibody (Fig. 6A, bottom). To determine the optimal amount of protein input to use for the IPs, a constant amount (3 μg) of the mαPanNav antibody was used to IP proteins from varying amounts of WT cerebellar lysates (Fig. 6B). Analyses of Western blots with the mαPanNav antibody revealed that increasing the amount of lysate resulted in increased immunoprecipitated Nav α subunit proteins, with a plateau at 1 mg of lysate. Immunoblotting of the post IP supernatants revealed that 3 μg mαPanNav antibody does not completely clear the Nav α subunit proteins even at the lowest (0.25 mg) of lysate tested (Fig. 6B, bottom): approximately 70% depletion of Nav α subunit proteins from 1 mg of total cerebellar lysate was achieved. These experiments revealed that 1 mg of cerebellar lysate and 3 μg of mαPanNav antibody (or mIgG) are optimal for robust IP of native Nav channel complexes from mouse cerebellum.
Figure 6.
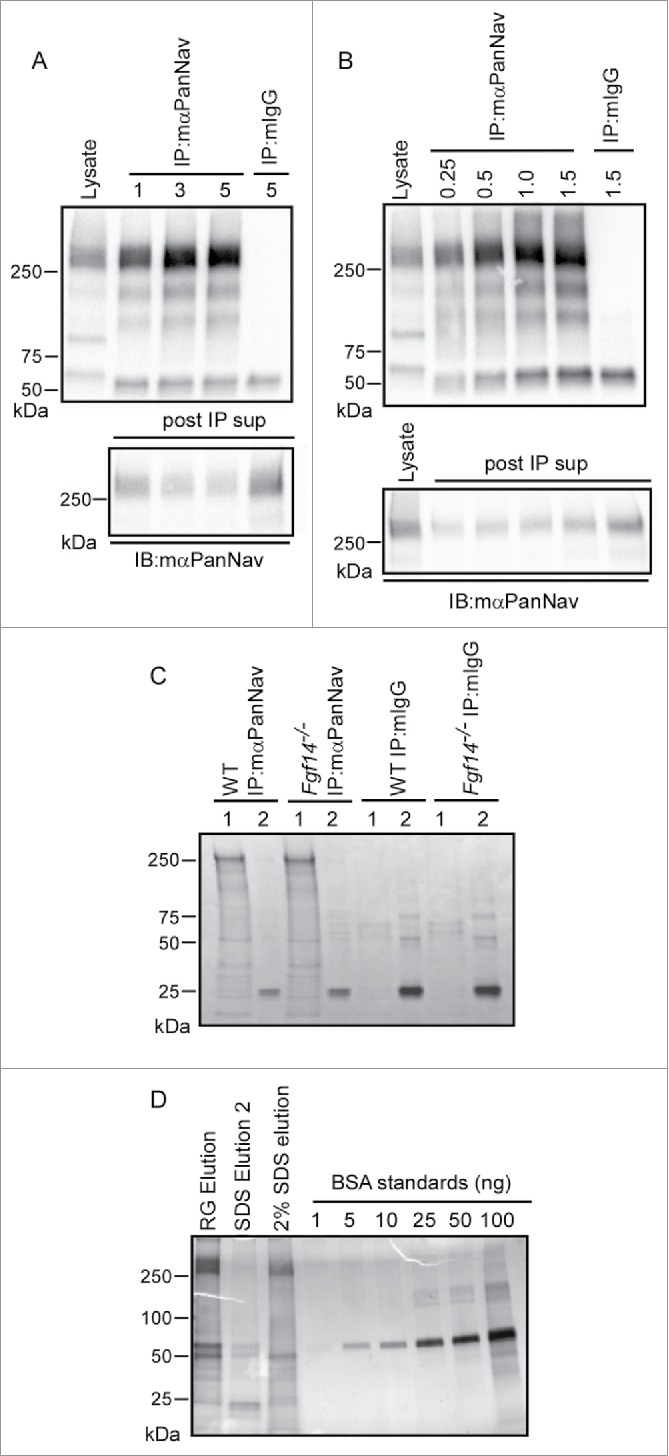
Optimization of mαPanNav Immunoprecipitations. A. Western blots of WT cerebellar lysates and proteins immunoprecipitated from 0.5 mg of WT cerebellar lysates with variable amounts of the mαPanNav antibody or normal mouse IgG (mIgG). Immunoblotting (IB) with the mαPanNav antibody showed no apparent increase in the amount of Nav α subunit proteins precipitating with 3 or 5 μg (compared with 1 µg) of the mαPanNav antibody. Analysis of the corresponding post IP supernatants (post IP sup) revealed that approximately 70% depletion of the Nav α subunit proteins was achieved with 3 μg of antibody. B. Western blots of WT cerebellar lysate and proteins immunoprecipitated with 3 μg of the mαPanNav antibody or mIgG from variable starting amounts of cerebellar protein. IB with the mαPanNav antibody revealed increasing amounts of Nav α subunit proteins precipitating from samples ranging from 0.25 mg to 1 mg total protein, but no further increase when the starting sample was increased to 1.5 mg protein. Analysis of the corresponding post IP supernatants (post IP sup) revealed that approximately 70% depletion of Nav α subunit proteins from the 1 mg protein sample was achieved. C. SYPRO Ruby stained gel of proteins immunoprecipitated with the mαPanNav antibody or mIgG from WT and Fgf14−/− cerebellar lysates. Beads were eluted first with (1) 2% Rapigest, followed by (2) elution with 1% SDS. Proteins running at the molecular weight corresponding to the Nav α subunits (˜250 kDa) are clearly evident in mαPanNav-IPs from WT and Fgf14−/− cerebella. D. Silver stained gel of proteins immunoprecipitated with the mαPanNav antibody from WT cerebellum. Precipitated proteins were analyzed from beads eluted first with 2% Rapigest followed by 1% SDS elution or beads eluted only with 2% SDS. To estimate the amount of Nav α subunit proteins in each IP sample, a bovine serum albumin (BSA) standard curve was also run. Proteins running at the molecular weight corresponding to the Nav α subunits (˜250 kDa) are clearly evident in the Rapigest and 2% SDS elutions. An estimated 50-100 ng of Nav α subunit proteins are present in the Rapigest elution.
To determine whether distinct proteins could be identified in IPs from WT and Fgf14−/− cerebellum, immunoprecipitated proteins were fractionated on 1D-gels and visualized using SYPRO Ruby (Fig. 6C). Proteins were first eluted from mαPanNav- or mIgG-conjugated beads using Rapigest (“1”) followed by a 1% SDS elution (“2”). Analysis of the SYPRO Ruby stained gel revealed several bands that were specific to the mαPanNav-IP and absent from the control mIgG-IP Rapigest elutions. The most intense band ran at a high molecular weight, consistent with the size of Nav α subunits and was present in IPs from both WT and Fgf14−/− cerebellar lysates. No obvious differences in the putative Nav α subunit bands or in the many other bands present in WT and Fgf14−/− mαPanNav-IPs were observed. To estimate the amount of the Nav α subunit proteins present in the mαPanNav-IPs, immunoprecipitated proteins from a WT IP eluted with Rapigest (RG) followed by 1% SDS (SDS Elution2) or a WT IP eluted with 2% SDS only were fractionated on a 1D-gel and visualized by silver staining (Fig. 6D). For estimation of protein content, BSA standards were also run. As illustrated in Fig. 6D, the silver staining revealed robust bands at a molecular weight (∼250 kDa) corresponding to Nav α subunit proteins. The Rapigest appears to be more efficient at eluting proteins from mαPanNav-beads than the 2% SDS. Comparison of the band intensities of the BSA standards with the IP samples provided an estimate of 50 ng of Nav α subunit protein (∼250 kDa band) in the 10% of sample run on the gel, or a total of 0.5 μg of Nav α subunit proteins in the sample.
Protein components of WT and Fgf14−/− cerebellar Nav channel complexes
To identify and compare the protein components of native Nav channel complexes in WT and Fgf14−/− cerebellum, samples were analyzed using 2D-LC-MS/MS. The most abundant protein identified in the mαPanNav-IPs was Nav1.2 (Table 2). A representative fragmentation spectrum of a Nav1.2 tryptic peptide, as well as the amino acid sequence derived from this spectrum, is illustrated in Figure 7A. This analysis yielded a total of 18 unique Nav1.2 peptides in the WT IP and 26 unique peptides in the Fgf14−/− IP, equivalent to 12% or 16% amino acid sequence coverage of the Nav1.2 protein (Fig. 7B and Table 2). Notably, all identified Nav1.2 peptides are located in the intracellular cytoplasmic domains; no peptides from the transmembrane domains were found. These analyses also identified Nav1.1, previously identified Nav channel interacting proteins, including Navβ2 and calmodulin, and a number of potentially novel Nav channel interacting proteins (Table 2). None of these proteins, with the exception of calmodulin, were identified in the mIgG control IPs.
Table 2.
Proteins identified in native WT and Fgf14−/− cerebellar Nav channel complexes.
| Identified Protein | Gene | Number of unique peptides (total spectra) | % amino acid sequence coverage from WT (Fgf14−/−) IPs | |
|---|---|---|---|---|
| WT | Fgf14−/− | |||
| Voltage-gated Na+ channel α subunit Nav1.2 | Scn2a | 18 (28) | 26 (44) | 12 (16) |
| Calmodulin* | Calm | 8 (27) | 8 (29) | 31 (31) |
| Cofilin-1 | Cfl1 | 6 (11) | 7 (12) | 46 (51) |
| Microtubule-associated protein 6 | Map6 | 6 (8) | 11 (13) | 8 (14) |
| Reticulocalbin-2 | Rcn2 | 4 (4) | 5 (7) | 20 (25) |
| Purkinje cell protein 2 | Pcp2 | 3 (5) | 3 (4) | 32 (32) |
| Voltage-gated Na+ channel β2 subunit | Scn2b | 3 (3) | 3 (3) | 16 (16) |
| Polymerase delta-interacting protein 3 | Poldip3 | 3 (3) | 9 (11) | 11 (35) |
| Myosin light polypeptide 6 | Myl6 | 3 (3) | 5 (5) | 24 (38) |
| Voltage-gated Na+ channel α subunit Nav1.1 | Scn1a | 2 (3) | 4 (4) | 2.9 (5) |
| Tubulin α−1B chain | Tuba1b | 2 (3) | 3 (5) | 6 (10) |
| ATP synthase-coupling factor 6, mitochondrial | Atp5j | 2 (3) | 2 (3) | 35 (35) |
| ADP/ATP translocase 1 | Slc25a4 | 2 (3) | 2 (3) | 8 (8) |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 3 | Chchd3 | 2 (2) | 0 | 10 (0) |
| Translation initiation factor IF-2, mitochondrial | Mtif2 | 1 (1) | 2 (3) | 2 (4) |
| 78 kDa glucose-regulated protein | Hspa5 | 1 (1) | 5 (6) | 2 (9) |
| 2-oxoisovalerate dehydrogenase subunit α, mitochondrial | Bckdha | 0 | 9 (12) | 0 (26) |
| ATP synthase subunit d, mitochondrial | Atp5h | 0 | 3 (3) | 0 (28) |
| RNA-binding protein FUS | Fus | 0 | 2 (2) | 0 (5) |
Notes. The number of exclusive unique peptides and total spectra as well as the percent amino acid sequence coverage obtained for each protein are presented. Rows in bold indicate voltage-gated Na+ (Nav) channel α subunits or previously identified Nav channel interacting proteins.
There were also calmodulin peptides identified in the mIgG-IPs from WT (n = 3 peptides) and Fgf14−/− (n = 2 peptides) cerebellar lysates.
Figure 7.
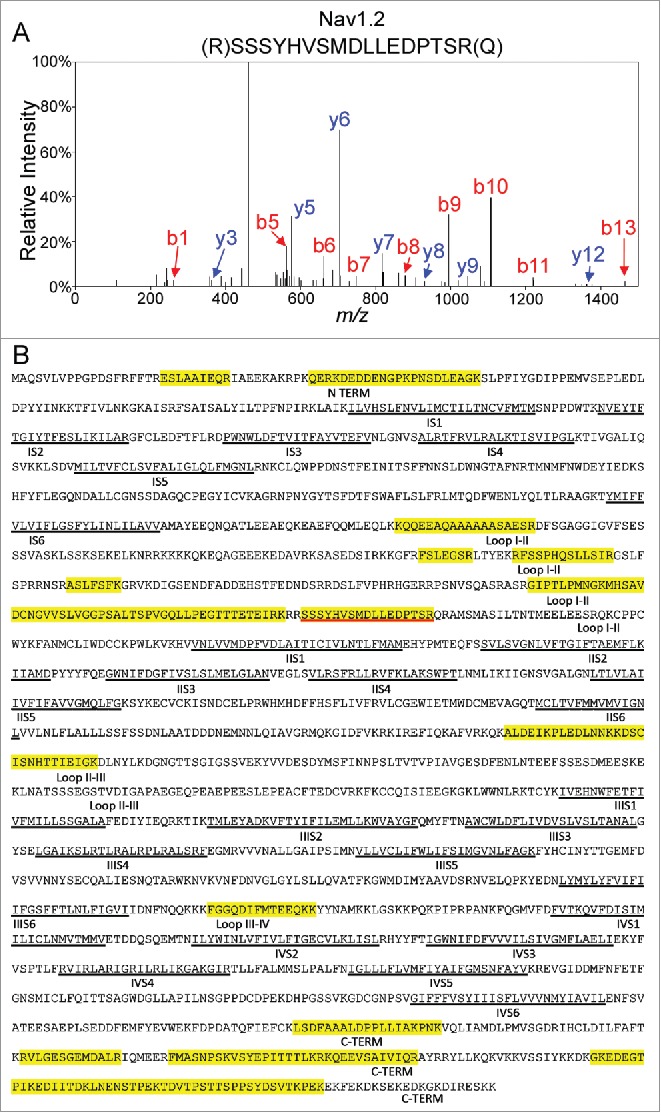
Mass Spectrometric Identification of Nav1.2 Using In-Solution 2D-LC-MS/MS. A. Representative MS2 fragmentation spectrum of one of the identified Nav1.2 tryptic peptides, corresponding to the sequence NH2-(R)SSSYHVSMDLLEDPTSR(Q) with the y- (in blue) and b- (in red) ions highlighted. B. Amino acid sequence coverage obtained for the mouse Nav1.2 protein following IP of cerebellar Nav channel complexes and LC-MS/MS proteomic analysis. Detected peptides are highlighted in yellow. The peptide for which the fragmentation spectrum is shown (in A) is underlined in red. Transmembrane segments (S1-S6) in each domain (I-IV) are underlined in black. Interdomain cytoplasmic loops I-II, II-III, and III-IV, as well as the N-terminal and carboxyl terminal domains, are also indicated.
The numbers of unique peptides and total spectra obtained for each protein in the WT and Fgf14−/− mαPanNav-IP samples, as well as the amino acid sequence coverages obtained for each, are provided in Table 2 (and Table S2). As is evident, the numbers of peptides identified in the mαPanNav-IPs from the WT and Fgf14−/− samples are similar. Interestingly, and in contrast with the MS-based proteomic analysis of the complexes immunoprecipitated from WT cerebellar lysates with the mαFGF14 antibody, Nav1.6 was not identified in the mαPanNav-IPs from the WT (or the Fgf14−/−) cerebellar lysates (see Discussion).
Discussion
The experiments here used an anti-iFGF14 mouse monoclonal antibody that provides efficient immunoprecipitation of the iFGF14 protein from native tissue to enable the first MS-based analysis of native iFGF14 protein complexes. The results of these experiments reveal that Nav α subunits and multiple previously identified and putative Nav α interacting proteins co-immunoprecipitate with iFGF14 from adult WT mouse cerebellum. In contrast, no pore-forming α subunits or accessory subunits of other types of ion channels were revealed in these experiments. Further proteomic experiments here revealed that the protein components of native cerebellar Nav channel complexes in the presence and absence of iFGF14 are quite similar, suggesting that iFGF14 does not measurably impact the association between Nav α subunits and Nav channel interacting proteins.
Mass spectrometry-based identification of native cerebellar iFGF14 complexes
The IP approach of purifying iFGF14 complexes from mouse cerebellum was efficient, and the optimization steps allowed us to maximize the amount of iFGF14 protein obtained. Although we were not able to clear 100% of the iFGF14 protein from mouse cerebellar lysates with a single IP, we routinely achieved 70-80% depletion. The iFGF14 in the supernatant could be further depleted with sequential IPs from the same lysate. The proteomic analyses of native cerebellar iFGF14 complexes presented here revealed that the vast majority (87%) of the peptides identified in adult WT mouse cerebellar iFGF14 complexes reflect Nav channel α subunits or proteins previously shown, or suggested, to interact directly with Nav α subunits.22-27 For example, we identified 3 of the 4 major central nervous system Nav channel α subunits,32 Nav1.1, Nav1.2, and Nav1.6. These findings are in accordance with previous interaction data13,14,17 and Nav-iFGF crystal structures15,16 demonstrating that iFGFs interact directly with the C-terminal domain of Nav channel α subunits. On a functional point of view, this large relative representation of Nav channel α subunits in cerebellar iFGF14 immunoprecipitates is consistent with the key role of iFGFs in the regulation of spontaneous and evoked action potential firing in adult cerebellar granule and Purkinje neurons.11,12 We also identified Nav1.4, the skeletal muscle Nav channel α subunit,33 suggesting a role for Nav1.4-encoded channels in the cerebellum. Interestingly, the finding that Nav1.4 co-immunoprecipitates with iFGF14 could also be important in light of the muscle weakness phenotype seen in Fgf14−/− animals.8,9
Native cerebellar iFGF14 complexes also contained numerous proteins previously found to interact with Nav α subunits, including Navβ1, Navβ2, Navβ4, synaptotagmin-2, calmodulin, ankyrin G, casein kinase II and 14.3.3.22-27 Further experiments will be needed to determine whether these proteins immunoprecipitate with iFGF14 as passenger proteins bound to Nav α subunits or interact directly with iFGF14. Several novel, putative iFGF14 interacting proteins were also identified in native cerebellar iFGF14 complexes, although peptides from these proteins were much less abundant that those from Nav α subunits and other Nav channel associated proteins. Additional biochemical analyses will be necessary to characterize these interactions and functional assays will be needed to determine the potential physiologic role of these interactions.
It was recently reported that iFGF14 regulates cerebellar granule neuron to Purkinje neuron synaptic transmission by modulating the functional density of presynaptic voltage-gated Ca2+ (Cav) channels, although a direct interaction between iFGF14 and Cav channels could not be demonstrated.19 It was also recently reported that iFGF13 regulates the functional cell surface expression of Cav channels in mouse ventricular myocytes, although, again, no direct interaction between iFGF13 and Cav channel subunits was found.18 No Cav channel subunits were identified in the MS-based proteomic analysis of native mouse cerebellar iFGF14 complexes in the present study. Consistent with these findings, a recent proteomic study of neuronal Cav2 channel complexes isolated from adult rat/mouse brain did not identify iFGF14 or other iFGFs as components of native Cav2 channel complexes.34 It is possible that iFGF14 interacts directly with Cav channel subunits to regulate Cav current densities in the cerebellum, but that these interactions are low affinity and do not survive the protein isolation steps used. It is also possible that iFGFs interact transiently or indirectly with Cav channel subunits. Interestingly, synaptotagmin-2, one of the most abundant proteins identified in the iFGF14-IPs from WT cerebellum other than Nav α subunits, has been previously demonstrated to interact directly with brain Cav channels.35,36 Thus, a potentially interesting hypothesis is that iFGF14 regulates Cav channels via interaction with synaptotagmins.
Protein components of native WT and Fgf14−/− mouse cerebellar Nav channel complexes
The MS-based proteomic approach offers several advantages to conventional Western blot analyses including the fact that it is unbiased and that interactions with multiple channel subunits and accessory proteins can potentially be assessed at the same time. We speculated that this approach could also potentially enable determination of the effects of the loss of iFGF14 on the associations between Nav α subunits and Nav channel interacting proteins. In this second scenario, one would expect to identify some interacting proteins present only in Nav channel complexes from Fgf14−/− cerebella. The analyses identified the Nav1.2 and Nav1.1 α subunits, as well as Navβ2 and calmodulin, in the mαPanNav-IPs, but not in the control mIgG-IPs, from WT and Fgf14−/− cerebellar lysates. Interestingly, and in marked contrast with the MS-based proteomic analysis of the complexes immunoprecipitated from WT cerebellar lysates with the mαFGF14 antibody, Nav1.6 was not identified in the mαPanNav-IPs from the WT (or the Fgf14−/−) cerebellar lysates (Table 2). These findings suggest the interesting hypothesis that Nav1.6 is enriched in the mαFGF14-IPs because it is preferentially associated with iFGF14. It is certainly also possible, however, that the mαPanNav antibody has a higher affinity for Nav1.2 and Nav1.1 than for Nav1.6, resulting in more efficient IP of these subunits relative to Nav1.6. Additional experiments with other, perhaps Nav α subunit-specific antibodies, aimed at exploring these hypotheses directly will be of interest.
Relatively few differences in the protein compositions of Nav channel complexes isolated from WT vs. Fgf14−/− cerebella were found, suggesting that FGF14 does not recruit or displace other Nav α subunit interacting proteins. The Western Blot analyses here revealed that substantially more A-type iFGFs immunoprecipitate with Nav α subunits from Fgf14−/−, compared to WT, cerebellar lysates (Fig. 5), suggesting that the association of A-type iFGFs with Nav channels is increased with the loss of iFGF14. In addition, several proteins were identified in the Nav channel complexes immunoprecipitated from Fgf14−/−, but not WT, cerebella (Table 2), although these were mostly mitochondrial proteins or proteins involved in transcription and translation, and it is unclear whether they actually interact with and regulate neuronal Nav channels. Additional biochemical and functional studies will be required to determine the possible physiological roles of these novel proteins.
A potential limitation of these analyses is that the protein content of the IP samples obtained with the mαPanNav used here appears to be quite a bit lower than what was obtained in the mαFGF14-IP samples. For example, in the samples immunoprecipitated with the anti-iFGF14 antibody, we obtained 158 total spectra corresponding to Nav1.2 peptides, whereas we only obtained about 30 with the mαPanNav antibody. This lower IP yield also certainly explains the fact that no iFGF proteins were detected in the mαPanNav-IPs. Although the yield of Nav channel complexes could potentially have been increased by using additional cerebellar lysates, the high cost of the mαPanNav antibody was limiting. Generation of a monoclonal or polyclonal anti-Nav α subunit specific antibody that more efficiently immunoprecipitates native Nav channel complexes, however, might be a more productive experimental strategy for further analyses of the components of native Nav channel complexes.
Materials and methods
Animals
Wild type (WT) and Fgf14−/− mice8 in the C57BL/6J background were used in the experiments presented here. Animals were handled in accordance with the Guide for the Care and Use of Laboratory Animals (NIH), and all protocols were approved by the Washington University Animal Studies Committee.
Preparation of anti-iFGF14 or anti-PanNav α subunit antibody-coupled beads
A mouse monoclonal anti-iFGF14 antibody (mαFGF14, UC Davis/NIH NeuroMab Facility, clone N56/21), special ordered in Phosphate Buffered Saline (PBS) and coupled to CNBr-activated sepharose 4B beads (GE Healthcare) according to the manufacturer's instructions, was used for IP of iFGF14. Briefly, 1 g lyophilized CNBr-activated sepharose beads were suspended in 5 ml of dilute (1 mM) HCl, transferred to a sintered glass filter with medium porosity and washed with 200 ml dilute HCl over 15 min. 1.5 ml of washed sepharose beads were transferred to a 5 ml round bottom tube and incubated with 1 mg mαFGF14 antibody and 2 ml coupling buffer containing 0.1 M NaHCO3, 0.5 M NaCl, pH 8.3 overnight at 4°C with rotational mixing. Following incubation, beads were washed with 5 gel volumes of coupling buffer. Residual coupling sites were blocked by incubation in one gel volume of 0.1 M Tris-HCl (pH 8.0) for 4 h at 4°C with rotational mixing. Sepharose beads were then washed with 3 cycles (alternating pH) of buffer consisting of 5 gel volumes of 0.1 M acetic acid, 0.5 M NaCl, at pH 4.0 followed by 5 gel volumes of buffer containing 0.1 M Tris-HCl, 0.5 M NaCl, at pH 8.0. Sepharose beads were stored in IP buffer (20 mM Tris, 150 mM NaCl, pH 7.4 with 1% Triton X-100) containing 0.02% NaN3. For controls, the sepharose was hydrated with dilute HCl as above and coupling sites were quenched in 0.1 M Tris-HCl, pH 8.0 at 4°C. Control beads were washed and stored in IP buffer containing 0.02% NaN3.
A mouse monoclonal anti-PanNav α subunit-specific (mαPanNav, Sigma, #S8809) antibody was conjugated to Protein G Dynabeads (Thermo Fisher Scientific). Briefly, following washing with PBS, beads were suspended in 500 μl PBS containing the mαPanNav antibody and incubated with rotational mixing for 1 h at room temperature (RT). For Western blot experiments, 3 μg of the mαPanNav antibody and 20 μl protein G Dynabeads were used for each IP, unless noted otherwise. For proteomic experiments, 25 μg of the mαPanNav antibody and 166 μl of protein G Dynabeads were used. Control IgG beads were prepared using the same amounts (3 μg or 25 μg) of normal mouse IgG (mIgG, Santa Cruz Biotechnology, Inc.) instead of the mαPanNav antibody. Following incubation with the antibody or mIgG, beads were washed 3 times with 1 ml PBS containing 0.1% Tween-20 (PBST) and twice with 1 ml of cross-linking buffer containing 0.2 M triethanolamine, pH 8.2. Antibodies were cross-linked to beads by incubation with 20 mM dimethyl pimelimidate (DMP, Thermo Fisher Scientific) in cross-linking buffer for 30 min at RT with rotational mixing. The cross-linking reaction was stopped by the addition of 50 mM Tris-HCl (pH 7.5). After mixing for 15 min, the beads were washed as follows: 1) once with PBS; 2) once with 0.1 M citrate, pH 3.0; 3) once with 0.1 M Na-phosphate buffer, pH 8.1; 4) twice with PBST; and 5) twice with PBS. Beads were stored (up to 1 week) in PBS with 0.02% NaN3.
Immunoprecipitation of cerebellar iFGF14 or Nav channel complexes
Cerebella from 2-7 month-old male and female WT or Fgf14−/− mice were combined and homogenized in ice cold lysis buffer containing 20 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1:100 protease inhibitor cocktail (Protease Inhibitor Cocktail Set III, EDTA-Free, Merck Millipore). Homogenates were incubated for 15 min at 4°C with rotational mixing and centrifuged at 3000 rpm at 4°C for 10 min to remove the insoluble fraction. Protein concentrations were measured with the BCA assay (Thermo Fisher Scientific).
For mass spectrometric analysis of proteins immunoprecipitated with the mαFGF14-coupled beads, cerebella obtained from 25 WT and 25 Fgf14−/− mice were pooled. The pooled cerebellar lysates were precleared with 200 μl of control sepharose beads for 1 h at 4°C with rotational mixing. Each of the cleared lysates (8 mg) was added to 10 μl of mαFGF14-coupled sepharose beads in 4 replicates and incubated with rotational mixing overnight at 4°C. Beads were washed 4 times with ice cold lysis buffer and twice with ice cold PBS. Proteins were eluted from the beads in 2% Rapigest (Waters), 8 M urea (Sigma), 100 mM Tris (pH 8.5) at 37 °C for 30 min.
For mass spectrometric analysis of immunoprecipitated Nav channel proteins, cerebella from 4 WT and 4 Fgf14−/− mice were pooled. Protein lysates were precleared with 322 μl Protein G Dynabeads, and 8 mg of the cleared cerebellar protein lysate was added to 166 μl mαPanNav- or mIgG-coupled Protein G Dynabeads. IPs, washing, and elution of beads were performed as above.
For Western blot analyses of proteins co-immunoprecipitating with iFGF14 or Nav α subunits, 1-3 mg of cerebellar proteins was added to 10 μl of mαFGF14-coupled sepharose beads or 20 μl of mαPanNav-coupled Dynabeads for each IP, unless otherwise noted. Beads were washed 3 times with ice cold lysis buffer and proteins were eluted in 2% sodium dodecyl sulfate (SDS) at 55°C for 15-20 min or 85°C for 5 min.
Endoprotease digestions
Peptides were generated from proteins eluted from antibody bead IPs by endoprotease digestion under denaturing conditions. The eluted proteins were precipitated using the 2D protein clean up kit (GE Healthcare). The resulting pellets were dissolved in 8 M urea, 100 mM Tris (pH 8.5), reduced with 5 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride, pH 8.0) for 30 min at RT, and alkylated with 10 mM iodoacetamide (Bio-Rad) for 30 min at RT. Samples were then digested with 1 μg of endoproteinase Lys-C (Roche) overnight at 37°C, followed by trypsin (4 μg) (Sigma) overnight at 37°C. The digests were acidified with formic acid to a final concentration of 1%, extracted with NuTip porous graphite carbon wedge tips (Glygen), and eluted with aqueous acetonitrile (60%) containing formic acid (0.1%). The extracted peptides were dried, dissolved in aqueous acetonitrile/formic acid (1%/1%), stored at −80°C and analyzed using LC-MS.
Nano-liquid chromatography-mass spectrometry (LC-MS)
A 2D Plus (Eksigent) LC with a Nanoflex module and AS2 autosampler were coupled to a TripleTOF® 5600+ mass spectrometer (SCIEX). The 2D LC system was configured to load samples in tandem. The cHiPLC® columns (ChromXP C18 200 µm X 15 cm; particle size 3 µm, 120 Å) were equilibrated in aqueous acetonitrile (1%) containing 1% formic acid, solvent A (99% water/1% acetonitrile). The samples were loaded in a volume of 10 µl at a flow rate of 1.5 µl/min followed by organic gradient elution of peptides at a flow rate of 800 nl/min. The organic acetonitrile gradient was produced by increasing the proportion of solvent B (1% formic acid in 99% acetonitrile) relative to solvent A as follows: at 0 time, 98% solvent A, 2% solvent B; at 5 min, 2% A, 98% B; at 415 min, 65% A, 35% B; and, at 440 min, 20% A, 80% B. The initial chromatographic conditions were restored in 5 min and maintained for 20 min.
MS data acquisition was performed with a TripleTOF® 5600+ mass spectrometer (SCIEX) interfaced to the nano-chromatography with a Digital Picoview Nanospray source (New Objective, Inc.) via a 10 µm Silica PicoTip emitter (New Objective) and operated with a resolution of > 30,000fwhm for TOFMS scans. Data were acquired with the ion spray voltage at 2.9 kV, curtain gas at 10 PSI, nebulizer gas at 14 psi, and the interface heater temperature of 175 °C. For data-dependent acquisition, survey scans were acquired in 250 ms; from these, 50 product ion scans were selected for MS2 acquisition with a dwell time of 20 ms. Four time bins were summed for each scan at a frequency of 15.4 kHz (through monitoring of the 40 GHz multichannel TDC detector with 4-anode/channel detection). A rolling collision energy was applied to all precursor ions for collision-induced dissociation using the equation: CE = slope * m/z + intercept, where the slope for all charge states above +2 is 0.0625 and the intercept is −3,−5 and −6 for +2, +3 and +4, respectively.
MS data processing
Data were processed using the SCIEX MS Data Converter v 1.3 (SCIEX), converting the raw data files (*.wiff) to mgf files for protein database searching. The UNIPROT mouse protein database (downloaded April 21, 2011, with 105,706 sequences) was searched using Mascot software (ver. 2.2.04), allowing for up to 4 missed cleavages. The parent and product mass search tolerances were set at 0.8 Da and 20 millimass units, respectively. Carbamidomethyl was set as a fixed modification for cysteine residues and methionine residue oxidation was allowed as a variable modification. The protein database searches were analyzed using Scaffold (ver. 3_00_07) and proteins were identified using Prophet37,38 with protein and peptide thresholds of 99% and 90%, respectively. Tables S1 and S2 provide the complete list of identified peptides and proteins in the mαFGF14- and mαPanNav-IPs, respectively, and Figure S1 provides the annotated spectra of one hit proteins.
Gel electrophoresis and western blot analyses
Prior to electrophoresis, 50 mM TCEP (Thermo Fisher Scientific) was added and proteins were heated to 55°C for 15 min. Proteins were electrophoresed on 4-15% precast polyacrylamide gels (Bio-Rad) and analyzed by SYPRO Ruby staining (Thermo Fisher Scientific), silver staining (SilverQuest, Thermo Fisher Scientific) or Western blotting. SYPRO Ruby and silver staining were performed according to manufacturer's instructions. For Western blots, resolved proteins were transferred to PVDF membranes (Merck Millipore) for 1.5 h at 4°C and blocked for 1h at RT in PBS with 3% nonfat dry milk and 0.1% Tween-20. Membranes were incubated in primary antibodies diluted in PBS with 0.5% nonfat dry milk and 0.1% Tween-20 for 2 h at RT or overnight at 4°C. Washed membranes were incubated with goat-anti-mouse-HRP or goat-anti-rabbit-HRP (1:5000, Santa Cruz Biotechnology) and visualized with the Super Signal Femto chemiluminescence substrate (Thermo Fisher Scientific). In some Western blot experiments, membranes were stripped and reprobed with a second primary antibody.
The following primary antibodies were used: mouse monoclonal anti-panNav α subunit-specific (mαPanNav, 1:1000, Sigma, #S8809); mouse monoclonal anti-Nav1.1 (1:1000, UC Davis/NIH NeuroMab Facility, clone K74/71); mouse monoclonal anti-Nav1.2 (1:1000 UC Davis/NIH NeuroMab Facility, clone K69/3); rabbit polyclonal anti-Nav1.6 (1:300, gift from J. Trimmer); mouse monoclonal anti-znp1 (1:5000, Zebrafish International Resource Center) which recognizes mouse synaptotagmin-239; rabbit polyclonal anti-Navβ1 (1:5000)40; rabbit polyclonal anti-Navβ2 (1:1000)40; rabbit polyclonal anti-Navβ3 (1:1000)40; rabbit polyclonal anti-Navβ4 (1:4000)40; mouse monoclonal anti-transferrin receptor (1:500, Thermo Fisher Scientific, #13-6890); mouse monoclonal anti-pan-iFGF-A (1:1000, UC Davis/NIH NeuroMab, clone N253/22); and, rabbit polyclonal anti-FGF14 (1:1000). The rabbit polyclonal antibody against iFGF14 (RbαFGF14) was generated by inoculating rabbits with an iFGF14 peptide; raw antiserum was used for immunoblotting.14
Statistics
Results in Fig. 5C are expressed as means ± SEM, and the statistical analysis was performed using the Mann-Whitney test. Identification probabilities for proteins (in Tables S1 and S2) were obtained in Scaffold using Prophet.38
Supplementary Material
Abbreviations
- iFGF
intracellular Fibroblast Growth Factor
- IP
Immunoprecipitation
- MS
Mass Spectrometry
- mαFGF14
Anti-Fibroblast Growth Factor 14 monoclonal antibody
- mαPanNav
Anti-NaV α subunit monoclonal antibody
- mIgG
Mouse Immunoglobulin G
- NaV α subunit
Voltage-gated Na+ (NaV) channel pore-forming (α) subunit
- WT
Wild-Type
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Jun-Yang Lou for generating the polyclonal rabbit anti-FGF14 antibody used in the Western blot analyses described. We also thank L. Li for maintaining the Fgf14−/− mouse colony, P. Erdman-Gilmore and J.P. Malone for technical assistance.
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke (R01-NS065761 to DMO and JMN). MKB was supported by NIH Training Grants T32-GM007200 and T32-HL007275. The Proteomics Core Laboratory is supported by NIH CTSA Grant #UL1 TR000448. Financial support provided by the Marie Curie 7th Framework Program of the European Commission (PIRG06-GA-2009-256397 to CM) and the Fondation d'entreprise Genavie (to CM) is also gratefully acknowledged.
References
- [1].Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev 2005; 16:215-20; PMID:15863036; http://dx.doi.org/ 10.1016/j.cytogfr.2005.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem 2003; 278:34226-36; PMID:12815063; http://dx.doi.org/ 10.1074/jbc.M303183200 [DOI] [PubMed] [Google Scholar]
- [3].Brusse E, de Koning I, Maat-Kievit A, Oostra BA, Heutink P, van Swieten JC. Spinocerebellar ataxia associated with a mutation in the fibroblast growth factor 14 gene (SCA27): A new phenotype. Mov Disord 2006; 21:396-401; PMID:16211615; http://dx.doi.org/ 10.1002/mds.20708 [DOI] [PubMed] [Google Scholar]
- [4].Coebergh JA, Fransen van de Putte DE, Snoeck IN, Ruivenkamp C, van Haeringen A, Smit LM. A new variable phenotype in spinocerebellar ataxia 27 (SCA 27) caused by a deletion in the FGF14 gene. Eur J Paediatr Neurol 2014; 18:413-5; PMID:24252256; http://dx.doi.org/ 10.1016/j.ejpn.2013.10.006 [DOI] [PubMed] [Google Scholar]
- [5].Dalski A, Atici J, Kreuz FR, Hellenbroich Y, Schwinger E, Zuhlke C. Mutation analysis in the fibroblast growth factor 14 gene: frameshift mutation and polymorphisms in patients with inherited ataxias. Eur J Hum Genet 2005; 13:118-20; PMID:15470364; http://dx.doi.org/ 10.1038/sj.ejhg.5201286 [DOI] [PubMed] [Google Scholar]
- [6].Misceo D, Fannemel M, Baroy T, Roberto R, Tvedt B, Jaeger T, Bryn V, Stromme P, Frengen E. SCA27 caused by a chromosome translocation: further delineation of the phenotype. Neurogenetics 2009; 10:371-4; PMID:19471976; http://dx.doi.org/ 10.1007/s10048-009-0197-x [DOI] [PubMed] [Google Scholar]
- [7].van Swieten JC, Brusse E, de Graaf BM, Krieger E, van de Graaf R, de Koning I, Maat-Kievit A, Leegwater P, Dooijes D, Oostra BA, et al.. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia [corrected]. Am J Hum Genet 2003; 72:191-9; http://dx.doi.org/ 10.1086/345488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang Q, Bardgett ME, Wong M, Wozniak DF, Lou J, McNeil BD, Chen C, Nardi A, Reid DC, Yamada K, et al.. Ataxia and paroxysmal dyskinesia in mice lacking axonally transported FGF14. Neuron 2002; 35:25-38; PMID:12123606; http://dx.doi.org/ 10.1016/S0896-6273(02)00744-4 [DOI] [PubMed] [Google Scholar]
- [9].Wozniak DF, Xiao M, Xu L, Yamada KA, Ornitz DM. Impaired spatial learning and defective theta burst induced LTP in mice lacking fibroblast growth factor 14. Neurobiol Dis 2007; 26:14-26; PMID:17236779; http://dx.doi.org/ 10.1016/j.nbd.2006.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shakkottai VG, Xiao M, Xu L, Wong M, Nerbonne JM, Ornitz DM, Yamada KA. FGF14 regulates the intrinsic excitability of cerebellar Purkinje neurons. Neurobiol Dis 2009; 33:81-8; PMID:18930825; http://dx.doi.org/ 10.1016/j.nbd.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bosch MK, Carrasquillo Y, Ransdell JL, Kanakamedala A, Ornitz DM, Nerbonne JM. Intracellular FGF14 (iFGF14) Is Required for Spontaneous and Evoked Firing in Cerebellar Purkinje Neurons and for Motor Coordination and Balance. J Neurosci 2015; 35:6752-69; PMID:25926453; http://dx.doi.org/ 10.1523/JNEUROSCI.2663-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Goldfarb M, Schoorlemmer J, Williams A, Diwakar S, Wang Q, Huang X, Giza J, Tchetchik D, Kelley K, Vega A, et al.. Fibroblast growth factor homologous factors control neuronal excitability through modulation of voltage-gated sodium channels. Neuron 2007; 55:449-63; PMID:17678857; http://dx.doi.org/ 10.1016/j.neuron.2007.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Laezza F, Lampert A, Kozel MA, Gerber BR, Rush AM, Nerbonne JM, Waxman SG, Dib-Hajj SD, Ornitz DM. FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol Cell Neurosci 2009; 42:90-101; PMID:19465131; http://dx.doi.org/ 10.1016/j.mcn.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lou JY, Laezza F, Gerber BR, Xiao M, Yamada KA, Hartmann H, Craig AM, Nerbonne JM, Ornitz DM. Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J Physiol 2005; 569:179-93; PMID:16166153; http://dx.doi.org/ 10.1113/jphysiol.2005.097220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Goetz R, Dover K, Laezza F, Shtraizent N, Huang X, Tchetchik D, Eliseenkova AV, Xu CF, Neubert TA, Ornitz DM, et al.. Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J Biol Chem 2009; 284:17883-96; PMID:19406745; http://dx.doi.org/ 10.1074/jbc.M109.001842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang C, Chung BC, Yan H, Lee SY, Pitt GS. Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure 2012; 20:1167-76; PMID:22705208; http://dx.doi.org/ 10.1016/j.str.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang C, Hoch EG, Pitt GS. Identification of novel interaction sites that determine specificity between fibroblast growth factor homologous factors and voltage-gated sodium channels. J Biol Chem 2011; 286:24253-63; PMID:21566136; http://dx.doi.org/ 10.1074/jbc.M111.245803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hennessey JA, Wei EQ, Pitt GS. Fibroblast growth factor homologous factors modulate cardiac calcium channels. Circ Res 2013; PMID:23804213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yan H, Pablo JL, Pitt GS. FGF14 Regulates Presynaptic Ca(2+) Channels and Synaptic Transmission. Cell Rep 2013; 4:66-75; PMID:23831029; http://dx.doi.org/ 10.1016/j.celrep.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang Q, McEwen DG, Ornitz DM. Subcellular and developmental expression of alternatively spliced forms of fibroblast growth factor 14. Mech Dev 2000; 90:283-7; PMID:10640713; http://dx.doi.org/ 10.1016/S0925-4773(99)00241-5 [DOI] [PubMed] [Google Scholar]
- [21].Wang C, Hennessey JA, Kirkton RD, Graham V, Puranam RS, Rosenberg PB, Bursac N, Pitt GS. Fibroblast growth factor homologous factor 13 regulates Na+ channels and conduction velocity in murine hearts. Circ Res 2011; 109:775-82; PMID:21817159; http://dx.doi.org/ 10.1161/CIRCRESAHA.111.247957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Allouis M, Le Bouffant F, Wilders R, Peroz D, Schott JJ, Noireaud J, Le Marec H, Merot J, Escande D, Baro I. 14-3-3 is a regulator of the cardiac voltage-gated sodium channel Nav1.5. Circ Res 2006; 98:1538-46; PMID:16728661; http://dx.doi.org/ 10.1161/01.RES.0000229244.97497.2c [DOI] [PubMed] [Google Scholar]
- [23].Brechet A, Fache MP, Brachet A, Ferracci G, Baude A, Irondelle M, Pereira S, Leterrier C, Dargent B. Protein kinase CK2 contributes to the organization of sodium channels in axonal membranes by regulating their interactions with ankyrin G. J Cell Biol 2008; 183:1101-14; PMID:19064667; http://dx.doi.org/ 10.1083/jcb.200805169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Isom LL. Sodium channel beta subunits: anything but auxiliary. Neuroscientist 2001; 7:42-54; PMID:11486343; http://dx.doi.org/ 10.1177/107385840100700108 [DOI] [PubMed] [Google Scholar]
- [25].Sampo B, Tricaud N, Leveque C, Seagar M, Couraud F, Dargent B. Direct interaction between synaptotagmin and the intracellular loop I-II of neuronal voltage-sensitive sodium channels. Proc Natl Acad Sci U S A 2000; 97:3666-71; PMID:10737807; http://dx.doi.org/ 10.1073/pnas.97.7.3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shao D, Okuse K, Djamgoz MB. Protein-protein interactions involving voltage-gated sodium channels: Post-translational regulation, intracellular trafficking and functional expression. Int J Biochem Cell Biol 2009; 41:1471-81; PMID:19401147; http://dx.doi.org/ 10.1016/j.biocel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- [27].Tan HL, Kupershmidt S, Zhang R, Stepanovic S, Roden DM, Wilde AA, Anderson ME, Balser JR. A calcium sensor in the sodium channel modulates cardiac excitability. Nature 2002; 415:442-7; PMID:11807557; http://dx.doi.org/ 10.1038/415442a [DOI] [PubMed] [Google Scholar]
- [28].Dover K, Solinas S, D'Angelo E, Goldfarb M. Long-term inactivation particle for voltage-gated sodium channels. J Physiol 2010; 588:3695-711; PMID:20679355; http://dx.doi.org/ 10.1113/jphysiol.2010.192559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu CJ, Dib-Hajj SD, Renganathan M, Cummins TR, Waxman SG. Modulation of the cardiac sodium channel Nav1.5 by fibroblast growth factor homologous factor 1B. J Biol Chem 2003; 278:1029-36; PMID:12401812; http://dx.doi.org/ 10.1074/jbc.M207074200 [DOI] [PubMed] [Google Scholar]
- [30].Rush AM, Wittmack EK, Tyrrell L, Black JA, Dib-Hajj SD, Waxman SG. Differential modulation of sodium channel Na(v)1.6 by two members of the fibroblast growth factor homologous factor 2 subfamily. Eur J Neurosci 2006; 23:2551-62; PMID:16817858; http://dx.doi.org/ 10.1111/j.1460-9568.2006.04789.x [DOI] [PubMed] [Google Scholar]
- [31].Wittmack EK, Rush AM, Craner MJ, Goldfarb M, Waxman SG, Dib-Hajj SD. Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J Neurosci 2004; 24:6765-75; PMID:15282281; http://dx.doi.org/ 10.1523/JNEUROSCI.1628-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Goldin AL. Resurgence of sodium channel research. Annu Rev Physiol 2001; 63:871-94; PMID:11181979; http://dx.doi.org/ 10.1146/annurev.physiol.63.1.871 [DOI] [PubMed] [Google Scholar]
- [33].Kraner SD, Novak KR, Wang Q, Peng J, Rich MM. Altered sodium channel-protein associations in critical illness myopathy. Skelet Muscle 2012; 2:17; PMID:22935229; http://dx.doi.org/ 10.1186/2044-5040-2-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Muller CS, Haupt A, Bildl W, Schindler J, Knaus HG, Meissner M, Rammner B, Striessnig J, Flockerzi V, Fakler B, et al.. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci U S A 2010; 107:14950-7; PMID:20668236; http://dx.doi.org/ 10.1073/pnas.1005940107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Leveque C, Hoshino T, David P, Shoji-Kasai Y, Leys K, Omori A, Lang B, el Far O, Sato K, Martin-Moutot N, et al.. The synaptic vesicle protein synaptotagmin associates with calcium channels and is a putative Lambert-Eaton myasthenic syndrome antigen. Proc Natl Acad Sci U S A 1992; 89:3625-9; PMID:1314395; http://dx.doi.org/ 10.1073/pnas.89.8.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim DK, Catterall WA. Ca2+-dependent and -independent interactions of the isoforms of the alpha1A subunit of brain Ca2+ channels with presynaptic SNARE proteins. Proc Natl Acad Sci U S A 1997; 94:14782-6; PMID:9405690; http://dx.doi.org/ 10.1073/pnas.94.26.14782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 2002; 74:5383-92; PMID:12403597; http://dx.doi.org/ 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- [38].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003; 75:4646-58; PMID:14632076; http://dx.doi.org/ 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- [39].Fox MA, Sanes JR. Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol 2007; 503:280-96; PMID:17492637; http://dx.doi.org/ 10.1002/cne.21381 [DOI] [PubMed] [Google Scholar]
- [40].Wong HK, Sakurai T, Oyama F, Kaneko K, Wada K, Miyazaki H, Kurosawa M, De Strooper B, Saftig P, Nukina N. beta Subunits of voltage-gated sodium channels are novel substrates of beta-site amyloid precursor protein-cleaving enzyme (BACE1) and gamma-secretase. J Biol Chem 2005; 280:23009-17; PMID:15824102; http://dx.doi.org/ 10.1074/jbc.M414648200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


