Abstract
Given the increasing emphasis on delivering high-quality, cost-efficient healthcare, improved methodologies are needed to measure the accuracy and utility of ordered diagnostic examinations in achieving the appropriate diagnosis. Here, we present a data-driven approach for performing automated quality assessment of radiologic interpretations using other clinical information (e.g., pathology) as a reference standard for individual radiologists, subspecialty sections, imaging modalities, and entire departments. Downstream diagnostic conclusions from the electronic medical record are utilized as “truth” to which upstream diagnoses generated by radiology are compared. The described system automatically extracts and compares patient medical data to characterize concordance between clinical sources. Initial results are presented in the context of breast imaging, matching 18 101 radiologic interpretations with 301 pathology diagnoses and achieving a precision and recall of 84% and 92%, respectively. The presented data-driven method highlights the challenges of integrating multiple data sources and the application of information extraction tools to facilitate healthcare quality improvement.
Keywords: quality improvement, radiology, pathology, information storage and retrieval, mammography
INTRODUCTION
The changing landscape of healthcare delivery and reimbursement has underscored the need for measures of quality. In radiology, as in many disciplines, current methods of quality assessment involve a blind peer review. Past studies have demonstrated that errors exist in approximately 4% of radiological interpretations reported during daily practice.1 Furthermore, variability in interpretations may exceed 45% among radiologists, as shown in a study that compared breast recommendations at accredited medical centers.2 Variation may occur due to radiologists’ varying levels of experience or differences in image quality based on scanning protocols and available hardware. While these variations in interpretation typically do not negatively impact a patient’s diagnosis or subsequent care, some instances may result in an abnormality being identified incorrectly (false positive) or missed (false negative). Current tools for reviewing diagnostic accuracy include RADPEER and RadReview, which are online peer-review systems that score clinical performance based on the completeness of findings, interpretation of the findings, and significance of omissions.3 Nevertheless, several shortcomings are noted: 1) both peer-review approaches are susceptible to systematic error in the interpretation task because other radiologists serve as the “reference standard”; 2) the process is time-consuming, resulting in lost productivity; 3) the criteria for grading may not be clearly defined or (consistently) followed by reviewers; and 4) the re-interpretation is limited by the same constraints as the original interpretation (e.g., poor image quality). Improved methods for using documented observations in place of than peer review to assess diagnostic accuracy and pinpoint potential sources of error or variability provides opportunities for improving the overall quality of information delivered to providers.
Here, we present an automated, objective approach to measuring the quality of radiology reports by comparing radiology findings with diagnoses provided by other clinical data sources (e.g., pathology). The goal is to establish a method for measuring the accuracy of a health system at multiple levels of granularity, from individual radiologists to subspecialty sections, modalities, and entire departments. The approach utilizes downstream diagnostic conclusions captured in the data provided by other departments, such as pathology and surgery, as “truth” to which earlier diagnoses generated by radiology are compared. We demonstrate this approach initially in the area of breast cancer screening as existing legislation such as The Mammography Quality Standards Act (MQSA),4 requires routine audits to calculate the positive predictive value (PPV) for individual radiologists based on concordance with pathology. Therefore, a reference standard exists for matching breast screening cases to pathology, and multiple studies have previously examined the PPV of Breast Imaging-Reporting and Data System (BI-RADS) assessments at different institutions using retrospective datasets of varying sizes.5,6
MATERIALS AND METHODS
Overall Architecture
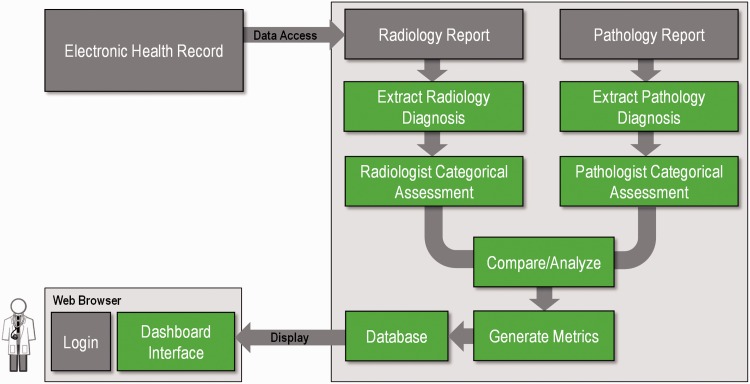
The overall software architecture is illustrated in Figure 1. Pulling data from the electronic health record, our system examines all downstream information (e.g., clinical reports) for a given imaging study relevant to diagnosis. Once the information is pulled, an information extraction pipeline implemented using the Apache Unstructured Information Management Architecture structures information from the narrative text of both reports. In the case of radiology reports, the diagnostic statement typically occurs within the “impressions” or “conclusion” section. In a pathology report, diagnostic information may be found in the “final diagnosis” section. Each report has its own structure and variations: a series of annotators and aggregate engines7 extract key data elements such as BI-RADS category, laterality, and pathologic diagnosis. These components are described further in the following sections.
Figure 1:
Overview of the system architecture. Radiology and pathology reports are retrieved from the electronic health record. Given that these reports are semi-structured, natural language processing is used to extract and categorize relevant diagnostic information from each report. This information is then matched and scored based on agreement of the information between reports and presented as part of an interactive dashboard.
Information Extraction and Scoring
Information Extraction
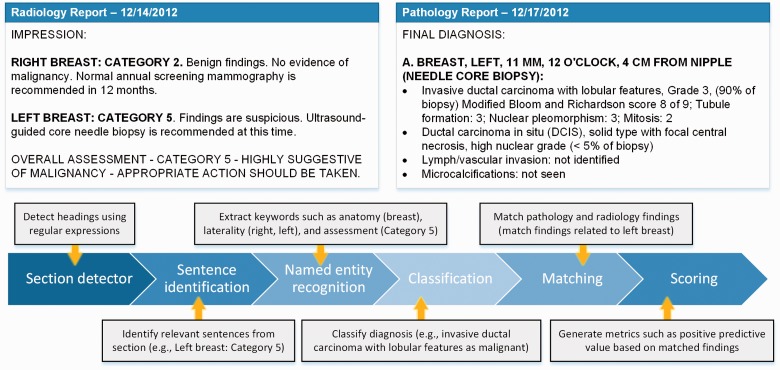
The process (summarized in Figure 2) starts with identifying relevant sections: an annotator that consists of regular expressions identifies relevant sections based on headings. In the radiology reports for breast imaging, assessments are almost always found in the “impressions” section. In pathology reports, diagnostic information about each specimen is reported in the “final diagnosis” section. Subsections that describe unique findings for each specimen (e.g., if multiple specimens were taken during a biopsy) are further characterized based on the structure of the report. Such information is typically identifiable based on formatting (e.g., capitalization, colon use) and paragraph breaks. Once the relevant section/paragraph has been identified, each sentence is then tokenized based on punctuation and categorized as to whether relevant information appears. Specifically, anatomy (i.e., breast), side (e.g., right, left, bilateral), and assessment (e.g., Category 4A) are extracted from radiology report sentences. Similar information is then extracted from the pathology report. From the specimen label (e.g., “A. Breast, Left, 11 mm, 12 o’clock, 4 cm from nipple”) information such as anatomy and side are identified. From the findings listed immediately below the label, diagnostic information (e.g., ductal carcinoma in situ) is extracted. Named entity recognition is performed using a dictionary lookup approach, given the small number of variations that occur in the targeted information in breast imaging.
Figure 2:
Process of information extraction, matching, and scoring.
Matching
Associations between radiology and pathology findings are currently not explicitly documented in clinical reports. We define a matching algorithm based on three assumptions: 1) the pathology diagnosis chronologically follows the radiology examination; 2) the pathology findings are reported within 90 days after the imaging examination; and 3) contextual information such as laterality is used to ensure that findings from multiple suspicious lesions are matched correctly. These assumptions are modeled after the current workflow for generating MQSA audit data where breast radiologists and pathologists routinely meet to review cases that are sent for biopsy.
Scoring
To generate the score, the radiology and pathology findings are classified into preset categories. In the case of breast imaging, the radiologic assessments are defined by the BI-RADS score. Cases with a BI-RADS category between 1 and 3 are assumed to be benign and are expected to have low PPV, while BI-RADS 4A, 4B, 4C, and 5 should have monotonically increasing PPVs. Conversely, pathology is less structured and provides a variety of diagnostic information (e.g., histology, grade) that must be classified into the appropriate category (e.g., benign, malignant). A statistical classifier based on conditional random fields (CRF)8 is used. A CRF was trained using the Mallet toolkit9 on a collection of 4160 pathology reports that were already manually reviewed as part of the existing mammography auditing process at our institution prior to 2010. The CRF categorized free-text descriptions drawn from pathology diagnosis sections into one of three labels: benign (e.g., benign breast tissue with biopsy site), benign but high risk (e.g., atypical lobular hyperplasia/lobular carcinoma in situ), or malignant (e.g., invasive lobular carcinoma, classic type).
User Interface
Dashboard
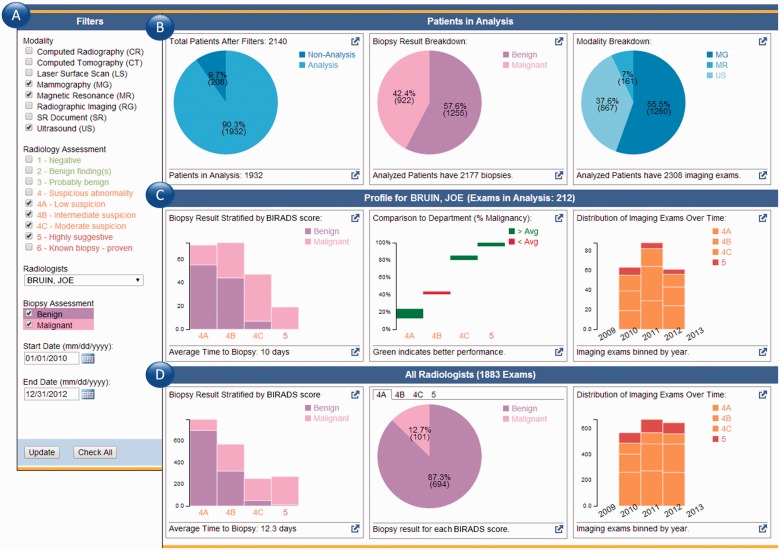
Figure 3 depicts the populated dashboard that is displayed after a user logs into the system. While the application utilizes data from the electronic health record (EHR), it runs independently as a web interface consisting of two primary components: the filter and data summary pane. Initially, the user is presented with a data summary generated based on all examinations in the radiology information system associated with that individual. Filters (Figure 3a) are used to further refine the display by specifying date ranges (e.g., month vs year) and desired modalities (e.g., mammography vs ultrasound). The data summary pane (Figure 3b) generates a visual representation of the underlying data to give users context as to the examinations included in the analysis and a breakdown of the analysis results. Graphs (Figure 3c) summarize the diagnostic performance for a selected radiologist: pathology diagnosis stratified by BI-RADS score, comparison of the individual’s PPV for each BI-RADS score in comparison to the department’s averages, and the distribution of BI-RADS scores given over time. Similarly, aggregate statistics across all breast radiologists (Figure 3d) are provided as a comparison point: breakdown of BI-RADS scores and benign/malignant pathology results, PPV for each BI-RADS score, and a breakdown of the BI-RADS scores given over time.
Figure 3:
Dashboard interface. (a) The filter pane allows users to constrain the types of examinations considered as part of the scoring mechanism. (b) A summary of the relevant radiology examinations based on the specified filters and the proportion that have matching pathology results. (c) Results for a given individual and his/her positive predictive rate when matched to pathology. Also shown is a comparison between that individual and the departmental average performance. In this example case, the individual outperforms the department average in his PPV for BI-RADS 4A. (d) A summary of the performance of the department as a whole.
RESULTS
Dataset
To evaluate the accuracy of the extraction and matching algorithm, the system was evaluated against a reference standard of 18 101 breast imaging examinations resulting in 301 pathological diagnoses that were performed in 2010 and 2011, following an institutional review board approved protocol. These cases were chosen because they had already been manually reviewed as part of the standard auditing process.
Information Extraction and Matching Performance
The performance of the Unstructured Information Management Architecture pipeline is summarized in Table 1. We evaluated the performance of our matching algorithm by generating a linked dataset between the extracted radiology and pathology assessments. Our results showed that 84.7% of the radiology-pathology matches made automatically were in agreement with those defined in the reference dataset. The primary sources of error were: 1) biopsies occurring outside of the 90-day window defined by our algorithm; 2) not all biopsies were performed at our institution, making this information unavailable in the medical record but was obtained by a patient navigator and entered in the reference dataset, as required by MQSA; and 3) in a small number of cases (<5%), findings documented in the pathology report did not match what was captured in the reference dataset. Further study is necessary to assess the reason behind the conflicting information (e.g., additional factors were considered beyond what was documented in the record).
Table 1:
Summary of extraction performance for identifying relevant information from narrative text
| Recall | Precision | Accuracy | |
|---|---|---|---|
| Radiology assessment (e.g., identification of BI-RADS score) | 0.914 | 0.986 | 0.902 |
| Pathology assessment (e.g., identification of diagnosis) | 0.887 | 0.981 | 0.873 |
| Pathology diagnosis classifier (e.g., benign/malignant) | Benign: 0.97 | Benign: 0.97 | 0.953 |
| High risk: 0.66 | High risk: 0.81 | ||
| Malignant: 0.983 | Malignant: 0.95 | ||
| Radiology-Pathology matching | 0.847 | 0.929 | 0.796 |
The CRF classifier created to categorize pathology diagnoses as benign, benign but high risk, or malignant was evaluated using sentences extracted from the pathology narrative and compared against assignments specified in the reference. The CRF classifier achieved an accuracy of 95.3%. The primary source of error occurred in instances wherein sections containing the pathology diagnosis were not tokenized properly, mixing information across several specimens.
DISCUSSION
Our system demonstrates an automated approach to performing quality assurance and evaluating the diagnostic accuracy of radiology reports. The informatics approach described in this paper illustrates a data-driven, objective method for comparing radiology results with downstream clinical information. Nevertheless, several limitations are noted. The use of pathology as the reference diagnosis permits the assessment of specificity but not sensitivity. The system cannot assess the accuracy of a diagnosis if the patient does not have a subsequent biopsy, hence overlooking cases where a radiologist may have missed an abnormal finding. In addition, pathology findings may also inherently have errors, as shown in a recent study that demonstrated a discordance rate of 24.7% among three pathologist interpretations with the highest variability in ductal carcinoma in situ and atypia.10 One approach to resolving conflicting information is incorporating data from additional clinical sources such as surgical/oncologist notes and functional tests. In addition, the scoring metric needs to handle uncertainty and ambiguity that are inherent in clinical diagnosis. In breast imaging, assessment is aided by the structured nature of BI-RADS, but in other contexts such as diagnosing patients with pneumonia, scoring the accuracy is more difficult. An example would be if a patient is diagnosed with pneumonia and receives antibiotics, but the sputum and blood cultures are found to be negative. In this scenario, the downstream information is inconclusive to determine whether the radiologist provided the correct diagnosis or even if incorrect, whether the information resulted in the correct course of treatment.
As part of the ongoing work, we are conducting pilot studies with attending radiologists and fellows to evaluate the utility of the information presented through the dashboard. Data is also being collected to assess how users respond to their scores and whether the system improved their abilities to discriminate cases that are similar to ones found to be discordant. Additional annotators will be developed to handle a broader range of reports. In addition, the dashboard can be extended to allow users (e.g., residents, attending physicians) to review discordant cases for continuing education. Finally, we will explore additional metrics that can be generated through the integration of patient record data. For instance, the number of imaging and other diagnostic tests ordered before a definitive diagnosis is reached can be characterized. Combining this information with cost and time, the optimal diagnostic pathways can be identified based on an analysis of patients in the database. As data from the electronic health record becomes increasingly integrated, additional data sources may be used to characterize the downstream impact of the radiologist’s interpretation on patient outcomes. Ultimately, this information will be useful in assessing the value of imaging information and improving the quality of information that radiology contributes to the patient’s process of care.3
FUNDING
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
W.H. designed the study, supervised implementation of the work, performed the data analysis and interpretation of the results, and drafted the paper. S.X.H. contributed to the implementation and revised the paper. C.W.A. and A.A.T.B. contributed to the study design and made substantive revisions to the paper. D.R.E. provided the basis for the study, contributed to the study design, and provided feedback of the paper.
REFERENCES
- 1.Borgstede JP, Lewis RS, Bhargavan M, Sunshine JH. RADPEER quality assurance program: a multifacility study of interpretive disagreement rates. J Am College Radiol. 2004;1(1):59–65. [DOI] [PubMed] [Google Scholar]
- 2.Johnson CD, Krecke KN, Miranda R, Roberts CC, Denham C. Developing a radiology quality and safety program: a primer. Radiographics. 2009;29(4):951–959. [DOI] [PubMed] [Google Scholar]
- 3.Mahgerefteh S, Kruskal JB, Yam CS, Blachar A, Sosna J. Peer review in diagnostic radiology: current state and a vision for the future. Radiographics. 2009;29(5):1221–1231. [DOI] [PubMed] [Google Scholar]
- 4.Monsees BS. The Mammography Quality Standards Act: an overview of the regulations and guidance. Radiol Clin North America. 2000;38(4):759–772. [DOI] [PubMed] [Google Scholar]
- 5.Liberman L, Abramson AF, Squires FB, Glassman J, Morris E, Dershaw D. The breast imaging reporting and data system: positive predictive value of mammographic features and final assessment categories. Am J Roentgenol. 1998;171(1):35–40. [DOI] [PubMed] [Google Scholar]
- 6.Lacquement MA, Mitchell D, Hollingsworth AB. Positive predictive value of the breast imaging reporting and data system. J Am College Surgeons. 1999;189(1):34–40. [DOI] [PubMed] [Google Scholar]
- 7.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10(3–4):327–48. [Google Scholar]
- 8.Lafferty J, McCallum A, Pereira FCN. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data, 2001. [Google Scholar]
- 9.McCallum A.MALLET: A Machine Learning for Language Toolkit. Amherst: UMass, 2002. [Google Scholar]
- 10.Elmore JG, Longton GM, Carney PA, et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA. 2015;313(11):1122–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]