Abstract
Background and objective: There is an increasing desire to share de-identified electronic health records (EHRs) for secondary uses, but there are concerns that clinical terms can be exploited to compromise patient identities. Anonymization algorithms mitigate such threats while enabling novel discoveries, but their evaluation has been limited to single institutions. Here, we study how an existing clinical profile anonymization fares at multiple medical centers.
Methods: We apply a state-of-the-art k-anonymization algorithm, with k set to the standard value 5, to the International Classification of Disease, ninth edition codes for patients in a hypothyroidism association study at three medical centers: Marshfield Clinic, Northwestern University, and Vanderbilt University. We assess utility when anonymizing at three population levels: all patients in 1) the EHR system; 2) the biorepository; and 3) a hypothyroidism study. We evaluate utility using 1) changes to the number included in the dataset, 2) number of codes included, and 3) regions generalization and suppression were required.
Results: Our findings yield several notable results. First, we show that anonymizing in the context of the entire EHR yields a significantly greater quantity of data by reducing the amount of generalized regions from ∼15% to ∼0.5%. Second, ∼70% of codes that needed generalization only generalized two or three codes in the largest anonymization.
Conclusions: Sharing large volumes of clinical data in support of phenome-wide association studies is possible while safeguarding privacy to the underlying individuals.
Keywords: privacy, generalization, secondary use, anonymization, clinical codes
INTRODUCTION
Lower costs for computing and high-throughput technologies enable healthcare institutions to amass large volumes of clinical and genomic data.1 While these data can enable personalized medical systems and save costs,2 it is increasingly recognized that data can also be repurposed to support the discovery of novel biomedical associations and facilitate comparative effectiveness investigations.3–5 Given that many of these studies are sponsored by federal funding agencies, various policies, including the recent Genome Sequence Data Sharing Policy of the National Institutes of Health (NIH),6 require public sharing of such data for reuse and transparency.6 Concurrently, it is critical that the privacy of participants is maintained. In support of this goal, the NIH policies recommend de-identifying data before dissemination.7–9
Care must be taken when sharing such data from electronic health record (EHR) systems because it can be exploited to re-identify the patients from whom the data was derived.10 For instance, it was shown that the combination of a patient’s billing codes, which are often invoked in biomedical research, can be leveraged to re-identify a patient.11 Yet demonstrations of such attacks have also led to the development of a range of data-based protections that support secondary research with clinical data.12 In particular, to mitigate attacks on such codes, several mechanisms have been introduced to support genome-phenome association studies with rigorous guarantees of privacy.13,14 These methods are differentiated by how they account for the knowledge available to the recipient of the data (e.g., if they know that a targeted patient is in a specific study cohort as opposed to the general population who received treatment at a medical institution).
However, the development of such methods is insufficient to demonstrate the extent to which they scale or are useful across medical centers. Towards this goal, Heatherly and colleagues15 leveraged data from a single EHR to simulate how the size of the available patient population at other academic medical institutions across the United States can preserve privacy while increasing data utility when compared to study-specific data sets. It was hypothesized that the larger the dataset anonymized, the more accurate the results would be after anonymization. The findings, however, indicated that while this was true when the full EHR was anonymized, when the biorepository was anonymized, there was a net loss in the quantity and specificity of clinical codes shared vs anonymizing only the study cohort.14 It was observed that the criteria used for incorporating patients into the subset may have biased the result.
In the present paper, we report on an evaluation of clinical data anonymization algorithm at three academic medical centers. In particular, we focus on an existing k-anonymization algorithm, where k = 5, because its scalability has been posited in prior investigations. We show that each medical institution can leverage all patients in the EHR system to protect a select group of patients involved in a research program (e.g., a study cohort). We further show that even when the protection is restricted to only the subset of EHR patients in the biorepository of a medical center, a significantly greater volume of clinical data can be released than if the organization simply protects a specific cohort. Most importantly, we observe that as the size of the population grows, the amount of data that may be shared in a privacy-preserving manner grows as well, which validates the original hypothesis.
BACKGROUND
Privacy in medical research
In the United States, the exact strategy for achieving de-identification is based on guidance articulated in the Privacy Rule of the Health Insurance Portability and Accountability Act.8 Specifically, de-identification under the Privacy Rule can be achieved through 1) safe harbor or 2) an expert determination. The former approach states that certain attributes about a patient must be removed (e.g., personal name and medical record number) or abstracted (e.g., 5-digit ZIP codes into 3-digit ZIP codes).8 The latter approach permits institutions to share any data provided that an expert certifies the risk of re-identification is very small.
Despite advancements in data sanitization algorithms,11 the past decade has witnessed the development of a variety of linkage-based re-identification attacks against biomedical data, including genomics, (with varying degrees of success).10,16–19 These incidents demonstrate the need to develop alternative models of privacy. For instance, an increasingly prevalent approach is to adopt data manipulation models that use randomization (e.g., ε-differential privacy) to make it difficult to ascertain whether a patient’s data was incorporated in a dataset.20,21 It has also been demonstrated that, in theory, such randomization approaches can be applied to set-based data systems (i.e., where an individual is assigned a variable number of values from a domain, such as a set of diagnosis codes, as we study in this work).22
In recognition that such models cannot sufficiently address all data sharing situations, it has been suggested that healthcare organizations approve research participants for a broad range of research topics23 and accept that there are no guarantees made as to their privacy outcomes.24
Clinical profile anonymization
It has been shown that fewer than 10 International Classification of Disease, ninth edition (ICD-9) codes could, in many instances, uniquely identify a patient.11 However, this observation was based on the assumption that the anticipated recipient of the data would have knowledge equivalent to that of an insider (i.e., an individual with unlimited access to the full records of the institution). In a subsequent study, it was shown that when the anticipated recipient had less knowledge than an insider might (e.g., a researcher from another institution), there is a much greater ability to release clinical details.14 In this case, data sets can be anonymized, using generalization and suppression of codes, such that they enable the discovery of genome-phenome associations that are virtually identical to those mined from the original, non-anonymized data.14 Most recently, investigators at Kaiser Permanente showed that a similar approach to k-anonymization (further detailed below) yielded safer datasets with greater research potential than a more naïve approach for approximately 70 000 EHR-derived records submitted to the Database of Genotypes and Phenotypes (dbGaP) at the NIH.25 We note that in this work, we focus on k-anonymization, as opposed to alternative privacy models, such as ε-differential privacy, because their scalability has been theoretically posited, but not empirically assessed with data from multiple healthcare institutions.
METHODS
Materials
To perform the analysis, data were collected from three medical centers: 1) the Marshfield Clinic; 2) Northwestern University; and 3) Vanderbilt University Medical Center. Data from each institution were classified into three telescoping populations. The first is the component of the EHR that consists of all individuals with clinical codes. For the purposes of the analysis, we simply refer to this as an institution’s EHR. The second is the subset of the EHR for which the institution has available biospecimens or for which they have already completed genotyping (e.g., Marshfield’s Personalized Medicine Research Project biobank,26 Northwestern’s NUgene,27 and Vanderbilt’s BioVU28). Henceforth, this group is referred to as the institution’s biorepository. The third is a specific study cohort taken from each biorepository. Specifically, each of the three sites collaborated in a genome-wide association study for hypothyroidism for the Electronic Medical Records and Genomics network.29 And, for this evaluation, the records that each site submitted to the consortium project are referred to as the study cohort. Summary record counts for each data set are provided in Table 1.
Table 1:
Number of patients with records and assigned codes in the various datasets of this study pre- and post-anonymization
| Dataset | Number of Patients and Code Occurrences |
||
|---|---|---|---|
| Marshfield | Northwestern | Vanderbilt | |
| Original | 1684 | 742 | 5994 |
| 1 298 732 | 29 161 | 272 080 | |
| After anonymization according to: | |||
| Study Cohort, n (%) | 1681 (99.8) | 737 (99.3) | 5971 (99.6) |
| 1 291 294 (99.4) | 28 922 (99.2) | 270 867 (99.6) | |
| Biorepository, n (%) | 1684 (100) | 742 (100) | 5595 (93.3) |
| 1 297 723 (99.9) | 29 154 (99.98) | 248 925 (91.5) | |
| EHR, n (%) | 1684 (100) | 742 (100) | 5994 (100) |
| 1 298 732 (100%) | 29 161 (100) | 272 043 (99.99) | |
Original corresponds to original study (i.e., no anonymization); Study Cohort corresponds to anonymization over the hypothyroidism research set; Biorepository corresponds to anonymization over all records for which genetic information is available; and EHR corresponds to anonymization over all patient records available at the study site.
Data protection model
In this analysis, we assume that the attacker has access to medical data in the form of clinical codes (e.g., 5-digit ICD-9 codes). Specifically, the attacker has gained access to an individual’s discharge summary, which includes all of the ICD-9 codes from a single visit, and is curious to discover any other codes associated with that person from other visits, a standard assumption applied in prior studies.13–15
To protect the data, we apply an existing implementation of the k-anonymization principle,30 where k was set to a best practice of 5, for clinical codes generated via visits made by a patient to a medical institution.14 For context, here we provide a high-level overview of the principle and corresponding algorithm to realize this principle in a dataset, and direct the reader to the appendix for a detailed description and example. In this setting, each visit is represented as a set of one or more ICD-9 codes. k-anonymization is applied such that, for each visit associated with the patient, there exist at least k − 1 other records with those diagnosis codes across all visits (this process is explained in further detail below). Primarily, this is accomplished through the process of generalization. Specifically, we used the hierarchy of ICD-9 codes developed originally for phenome-wide association studies (PheWAS) to convert sensitive 5-digit codes into less sensitive groups of codes.29,33 The PheWAS hierarchy was chosen because it has been shown to provide greater utility in association studies that reuse data from EMRs.31,32 We note here that while the hierarchy was originally used for proof-of-concept for an initial PheWAS of five SNPs, it was created to be used for arbitrary studies.
RESULTS
Table 1 summarizes the number of records available in the hypothyroidism cohort. In each dataset, we see that anonymizing ICD-9 codes at the study-level suppresses (i.e., they could not be released with a guarantee of 5-anonymity) more records in comparison to anonymizing at the full EHR level. However, we find in the Vanderbilt dataset that biorepository-level anonymization also results in a reduction in the number of records of about 400 patients. It should be noted that both Northwestern and Marshfield exhibit the hypothesized behavior alluded to in the introduction.14 Specifically, the size of the available dataset directly correlates with better anonymization results. We believe that the anomaly associated with Vanderbilt transpires because the biorepository population is moderately large and, as an artifact, incorporates a larger number of rare diagnoses that the anonymization aims to protect. This problem is not exhibited in the smaller cohorts and biorepositories because they have less rare diagnoses (because they have a frequency of zero) or the larger EMR populations (because the majority of diagnosis codes have sufficient frequency).
Table 1 also reports the total number of (Record, Diagnosis) pairs in the dataset at each level of anonymization. We again see that anonymization at the EHR level leads to the highest degree of fidelity. For Northwestern and Marshfield, this implies that every diagnosis code in the original data is still represented in the anonymized version. For Vanderbilt, there is some suppression, but the EHR level remains the closest to the original dataset.
Table 2 provides a summary of the extent to which datasets required codes to be generalized. Original indicates the number of distinct 5-digit ICD-9 codes that were originally included in the demonstration cohort without anonymization. It should be noted that there is substantial variability in the number of codes generalized values and regions, which may be due to different business and documentation processes at each institution. Nonetheless, it was again found that even though each EHR required some generalization, the rate was substantially lower than for the biorepository and demonstration level. In each dataset, fewer than 1% of codes needed to be generalized.
Table 2:
Number of generalized and suppressed codes per study site with the number of PheWAS regions which required generalization
| Anonymization Operation | Marshfield | Northwestern | Vanderbilt | |
|---|---|---|---|---|
| Original | – | 8052 | 8734 | 2910 |
| Study Cohort, n (%) | Generalized | 522 (6.4) | 1663 (19.0) | 381 (13.0) |
| Regions | 341 (33.0) | 271 (26.2) | 447 (43.2) | |
| Suppressed | 1 | 3 | 2 | |
| Biorepository, n (%) | Generalized | 206 (2.5) | 258 (2.9) | 216 (7.4) |
| Regions | 149 (14.4) | 165 (13.1) | 174 (16.8) | |
| Suppressed | 0 | 0 | 1 | |
| her, n (%) | Generalized | 18 (0.2) | 66 (0.7) | 10 (0.3) |
| Regions | 14 (1.4) | 9 (0.9) | 31 (3.0) | |
| Suppressed | 0 | 0 | 0 |
Original shows the number of ICD9 codes originally in the study cohort. For each site, the top value corresponds to the number of generalized codes and the bottom value is the number of suppressed codes.
Phenotypic changes
To provide insight into how anonymization influences the utility of the data in a more context-specific manner, we report on the regions of the aforementioned PheWAS vocabulary where generalization transpired for each site at the demonstration- and the EHR-level.
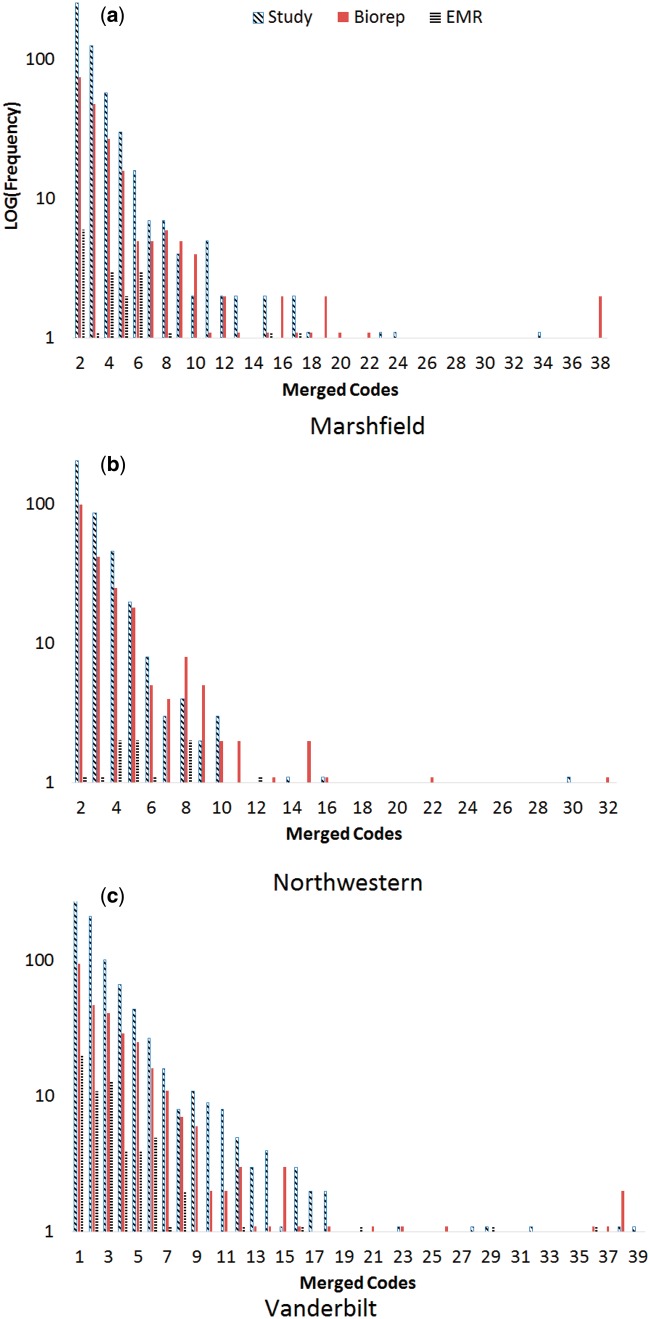
Figure 2 illustrates the distribution of codes within generalized regions of the vocabulary. That is, it shows how many codes are combined to ensure the required number of appearances allow it to meet the privacy requirement. As the figure illustrates, when codes are generalized, most are joined with only a few codes to achieve protection. However, at each site there is at least one group that requires at least 30 individual codes to be grouped in order to be generalized safely. Examples of these groups can be found in the annotated points in Figure 2. We also find that, at each level, the EHR anonymization requires fewer codes to be generalized, both overall and in each individual category.
Figure 1:
Distribution of generalizations required for each a) Marshfield, b) Northwestern, and c) Vanderbilt dataset. Note the y-axis is depicted in a log scale.
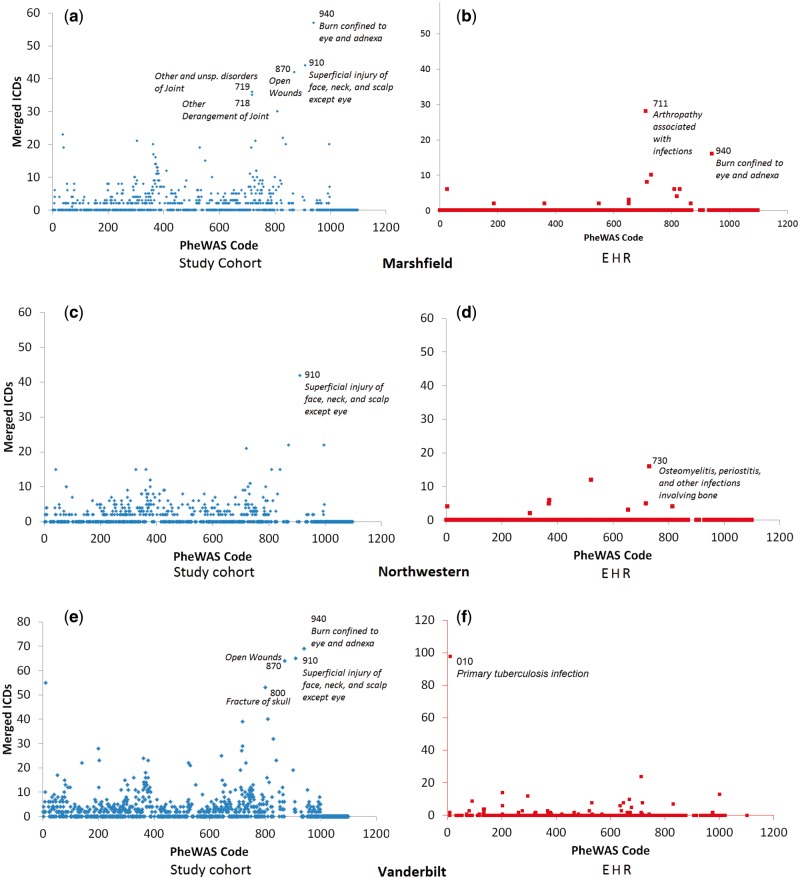
Figure 2:
Generalization in the datasets by PheWAS code. Each row corresponds to one of the three sites in the study. Notice that anonymizing at the EHR-level leads to fewer merged codes than anonymizing at the study-level.
Next, we illustrate which phenotypic regions required generalization. In Figure 2a and b, we show the results for the Marshfield anonymization. Here, it can be seen that approximately 55 codes relating to PheWAS topic 940 (Burn confined to eye and adnexa) were generalized. Note that this does not necessarily mean that they were generalized into a single code. Rather, it indicates the overall number of codes in that area that required generalization.
Figures 2c–f reveal similar trends for the other institutions. At Vanderbilt, the EHR anonymization has 31 regions of generalized codes to satisfy 5-anonymization, but the study cohort-level anonymization required 447 of these regions requiring generalization. This implies that there are fewer than five instances of each of many of the specific 5-digit ICD-9 representation, but that when considered slightly less specifically (i.e., 3-digit code) there are enough instances to allow the data to be released. Also in Table 2, we provide the counts of the PheWAS regions that require generalization for each site. Here, we find that each site has the same general trend that we described for Vanderbilt.
Table 3 provides general insight into the distributions of the generalizations in each dataset. For example, the mean cell in Marshfield’s Study-level statistics indicates that, for codes which were grouped, the average length of this grouping (i.e., number of codes placed together) was 3.5. As a condition, we ignore groupings of size 1 due to the fact that no grouping is necessary). It can be seen that the overall trend is highly dependent upon the specific dataset anonymized. Specifically, Marshfield and Northwestern follow a similar pattern of an increasing mean with the dataset size, but a drastically dropping max value. Vanderbilt, however, follows an opposite pattern, where the study-level has the highest mean value, but the difference between the study-level max and the EMR-level max is minimal.
Table 3:
Distribution of generalized PheWAS codes in the datasets
| Marshfield | Northwestern | Vanderbilt | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study | Biorepository | EMR | Study | Biorepository | EMR | Study | Biorepository | EMR | |
| Mean | 3.47 | 4.69 | 5.39 | 3.06 | 3.87 | 5.70 | 8.63 | 7.23 | 5.79 |
| Median | 3.00 | 3.00 | 4.00 | 2.00 | 3.00 | 5.00 | 7.00 | 7.00 | 6.00 |
| St. Dev | 2.90 | 4.96 | 4.47 | 2.17 | 3.41 | 2.79 | 3.85 | 3.11 | 2.46 |
| Max | 34.0 | 38.0 | 18.0 | 30.0 | 32.0 | 12.0 | 39.0 | 38.0 | 36.0 |
DISCUSSION
The results indicate that a greater quantity of data sharing is possible if an institution leverages the entire population in its EHR as a protecting population when data on a specific cohort is shared. This is similar to the findings first observed in the simulated analysis using data from the Vanderbilt EHR.15 The results lend credence to the possibility of sharing large volumes of clinical information when a dataset must be distributed. We believe the finding is notable because many groups of attackers will not know whether a specific targeted individual is actually in a released dataset. Thus, even if a specific matching record is found within the dataset, there is at most a 1/k chance that the corresponding patient is, in fact, the correct re-identification.
We also note that the bias originally shown in the Vanderbilt biorepository (i.e., higher rates of generalization required) did not reproduce within the other sites’ biorepositories. This suggests that the original observation noted in14 may be an artifact of an investigation with a single institution and is not necessarily an inherent property of all biorepositories.
It is also worth noting that, in this study, we conducted the analysis using three sites with very different data models. At the Marshfield Clinic, there is a high average number of diagnoses per patient, while at Northwestern there are fewer patients and diagnoses. However, each site sees benefit to our strategy of anonymization, which suggests that it does not rely upon a specific data model in order to function effectively.
We acknowledge that there are several limitations to this study. Foremost is that the study was performed using a greedy selection algorithm. It is entirely possible that heuristics could be developed to allow anonymization with greater utility in the end. Further, this was a study using only three sites. However, since the study began, Kaiser Permanente performed a similar analysis on their data in preparation for submission to dbGaP and found similar results.25 While it can be seen that one specific data model is not a requirement for success through this algorithm, it is possible that there are certain data models for which it will not work at all. That is, collections which have been selected to represent a certain set of conditions may have significantly different ICD9 statistics than we observe through the institutions represented here. In these cases, we have no evidence to suggest how an anonymize-large strategy would fare. We also note that this study involves clinical profile anonymization only. When data is fully released, it generally also includes demographic information. Further inquiry needs to be performed in order to determine the best way to privately share this information as well.
Additionally, there is no clear trend in the summary statistics presented in Table 3. There are two possible reasons for this phenomenon. First, as noted in prior studies,14,15 there may be a systematic bias present in the selection of participants in the Vanderbilt biorepository, although it dramatically subsides through selection of either a cohort or the entire EMR. The finding in this work may be an additional manifestation of this behavior. Second, we note that the anonymization algorithm is a heuristic and does not seek to provide any guarantee of optimality. It is possible that some underlying trend could emerge, if the anonymization algorithm aimed to provide optimal groupings or an approximation of such a solution.
CONCLUSIONS
This work shows that it is possible to share considerably more clinical information about research participants than may have been considered feasible while still protecting their privacy. This finding has notable implications for the submission of data to third-party-controlled repositories (e.g., dbGaP), namely that an institution may be able to contribute larger quantities of data (i.e., more clinical codes for a study cohort) for general research usage than they may previously have realized. Although this analysis does not consider the demographic attributes of the patient population, we believe that this is a significant advance toward the more-detailed release of data with large-scale genome-phenome associations.
FUNDING
This work was supported, in part, by National Science Foundation grant number [CCF0424422] and National Institutes of Health grant numbers [U01HG006385, U01HG006378, UL1TR000135, R01HG006844, R01LM010685, R01GM105688, U01HG006389, U01HG006388, 8UL1TR000150-05].
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
R.H. designed the algorithm, wrote the code, analyzed the results of the experiments, and wrote the paper. J.P., L.R., and P.P. performed data analysis and revised the paper. B.M. designed and supervised the study, analyzed the data, and revised the paper. J.D. and P.H. revised the paper.
ACKNOWLEDGEMENTS
We thank the members of the Electronic Medical Records and Genomics (eMERGE)Network for useful feedback during the development of this work. We would like to thank the principal investigators from each of the sites: Rex Chisolm, Cathy McCarty, and Maureen Smith; as well as Melissa Basford with the eMERGE Coordinating Center.
REFERENCES
- 1.Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA. 2013;309:1351–1352. [DOI] [PubMed] [Google Scholar]
- 2.Bates DW, Saria S, Ohno-Machado L, Shah A, Escobar G. Big data in health care: using analytics to identify and manage high-risk and high-cost patients. Health Aff. 2014;33:1123–1131. [DOI] [PubMed] [Google Scholar]
- 3.Richesson RL, Hammond RE, Nahm M, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. JAMIA. 2013;e2:e226–e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J Am Med Inform. 2013;e2:e206–e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. JAMIA. 2014;21(4):576–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Institutes of Health. NIH genomic data sharing policy. NOT-OD-14-124. August 27, 2014. [Google Scholar]
- 7.National Institutes of Health. Final NIH Statement on Sharing Research Data, NOT-OD-03-032. 2003. [Google Scholar]
- 8.National Institutes of Health. Policy for Sharing of Data Obtained in NIH Supported or Conducted Genome-Wide Association Studies (GWAS). NOT-OD-07-088. 2007. [Google Scholar]
- 9.U.S. Department of Health and Human Services. Standards for privacy of individually identifiable health information, Final Rule. 45 CFR Parts 160 and 164. August 14, 2002. [PubMed] [Google Scholar]
- 10.El Emam K, Jonker E, Arbuckle L, Malin B. A systematic review of re-identification attacks on health data. PLoS One. 2011;6:e28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loukides G, Denny J, Malin B. The disclosure of diagnosis codes can breach research participants’ privacy. JAMIA. 2010;17:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gkoulalas-Divanis A, Loukides G, Sun J. Publishing data from electronic health records while preserving privacy a survey of algorithms. J Biomed Inform. 2014;50:4–19. [DOI] [PubMed] [Google Scholar]
- 13.Loukides G, Gkoulalas-Divanis A, Malin B. Anonymization of electronic medical records for validating genome-wide association studies. Proc Natl Acad Sci USA. 2010;107:7898–7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heatherly R, Loukides G, Denny J, Haines J, Roden D, Malin B. Enabling genomic-phenomic association discovery without sacrificing anonymity. PLoS One. 2013;8:e53875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heatherly R, Denny JC, Haines JL, Roden DM, Malin B. Size matters: how population size influences genotype-phenotype association studies in anonymized data. J Biomed Inform. 2014;52:243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science. 2013;339:3l21–3124. [DOI] [PubMed] [Google Scholar]
- 17.Homer N, Szelinger S, Redman M, et al. Resolving individuals contributing trace amounts of dna to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Im HK, Gamazon ER, Nicolae DL, Cox NJ. On sharing quantitative trait GWAS results in an era of multiple-omics data and the limits of genomic privacy. Am J Hum Genet. 2012;90:591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Z, Owen AB, Altman RB. Genomic research and human subject privacy. Science. 2004;305:183. [DOI] [PubMed] [Google Scholar]
- 20.Dwork C, Pottenger Toward practicing privacy. JAMIA. 2013;20:102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner J, Xiong L, Xiao Y, et al. SHARE: system design and case studies for statistical health information release. JAMIA. 2014;20:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammed N, Jiang X, Chen R, Fung BC, Ohno-Machado L. Privacy-preserving heterogenous health data sharing. JAMIA. 2013;20:426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohane IS, Altman RB. Health-information altruists - a potentially critical resource. N Engl J Med. 2005;353:2074–2077. [DOI] [PubMed] [Google Scholar]
- 24.Lunshof JE, Chadwick R, Vorhaus DB, Church GM. From genetic privacy to open consent. Nat Rev Genet. 2008;9:406–411. [DOI] [PubMed] [Google Scholar]
- 25.Walter L, Sciortino S, Ranatunga D, et al. PS3-13: Re-identification risk associated with sharing linked genomic and phenotypic data from the Kaiser Permanente Research Program on Genes, Environment and Health (RPGEH). Clin Med Res. 2013;11:148. [Google Scholar]
- 26.McCarty CA, Garber A, Reeser JC, Fost NC, Personalized Medicine Research Project Community Advisory Group and Ethics and Security Advisory Board. Study newsletters, community and ethics advisory boards, and focus group discussions provide ongoing feedback for a large biobank. Am J Med Genet A. 2011;155A:737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf WA, Doyle MJ, Aufox SA, et al. DNA banking study in an ethnically diverse urban university hospital. Am J Hum Genet. 2003;73:423. [Google Scholar]
- 28.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharm Ther. 2008;84:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome-and phenome-wide studies. Am J Hum Genet. 2011;89:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney L. k-anonymity: a model for protecting privacy. Int J Uncertain, Fuzziness, Knowledge-based Sys. 2002;10:557–570. [Google Scholar]
- 31.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Namjou B, Marsolo K, Carroll, et al. Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development in IL5-IL13 to eosinophilic esophagitis. Front Genet. 2014;5:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Z, Mayer J, Ivacic L, et al. Phenome-wide association studies (PheWASs) for functional variants. Eur J Hum. 2015;23(4):523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]