Abstract
Background and Objective Electronic medical record (EMR) databases offer significant potential for developing clinical hypotheses and identifying disease risk associations by fitting statistical models that capture the relationship between a binary response variable and a set of predictor variables that represent clinical, phenotypical, and demographic data for the patient. However, EMR response data may be error prone for a variety of reasons. Performing a manual chart review to validate data accuracy is time consuming, which limits the number of chart reviews in a large database. The authors’ objective is to develop a new design-of-experiments–based systematic chart validation and review (DSCVR) approach that is more powerful than the random validation sampling used in existing approaches.
Methods The DSCVR approach judiciously and efficiently selects the cases to validate (i.e., validate whether the response values are correct for those cases) for maximum information content, based only on their predictor variable values. The final predictive model will be fit using only the validation sample, ignoring the remainder of the unvalidated and unreliable error-prone data. A Fisher information based D-optimality criterion is used, and an algorithm for optimizing it is developed.
Results The authors’ method is tested in a simulation comparison that is based on a sudden cardiac arrest case study with 23 041 patients’ records. This DSCVR approach, using the Fisher information based D-optimality criterion, results in a fitted model with much better predictive performance, as measured by the receiver operating characteristic curve and the accuracy in predicting whether a patient will experience the event, than a model fitted using a random validation sample.
Conclusions The simulation comparisons demonstrate that this DSCVR approach can produce predictive models that are significantly better than those produced from random validation sampling, especially when the event rate is low.
Keywords: electronic medical records, logistic regression, sudden cardiac arrest, validation sampling, design of experiments
1 INTRODUCTION
Enormous resources are devoted to compiling and integrating electronic medical record (EMR) databases. This has the potential for use in hypothesis generation, automatic identification of disease risk factors, and comparative effectiveness research. 1,2 The hypothesis generation and identification of disease risk factors involves fitting statistical models, such as logistic regression, that describe the relationship between the occurrence of a particular condition in a patient (represented by a binary response variable taking a value of 1 if the condition occurs in the patient and 0, otherwise) and clinical, phenotypical, and demographic data for the patient that are potential risk factors (represented by a vector of predictor variables, or predictors for short). Development of such models is more challenging when the events of interest are infrequent for a number of reasons, not the least of which is that the response values may be more likely to be miscoded in the administrative data due to unfamiliarity by less experienced data entry personnel.
Consider the following case study, which is used to illustrate concepts throughout this article. Sudden cardiac arrest (SCA) is an event that occurs in <0.125% of the population but is typically fatal. 3 An estimated 400,000 Americans die of SCA each year. A search using codes from the Ninth/Tenth Revision of the International Classification of Diseases (ICD-9/ICD-10) of the Northwestern Memorial Hospital EMR database between January 2006 and December 2010 yields 73 recorded SCA cases indicated by ICD-9 codes: 427.41, ventricular fibrillation; 427.42, ventricular flutter; and 427.5, cardiac arrest. The clinical measurements (blood tests, measurements from electro-cardiograms, physiological tests, etc.) on potential risk factors associated with SCA were available in medical records for N = 23 041 patients. 4 In EMR databases, although the predictors are usually entered quite reliably (since they are clinical measurements used for clinical decisions), there is often a surprisingly large percentage of errors in the recorded ICD-9/ICD-10 codes. This translates to errors in the response . A recent audit of ICD-10 coding of physicians’ clinical documentation showed error rates between 37% and 52% in various areas of specialties. 5 In our SCA data, 20 cases were randomly selected for review and validation from the set of cases that had an SCA event entered using the ICD-9 codes. Of these 20 selected cases, only 5 were true SCA events. If not handled properly, such high error rates render unreliable any statistical modeling approach.
High error rates are possibly due to the fact that for coding, hospitals often rely on less trained (relative to doctors) personnel and/or automated natural language processing methods that may be far from perfect, particularly for conditions that are not commonly encountered. 6–8 Considering that each record may take roughly 5–10 min to manually review, it is obviously cost prohibitive (virtually impossible) for doctors to manually review tens or hundreds of thousands of records to validate the response variables.
The problem considered in this article is now formally described as follows. Let denote the error-prone binary response variable, as recorded. Suppose there is a large database of N cases or records (which we subsequently refer to as “rows”), for each of which Y * and (the latter, without error) have been observed, and a reliable statistical model relating the true Y to the potential risk factors must be constructed. Existing approaches to the problem involve selecting a small random validation sample of rows from the complete data set and having an expert review and validate the response Y for each row in the validation sample. Notice that in this context, validation sampling refers to validating the correct Y values for a set of rows, as opposed to model validating. Lyles et al. 9 and Edwards et al. 10 combined the validation sample with the unvalidated error-prone data to fit bias-corrected models that account for the error rate. A drawback of this approach is that random sampling is inefficient when the event rate is low and the error rate is high. Moreover, the extreme error rates like the 75% error rate in our SCA case study may cause one to question the reliability of any model fit using the unvalidated data. For example, the approaches of Lyles et al. 9 and Edwards et al. 10 can correctly adjust for error-related biases only if the error mechanisms are correctly captured in the model. However, the error mechanisms may be too numerous or complex to do this.
In light of the preceding, this article considers a substantially different approach to fitting predictive models for risk assessment in large EMR data sets with unreliable response records (false positives and/or false negatives). Our approach is to collect a validation sample not randomly, but judiciously, chosen with the goal of giving us the most information on the relationships of interest. The intent is that the final predictive models will be fit using only the validation sample, ignoring the remainder of the unvalidated, error-prone data. The information in the error-prone data will only be used for judiciously choosing the validation sample, based on only the values for each row. Neither the Y nor the Y * values are considered when selecting the validation sample. Selecting the validation sample in this manner avoids causing a sampling bias in the fitted model, in the same way that conventional design of experiments (DOE) avoids a sampling bias. This will be referred as design-of-experiments–based systematic chart validation and review (DSCVR) with error-prone data hereafter.
2 METHODS
2.1 Description of DSCVR
Our DSCVR approach is akin to designing a small but powerful experimental study, aided by information extracted from the much larger, error-prone set of observational data. Figure 1 illustrates the analogies between DSCVR (judiciously choosing a small sample of rows to validate from among a large error-prone set of data) and the classical DOE problem. In the machine learning literature, the active learning paradigm 11–15 refers to the situation in which one has a large set of cases that are unclassified, a subset of which will be selected and classified by a human expert and used as the training data on which to fit a model for classifying future cases. The goal is to select the subset of cases to label, usually in a sequential manner, in order to reduce some measure of uncertainty in the fitted classification model. As such, our DSCVR approach is related to what can be viewed as a form of active learning.
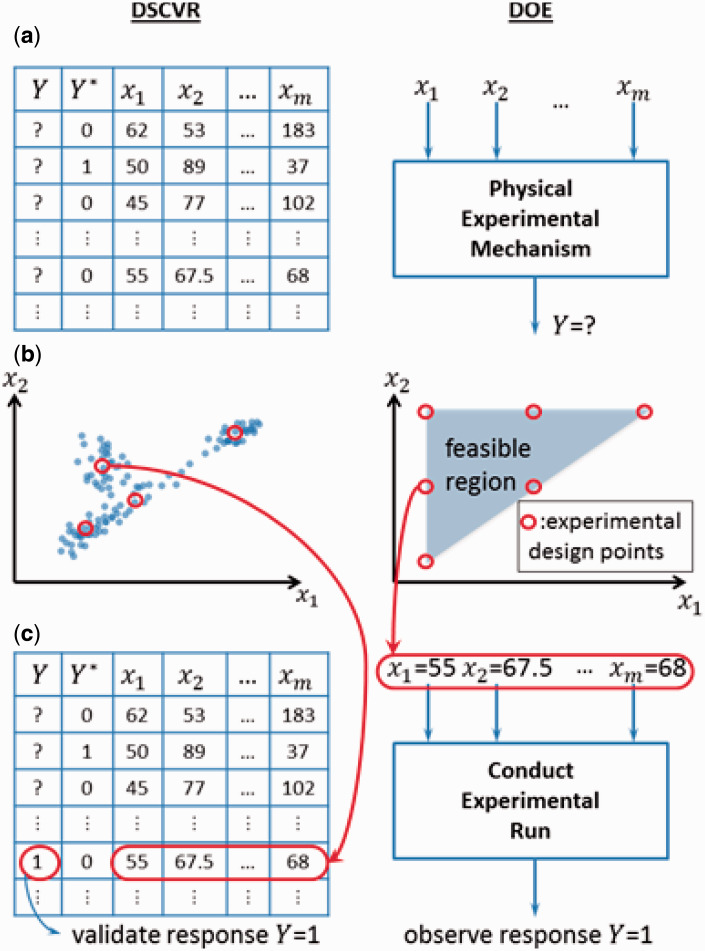
Figure 1:
Analogies between DSCVR (left column) and DOE (right column). ( a ) Mechanisms for generating an observation of the true response Y . Little is known of Y for a specific value of x until a case having those x is chosen from the error-prone data set (left) and validated, or until an experimental run at those x values is designed (right) and conducted. ( b ) Red circles represent the x values for the chosen set of cases to be validated (left) or the chosen set of x values at which to conduct the experimental runs (right) The complete feasible set of x values from which the red circles can be chosen are the set of all x values in the error-prone data set (blue dots; left) or the feasible experimental region (shaded area; right). ( c ) After choosing the x values for one case to be validated (left) or at which to conduct one experimental run (right), we observe the Y value corresponding to the chosen x values.
In the DOE literature, different methods have been developed to select the values to optimize some measure of quality of the resulting fitted model, and the most common optimal design criteria when fitting linear regression models are the so-called “alphabetic” optimality criteria. 16,17 This article focuses on D-optimality—that is, maximizing the determinant of Fisher information matrix—which is perhaps the most common and popular criterion.
In our problem, a logistic regression model is assumed to represent the true relationship between predictors and response Y , where 1 is included as the first element of , and are the parameters to be estimated. Let N denote the number of rows in the original error-prone data, the number of validation rows for which Y is observed without error, and the indices of the validated rows. For the case of a logistic regression model fit to the validated rows with row indices in J , it is straightforward to show that the log-likelihood is
| (1) |
and its Hessian (with respect to ) is
| (2) |
Because the Hessian does not depend on the response observations, it equals its expectation, the negative of which is the Fisher information 18
| (3) |
where
Thus, our DSCVR strategy is to choose the cases to be validated as the rows whose values maximize , with given by Eq. (3) , and the symbol denoting the determinant of a matrix.
2.2 DSCVR Design Algorithm for Validation Sample Selection Using the Fisher Information
In the traditional DOE literature, algorithmic design optimization is quite well developed 8,19–21 and is now standard in many commercial DOE software packages. However, in spite of the analogies illustrated in Figure 1 , existing optimal DOE algorithms are not directly applicable to optimal design for DSCVR for a number of reasons. First, the design space (the set of feasible values for ) is neither a continuous domain (common in traditional DOE when the input factors are continuous variables), nor a region or subregion of a grid (common in traditional DOE when the input factors have discrete settings). In DSCVR, the design space is the set of existing values in the rows of the error-prone database. For large data sets there are a great many possible values to consider (in the SCA example 23 041 data points), and these data points typically have quite irregular structure due to issues such as multicollinearity, clusters, outliers, etc. in the -space. Second, when considering whether/how to modify an value in the DSCVR design, its elements cannot be modified independently, as in the coordinate exchange algorithm of Meyer and Nachtsheim 21 (a popular heuristic for design optimization), because only the fixed set of values in the error-prone data set are permissible. Hence, our problem is first formulated as a binary integer optimization problem, and then a heuristic approach for finding an (approximate) optimal solution is proposed. The heuristic is generally necessary, because state-of-art integer programming software has difficulty handling problems of a size typical for DSCVR problems.
Write the Fisher information matrix in Eq. (3) as , where has been introduced, and the 0/1 binary variables z = have been defined as (if the i th row is included in the validation sample) or (if the i th row is not included in the validation sample). Note that depends on which set of rows are selected for the validation sample, which are represented by the binary set z . The binary integer optimization formulation becomes
| (4) |
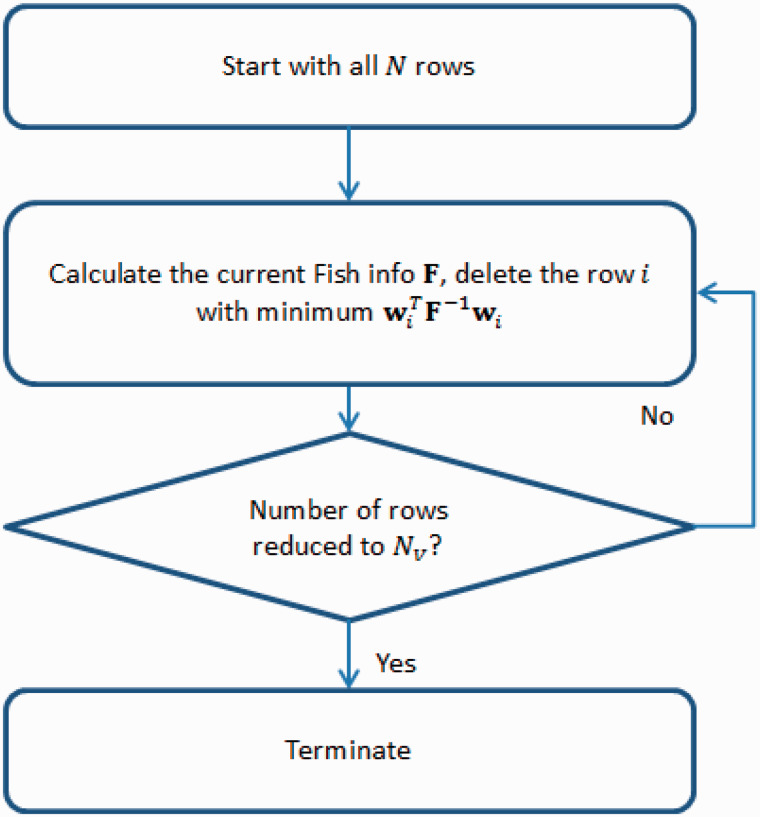
In light of the computational difficulty in solving the exact integer optimization, two heuristic approaches are considered, namely backward stepwise selection and hybrid backward/forward selection. As illustrated in Figure 2 , the backward selection algorithm starts with all N rows and, at each step, removes the single row that least reduces , where is the Fisher information matrix for the set of rows selected at the current stage of the algorithm. Because calculating directly for large N is time-consuming, is updated using the following result for the determinant of a rank-1 modification of a matrix , which is also used in the popular exchange algorithms for constructing D-optimal designs 19,21 :
| (5) |
In Eq. (5) , for the row (with index denoted by i ) that is deleted (or added in the hybrid backward/forward algorithm) at the current step. Hence, when updating , only needs to be calculated for each of the rows in the currently-selected validation sample. Notice that, from Eq. (5) , the row that least reduces is the row with the smallest . This step will be iterated and terminated when the number of rows selected is reduced to the desired size .
Figure 2:
Flow chart for backward stepwise selection.
In the hybrid forward/backward approach, at each step one also considers including rows that have been removed in previous steps. A number of variants of hybrid approaches have been tried. However, because the pure backward selection algorithm gave very similar results and is less computationally expensive, that is used for all of the examples in Section 3.
One complication is that the expression for
is a function of the
, which depend on the unknown parameters
. Consequently, in order to design the validation sample, a preliminary estimate
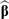 is needed. The examples in this article use the biased preliminary
is needed. The examples in this article use the biased preliminary
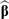 obtained by fitting the model to the original error-prone data set with
Y
* as the response. An alternative is to select the validation sample in two stages. In the first stage, a small pilot sample of rows would be selected (using the preceding algorithm with the preceding initial
obtained by fitting the model to the original error-prone data set with
Y
* as the response. An alternative is to select the validation sample in two stages. In the first stage, a small pilot sample of rows would be selected (using the preceding algorithm with the preceding initial
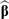 ) and validated. From this, an updated estimate
) and validated. From this, an updated estimate
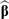 could be obtained and used when selecting the final sample of rows to validate.
could be obtained and used when selecting the final sample of rows to validate.
Another advantage of a two-stage design pertains to selecting the validation sample size
. As in any experimental design, one must balance the cost of experimentation with the quality of the fitted model. Larger
will result in a better fitted model, although the cost of chart review may be prohibitive. With a two-stage procedure, one can reassess the adequacy of
after the first-stage data are collected and analyzed. For example, the Fisher information matrix (updated using
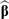 from the first-stage analysis) gives a quantitative assessment of model precision, which can be used as a criterion to decide whether a second stage is needed and, if so, how large should
be for the second-stage design.
from the first-stage analysis) gives a quantitative assessment of model precision, which can be used as a criterion to decide whether a second stage is needed and, if so, how large should
be for the second-stage design.
3 RESULTS AND DISCUSSION
3.1 SCA Example
For our case study, the testbed data described in Section 1 is used. There were a total of 70 different predictor variables, from which m = 10 variables that were believed to be the most important, based on the analysis in Mehrotra et al. 4 were retained. The 10 variables are age, average body mass index, history of congestive heart failure, history of myocardial infarction, maximal ejection fraction, maximal ventricular rate, minimal corrected QT interval, average P axis, recent diastolic blood pressure, and maximal low density lipoprotein. Binary predictors were coded as 0/1, and the continuous predictors were standardized, so that the rows and columns of have a common basis. Although and Y * exist for the N = 23 041 cases in this data set, the true Y values are missing, other than for the 20 randomly selected cases mentioned in Section 1 that had already been validated. Consequently, for the purpose of assessing the performance of the DSCVR approach, we use a Monte Carlo (MC) simulation in which we generate test simulation data sets with “true” response values generated via the approach (based on the actual SCA data) described in Appendix A.
A validation sample size of
N
v =
1000 was chosen. For each MC replicate, our backward selection algorithm of
Figure 2
was used to select the best 1000 rows to validate, out of the full 23 041 rows. For comparison, a validation sample was also chosen as a random sample from the 23 041 rows. Across the 100 MC replicates, the average value of
for the validation samples produced by our algorithm was 1.12e + 12, whereas the average of
for the randomly selected validation samples was only 1.04e + 4. Because the elements of
are the variances/covariances of
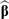 , and the dimension of
, and the dimension of
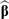 is 11, these results roughly correspond to the variances of the elements of
is 11, these results roughly correspond to the variances of the elements of
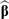 being on average
times smaller for the designed validation sample than for the randomly selected validation sample. This is a substantial improvement over random sampling.
being on average
times smaller for the designed validation sample than for the randomly selected validation sample. This is a substantial improvement over random sampling.
Another somewhat surprising and desirable characteristic of our DSCVR design strategy is that it tends to give a much higher proportion of true events in the validation sample than in the original data set. Recall that our true event rate is quite small, and in order to produce a reliable estimate of , obviously a sufficient number of events in our validation sample is needed. The average number of true events over the 23 041 rows in each MC replicate is 23 041 × 0.003 ≈ 70. In a randomly selected 1000-row validation sample, the average number of true events is only . In comparison, the average number of true events in our 1000-row DSCVR designed sample was 24, almost an order of magnitude larger.
For clinical purposes, an important consideration is how accurately the predictive model can identify patients’ risk of an event. With more accurate risk predictions, appropriate medical recommendations for preventive measures can be instituted, or better hypotheses can be generated for further testing. For assessing this aspect we compared the receiver operating characteristic (ROC) performance of our approach vs using a naive biased model (defined as a logistic regression model fit to all N rows with the error-prone Y * as the response), and vs existing validation methods that correct for the bias based on a random validation sample. The naive approach can be viewed as involving zero added cost, because no additional chart reviews or response validations are needed (hence, N v is irrelevant). Regarding the existing validation methods, since the multiple imputation method developed by Edwards et al. 10 and the joint likelihood method developed by Lyles et al. 9 gave quite similar results for this example, only the joint likelihood method is included in the comparisons. Notice that, when fitting the model, the joint likelihood approach uses the N v validated Y values, as well as the N unvalidated Y * values. In contrast, the DSCVR approach only uses the N v validated Y values and discards all of the Y * values.
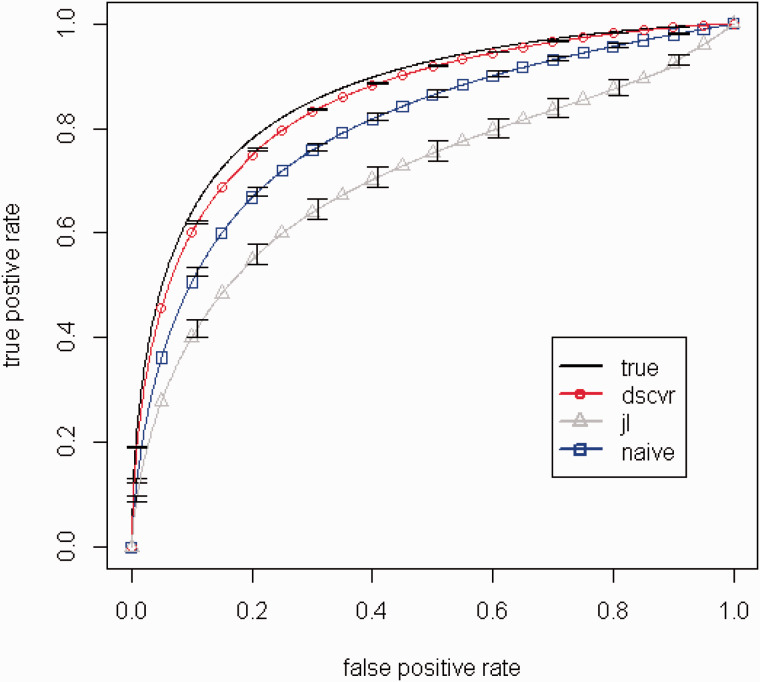
The ROC and the area under the ROC curve (AUC) were calculated for 1) our DSCVR approach, 2) a model using (which is a hypothetical benchmark, since is unknown in practice), 3) the naive biased model, and 4) the joint likelihood model. The ROCs were calculated for a very large test set of data (independent of the training sets on which the models were fit) that consisted of many copies of the values for the original 23 041 cases, but with a different set of values generated for each copy. Figure 3 shows the benchmark ROC using and the average (averaged across the 100 MC replicates) ROCs for the other three approaches (DSCVR, the naive biased model, and the joint likelihood approach), along with standard error bars for the average, and Table 1 shows the corresponding average AUCs and the standard errors of the averages. Note that a perfect classifier would have an AUC of 1.0, and a random classifier would have an AUC of 0.5 and an ROC that is a 45° diagonal line extending from the lower-left to the upper-right in Figure 3 . From Figure 3 , the DSCVR ROC is very close to the benchmark ROC curve using , and it is substantially better than the ROCs for either the naive model or the joint likelihood approach. Moreover, the DSCVR ROC has the smallest standard error bands (so narrow that they are barely visible in Figure 3 ), which means that it was the most consistent across the different MC replicates. It should be noted that the standard error bands reflect training variability, as opposed to test variability, because the training data to which the models were fit varied from replicate to replicate, and the set of test data was extremely large and did not change from replicate to replicate. It should also be noted that the standard error bands reflect uncertainty resulting from the random sampling variability introduced in the simulation for this particular set of data. This does not include other sources of uncertainty such as model biases (e.g., from neglecting important quadratic or interaction terms) or dependencies that can change over time, which may be present in practice. Error bands that reflect all possible sources of uncertainty would no doubt be wider than those shown in Figure 3 . Table 1 quantifies the differences via the AUC. The benchmark AUC using is 0.866. Remarkably, the DSCVR ROC has an average AUC of 0.850, which is nearly as good as the benchmark. Somewhat surprisingly, the joint likelihood had worse AUC than the naive method, which we discuss below.
Figure 3:
Benchmark ROC using and average ROCs for the three methods: “true” is the hypothetical benchmark using ; “dscvr” is the method of this article; “jl” is the joint likelihood approach; “naive” uses the model fit to the entire data with as the response.
Table 1:
Monte Carlo average AUCs for the four different approaches for various event rates sample sizes, and number of predictors. Standard errors of the average AUCs are in parentheses
| Using | Naive | DSCVR | JL | ||
|---|---|---|---|---|---|
| Event rate = 0.3% | 0.866 | 0.798(0.0055) | 0.850(0.0012) | 0.704(0.0161) | |
| Event rate = 1% | 0.862 | 0.834(0.0014) | 0.858(0.0002) | 0.826(0.0055) | |
| Event rate = 3% | 0.852 | 0.837(0.0007) | 0.850(0.0001) | 0.841(0.0006) | |
| Event rate = 0.3% | 0.866 | 0.798(0.0055) | 0.815(0.0049) | NA(NA) | |
| Event rate = 0.3% | 0.847 | 0.815(0.0033) | 0.837(0.0011) | 0.741(0.0153) | |
3.2 The Effects of Event Rate, Sample Size and Number of Predictors
The preceding is for a true event rate of 0.3%, which was based on the actual clinical data. The same MC simulation procedure was also used to investigate the performance of the DSCVR approach for true event rates of 1% and 3%. The true event rates were increased by adjusting the intercept parameter , while leaving the other elements of unchanged. The AUC results for the other true event rates are listed in Table 1 . When the event rate increases, all of the approaches clearly perform better (closer to benchmark AUC), but the DSCVR approach still comes out on top and has smallest standard errors for all three true event rates.
At the event rate of 3%, the differences between the three approaches are smaller. This is perhaps because the ROC and AUC only depend on the relative scoring, which only depends on the direction of the vector
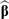 (as opposed to its magnitude), excluding
. In other words, as long as the estimated direction of the vector
is accurate, regardless of how poorly
and the magnitude of
are estimated, the ROC curve and AUC will still be good. However, accurate estimation of
and the magnitude of
, which impacts the absolute risk scores (as opposed to the relative scoring across patients), may still be of interest. To investigate this aspect, the root mean square error (RMSE) in estimating
was considered for the three different event rates in
Table 1
when
and
.
Table 2
shows the RMSE results, which were averaged over the 23 041 rows first and then averaged over the 100 replicates. Again, for all three event rates, the DSCVR approach did a better job at predicting the absolute risk probabilities than the other approaches. Notice that at the event rate of 3%, although the AUCs for three models are quite similar in
Table 1
, the RMSEs in
Table 2
are much smaller for the DSCVR approach than for the other approaches.
(as opposed to its magnitude), excluding
. In other words, as long as the estimated direction of the vector
is accurate, regardless of how poorly
and the magnitude of
are estimated, the ROC curve and AUC will still be good. However, accurate estimation of
and the magnitude of
, which impacts the absolute risk scores (as opposed to the relative scoring across patients), may still be of interest. To investigate this aspect, the root mean square error (RMSE) in estimating
was considered for the three different event rates in
Table 1
when
and
.
Table 2
shows the RMSE results, which were averaged over the 23 041 rows first and then averaged over the 100 replicates. Again, for all three event rates, the DSCVR approach did a better job at predicting the absolute risk probabilities than the other approaches. Notice that at the event rate of 3%, although the AUCs for three models are quite similar in
Table 1
, the RMSEs in
Table 2
are much smaller for the DSCVR approach than for the other approaches.
Table 2:
Average Root Mean Square Error (RMSE) in estimating for the 3 event rates in Table 1 with and .
| Event rate | DSCVR | JL | Naive |
|---|---|---|---|
| 0.3% | 0.006 | 0.027 | 0.008 |
| 1% | 0.009 | 0.021 | 0.022 |
| 3% | 0.012 | 0.027 | 0.052 |
One interesting observation is that although the joint likelihood approach had similar or better performance than the naive approach in the instances with higher true event rates, it performed substantially worse than the naive model when the true event rate was 0.3% and was also quite variable across the MC replicates. The most likely explanation is that when the event is rare, the random validation sample used in the joint likelihood approach will include very few true events. With too few true events, the bias correction inherent to the joint likelihood approach will clearly be unreliable. Moreover, the Hessian matrix of the joint log-likelihood function is poorly conditioned when there are too few true events, which makes optimization of the likelihood function a more erratic and poorly behaved problem with multiple local optima. This problem is exacerbated when the number of predictor variables is large, which is often the case in practical applications. In contrast to random validation sampling, our DSCVR approach resulted in almost an order of magnitude more true events in the validation sample. This, in turn, resulted in more reliable and stable estimation, as evident from Tables 1 and 2 .
A validation sample size of was also considered. Again, our DSCVR designed samples tend to include far more true events than random samples of the same size. The average number of true events in our DSCVR designed sample with was approximately 8, whereas the average number of true events in a random validation sample with is less than 1 ( ). Since the random sample often included not a single true event, the logistic regression model could not even be fit for the joint likelihood approach (hence the “NA” in Table 1 ). The DSCVR approach for resulted in an average AUC of 0.815, which is not drastically below the benchmark AUC of 0.866 using . Moreover, for the 0.3% event rate, the AUC for the DSCVR approach with is still far better than the AUC for the joint likelihood approach with .
A model with only the 5 most important variables was also considered, the results of which are shown in Table 1 ( , , event rate = 0.3%). The DSCVR designed sample again performs the best and has an average AUC that is the closest to the AUC of benchmark (0.837 vs of 0.847 for the benchmark AUC using ). Moreover, the standard error for the DSCVR AUC was substantially smaller than for the other methods, indicating that its performance was more consistent than the naive approach and joint likelihood approach.
3.3 Safeguarding Against Sampling Bias: DSCVR vs Oversampling Rows
Part of the reason behind the effectiveness of our DSCVR approach (in addition to selecting rows having advantageous values) is that it tends to select far more rows for the validation sample than does random sampling. In light of this, as an alternative to selecting the validation sample via DSCVR, one might consider oversampling the rows having Y * = 1 and undersampling the rows having . We will refer to this approach as oversampling observed positives (OOP). For example, with low event rates, one could select all of the rows having Y * = 1 and then randomly sample from the rows having Y * = 0 to complete the validation sample. For our SCA example ( and a true event rate of 0.3%), when the probability that depends only on the true response Y , we found that the OOP approach resulted in an average of 21 true events in the validation sample. Although the DSCVR approach resulted in even more true events (24 on average), the two approaches are quite close, and the average AUC for the OOP approach is only slightly worse than for the DSCVR approach (see the first row of Table 3 ).
Table 3:
Monte Carlo average AUCs (with their standard errors in parentheses) for DSCVR and OOP illustrating the performance degradation and bias for the OOP approach that can result when depends on . The DSCVR approach is unaffected by this.
| Case | Using | DSCVR | OOP |
|---|---|---|---|
| depends on only | 0.866 | 0.850 (0.0012) | 0.842 (0.0016) |
| depends on and | 0.866 | 0.850 (0.0012) | 0.186 (0.0042) |
However, the OOP approach should be used with caution. Unlike our DSCVR approach, the OOP approach can introduce a bias, and it is difficult to know whether the bias is present. To illustrate how the OOP approach can introduce a bias, reconsider the SCA example with , , and an event rate of 0.3%, but suppose the probability that Y * = 1 depends on both and Y via the model
where are the same parameters used in the logistic regression model for . This results in when either ( i ) or ( ii ) and is relatively large.
Using the preceding as the simulation model, the MC simulation was repeated, and the results are shown in the second row of
Table 3
. The performance of the OOP method has degraded dramatically, and its AUC is an abysmal 0.186 (an AUC of 0.5 can be achieved by pure random guessing). The reason for the degradation in OOP performance is that the specific manner in which
depends on both
and
results in a substantial bias, with
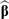 usually estimated in the opposite direction as
(hence, an AUC that is even worse than random guessing). In contrast, the DSCVR approach was not adversely affected, and its AUC remained unchanged (0.850) when
was allowed to depend on
. The reason is that the DSCVR approach selects the validation sample based only on their
values, without considering their
or
. Hence, it will not cause a sampling bias in the estimated coefficients, for the same reason that traditional DOE does not cause a bias.
usually estimated in the opposite direction as
(hence, an AUC that is even worse than random guessing). In contrast, the DSCVR approach was not adversely affected, and its AUC remained unchanged (0.850) when
was allowed to depend on
. The reason is that the DSCVR approach selects the validation sample based only on their
values, without considering their
or
. Hence, it will not cause a sampling bias in the estimated coefficients, for the same reason that traditional DOE does not cause a bias.
It should be noted that, in the preceding example in which
depends on
, the OOP approach resulted in virtually all true
events being selected in the validation sample. This is because
. In spite of this, the AUC for the OOP approach was degraded entirely because of the bias in
 .
.
4 CONCLUSIONS
With the objective of fitting reliable statistical models with highly error prone EMR data, this article presents a method that uses concepts from DOE to judiciously and effectively select the set of validation cases to have maximum information content. This is in stark contrast to existing methods based on random validation sampling. Furthermore, our DSCVR approach selects the validation sample based only on the values for each row without consideration of the Y or the Y * values, which need not be available at all when the validation sample is selected. It has been shown that our DSCVR approach has better ROC performance than existing methods in our cardiac event case study. Moreover, in the situations in which ROC performance was comparable, the DSCVR approach had better RMSE in estimating the response probabilities. It should be noted that our comparison results were from MC simulations based on the cardiac event case study. The predictor values were from the real data, but the response values were simulated. As such, there is no guarantee that the conclusions regarding ROC and RMSE performance will hold for actual future patients that are scored for cardiac risk.
In typical logistic regression applications with observational data, a common rule-of-thumb is that the number of true events in the sample required for the fitted model to have acceptable statistical precision should be roughly 10–20 per parameter. For our SCA data with parameters, this translates to a requirement of 100–200 true events in the validation sample. However, although we had on average only 24 true events in our validation samples with , the fitted models were still of high quality (the average AUC was 0.850, vs the hypothetical benchmark AUC of 0.866 using ). This underscores an inherent strength of DOE and using experimental, vs observational, data. Using experimental data from a well-designed experiment, the same statistical precision can be achieved with far smaller sample sizes. Our DSCVR approach inherits this strength, because of the manner in which the validation rows are chosen judiciously, based on their x values, similar to how experimental runs are chosen in DOE.
It is worth emphasizing that although our DSCVR approach produces a designed (as opposed to random) validation sample, it does not introduce a sampling bias in the estimated parameters. In general, choosing samples using methods other than random sampling may introduce substantial biases. For example, with highly imbalanced data, oversampling one response group and undersampling the other to give a more balanced training sample will introduce bias in the estimated parameters. It is straightforward to correct this bias (using Bayes rule), if the data were balanced based on the true response values by randomly sampling within each class. However, as demonstrated in Section 3.3, this is far more nuanced when the Y values are unknown and the data are balanced based on the error-prone Y * values. In this case, depending on the relationship between Y , Y *, and , substantial biases can be introduced that are difficult, or impossible, to correct. In contrast, our DSCVR approach avoids introducing a bias because the validation rows are chosen based solely on their values, without consideration of their response values.
One situation in which the DSCVR approach may perform poorly is when the wrong model is used. However, the naive biased approach and the joint likelihood approach suffer from the same deficiency. Our designed validation sample is optimal (heuristically) for a specific assumed model. If the actual model is different (say, with fewer variables, quadratic terms, etc.) the designed validation sample may no longer be optimal. In other words, our validation sample design may be sensitive to the assumed model structure. Using a two-stage design procedure, a more model-robust validation sample can be achieved. The first stage could be designed on the conservative side, including all relevant predictors. Then, variable selection can be done by analyzing the first-stage data and selecting the most important variables, based on which the second-stage design would be subsequently optimized (with the understanding that it will be combined with the first-stage data). We are currently investigating model-robust designs to be used in the first stage.
Missing data are commonly encountered in EMRs, and our DSCVR approach can automatically handle this, by design. In particular, when the DSCVR approach selects which subset of rows to validate, it does this based entirely on the x values for the rows. Hence, if some of the predictors are missing for a row, the DSCVR approach would simply not select that row for the validation sample. Moreover, all of the response values are treated as missing, since the DSCVR algorithm does not consider the Y or Y * values when selecting the validation sample; and when fitting the model, it only uses the validated cases, for which the outcome Y is determined by chart review.
The article has focused on the case of a binary categorical response variable. However, in empirical health risk modeling studies, it is also common to have a categorical response with more than two categories (e.g., indicating one of many subcategories of a particular disease experienced by the patient), a numerical count response (e.g., how many medical events did the patient experience), or a continuous numerical response (e.g., the ejection fraction). The DSCVR approach can be extended to these response modeling scenarios, perhaps to a broader class of generalized linear models, 22,23 which is currently under investigation. Finally, the DSCVR approach applies to the situation in which the only significant data errors are in the response variable. However, although the predictor variables are typically recorded more accurately (e.g., as clinical measurements), there are situations in which they may have significant errors as well (e.g., when patient records are mismatched). In these situations, the DSCVR approach is not applicable.
Funding
This work was supported by the National Science Foundation under Grant #CMMI-1436574, and institutional funding at Northwestern University through Center for Engineering and Health, and Department of Industrial Engineering and Management Science.
Competing Interests
None.
Contributors
All authors contributed to the conception of the approach and the algorithm development. L.O. conducted the simulation analyses, with feedback from D.A. and S.M. L.O. and S.M. compiled the data and conducted preliminary logistic regression analyses. All authors were involved in writing the article, have read and approved the final version, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
APPENDIX A: Description of the Monte Carlo Simulation and SCA Model
In order to assess the performance of the DSCVR approach, “true” response values have been generated via simulation. First, the parameters of a logistic regression model were estimated treating the actual and values from our SCA data as the predictors and response. In the simulation, these estimates were then treated as the “true” parameters, denoted by . Then, 100 MC replicates were conducted, and on each replicate a bootstrap sample of rows of values was drawn with replacement from the original set of rows. For each row of the bootstrap sample, the corresponding value was generated from a Bernoulli distribution with probability We let the error probability depend on Y via a Bernoulli model with estimated from our original data set with the 20 validated cases as follows. The actual marginalized event probabilities and error rate (marginalized across ) were roughly , and . From Bayes rule, and , which then were used as the Bernoulli probabilities when generating a value for each row of the bootstrap sample, based on the value generated for that row. We note that the values for and were only used to generate the simulation data sets, and they were not used in any way by our DSCVR algorithm when selecting the validation sample or subsequently fitting a logistic regression model to the validation sample.
REFERENCES
- 1. Batal I, Valizadegan H, Cooper GF, Hauskrecht M . A temporal pattern mining approach for classifying electronic health record data . ACM Trans Intell Syst Technol. 2013. ; 4 ( 4 ). doi :10.1145/2508037.2508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miriovsky BJ, Shulman LN, Abernethy AP . Importance of health information technology, electronic health records, and continuously aggregating data to comparative effectiveness research and learning health care . J Clin Oncol. 2012. ; 30 ( 34 ): 4243 – 4248 . [DOI] [PubMed] [Google Scholar]
- 3. SCA-AWARE . http://www.sca-aware.org/about-sca . February 10th 2014 . [Google Scholar]
- 4. Mehrotra S, Kim K, Liebovitz D, Goldberger J . Sudden cardiac arrest risk assessment using a multivariate model . J Am Coll Cardiol. 2012. ; 59 ( 13 Suppl 1 ): E564 . [Google Scholar]
- 5. ICD-10-STUDY . http://news.aapc.com/index.php/2013/09/aapc-releases-results-of-20000-record-icd-10-study/ . February 10th 2014 . [Google Scholar]
- 6. Jagannathan V, Mullett CJ, Arbogast JG, et al. . Assessment of commercial NLP engines for medication information extraction from dictated clinical notes . Int J Med Inform. 2009. ; 78 ( 4 ): 284 – 291 . [DOI] [PubMed] [Google Scholar]
- 7. Lussier YA, Shagina L, Friedman C . Automating icd-9-cm encoding using medical language processing: a feasibility study . Proc AMIA Symp. 2000. : 1072 . [PMC free article] [PubMed] [Google Scholar]
- 8. Pakhomov S, Weston SA, Jacobsen SJ, Chute CG, Meverden R, Roger VL . Electronic medical records for clinical research: application to the identification of heart failure . Am J Manag Care. 2007. ; 13 ( 6 Part 1 ): 281 – 288 . [PubMed] [Google Scholar]
- 9. Lyles RH, Tang L, Superak HM, et al. . Validation data-based adjustments for outcome misclassification in logistic regression: an illustration . Epidemiology . 2011. ; 22 ( 4 ): 589 – 597 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards JK, Cole SR, Troester MA, Richardson DB . Accounting for misclassified outcomes in binary regression models using multiple imputation with internal validation data . Am J Epidemiol. 2013. ; 177 ( 9 ): 904 – 912 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Settles B . Active legarning literature survey . Computer Sciences Technical Report 1648. Madison: University of Wisconsin; 2010 . [Google Scholar]
- 12. Zhang T, Oles FJ . A probability analysis on the value of unlabeled data for classification problems . In: Langley P , ed. Proceedings of the Seventeenth International Conference on Machine Learning . Morgan Kaufmann Publishers Inc; . San Francisco, CA, USA: , 2000. : 1191 – 1198 . [Google Scholar]
- 13. Hoi CH, Lin R, Lyu R . Large-scale text categorization by batch mode active learning . Proceedings of the 15th International Conference on World Wide Web , ACM, Edinburgh, Scotland, UK, 2006:633–642 . [Google Scholar]
- 14. Hoi CH, Jin R, Zhu J, et al. . Batch mode active learning and its application to medical image classification . Proceedings of the 23rd International Conference on Machine Learning, ACM , New York, NY, USA, 2006: 417–424 . [Google Scholar]
- 15. Schein AI, Ungar LH . Active learning for logistic regression: an evaluation . Mach Learn. 2007. ; 68 : 235 – 265 . [Google Scholar]
- 16. Montgomery DC . Design and Analysis of Experiments . 8th ed.New York: : John Wiley & Sons; ; 2012. . [Google Scholar]
- 17. Pukelsheim F . Optimal Design of Experiments (Classics in Applied Mathematics) . Society for Industrial and Applied Mathematic; , New York: ; 2006. . [Google Scholar]
- 18. Hosmer DW, Jr, Lemeshow S, Sturdivant RX . Applied Logistic Regression . 3rd ed.New York: : John Wiley & Sons; ; 2013. . [Google Scholar]
- 19. Fedorov V . Theory of Optimal Experiments (Probability and mathematical statistics) . New York: : Academic; ; 1972. . [Google Scholar]
- 20. Goos P, Jones B . Optimal Design of Experiments: A Case Study Approach . United Kingdom: : Wiley; ; 2011. . [Google Scholar]
- 21. Meyer RK, Nachtsheim CJ . The coordinate-exchange algorithm for constructing exact optimal experimental designs . Technometrics . 1995. ; 37 : 60 – 69 . [Google Scholar]
- 22. Agresti A . Categorical Data Analysis (Wiley Series in Probability and Statistics) . Hoboken, NJ, USA: : Wiley-Interscience; ; 2002. . [Google Scholar]
- 23. Mcculloch CE, Searle SR . Generalized, Linear, and Mixed Models (Wiley Series in Probability and Statistics) . Hoboken, NJ, USA: : Wiley-Interscience; ; 2001. . [Google Scholar]