Abstract
Objective Accurate food adverse sensitivity documentation in electronic health records (EHRs) is crucial to patient safety. This study examined, encoded, and grouped foods that caused any adverse sensitivity in a large allergy repository using natural language processing and standard terminologies.
Methods Using the Medical Text Extraction, Reasoning, and Mapping System (MTERMS), we processed both structured and free-text entries stored in an enterprise-wide allergy repository (Partners’ Enterprise-wide Allergy Repository), normalized diverse food allergen terms into concepts, and encoded these concepts using the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED-CT) and Unique Ingredient Identifiers (UNII) terminologies. Concept coverage also was assessed for these two terminologies. We further categorized allergen concepts into groups and calculated the frequencies of these concepts by group. Finally, we conducted an external validation of MTERMS’s performance when identifying food allergen terms, using a randomized sample from a different institution.
Results We identified 158 552 food allergen records (2140 unique terms) in the Partners repository, corresponding to 672 food allergen concepts. High-frequency groups included shellfish (19.3%), fruits or vegetables (18.4%), dairy (9.0%), peanuts (8.5%), tree nuts (8.5%), eggs (6.0%), grains (5.1%), and additives (4.7%). Ambiguous, generic concepts such as “nuts” and “seafood” accounted for 8.8% of the records. SNOMED-CT covered more concepts than UNII in terms of exact (81.7% vs 68.0%) and partial (14.3% vs 9.7%) matches.
Discussion Adverse sensitivities to food are diverse, and existing standard terminologies have gaps in their coverage of the breadth of allergy concepts.
Conclusion New strategies are needed to represent and standardize food adverse sensitivity concepts, to improve documentation in EHRs.
Keywords: food hypersensitivity, natural language processing, allergy and immunology, electronic health records, systematized nomenclature of medicine, vocabulary, controlled
OBJECTIVE, BACKGROUND, AND SIGNIFICANCE
Food allergies affect a significant portion of the global population, with a reported prevalence of between 1% and 10%.1,2 Food allergies are among the most common causes of Emergency Department visits, accounting for an estimated 525 600 Emergency Department visits annually in the United States.3,4 The International Codex Alimentarius guidelines and the US Food Allergen Labeling and Consumer Protection Act (FALCPA) require food manufacturers to label any product that contains an ingredient (eg, milk) or a food protein (eg, milk protein) derived from one or more of the eight major food allergen groups (eggs, fish, milk, peanuts, shellfish, soy, tree nuts, and wheat), which trigger an estimated 90% of food allergy reactions.5–11 Allergen labeling increases consumer awareness and is an important part of consumer-based avoidance strategies to prevent fatal anaphylactic and other types of allergic reactions from hidden allergens.5–9,12 Documenting food allergy information in electronic health records (EHRs) is important for patient care and safety. This documentation can trigger automated clinical decision support alerts in real time, to reduce adverse events13–15 caused by, for instance, food-derived products provided in the hospital.15 Although EHRs document different types of adverse sensitivities (including intolerances, idiosyncratic reactions, immune-mediated reactions, and other hypersensitivities), in this article, we use the term “food allergen” to represent food substances or sensitivities documented in the EHR’s allergy module.16
Representing and encoding allergy information using a common, standard terminology allows for data interoperability and continuity of patient care across heterogeneous systems and different organizations. The Systematized Nomenclature of Medicine – Clinical Terms (SNOMED-CT) is recommended by multiple organizations, including the National Quality Forum Quality Data Model, the Office of the National Coordinator Vocabulary Task Group, and the Individual Care Standards Collaborative Working Group (Canada) for encoding non-medication allergen concepts.17–21 In addition to SNOMED-CT, the Health Level 7 Consolidated Continuity of Care Document22,23 allows the use of the Federal Drug Administration (FDA) Unique Ingredient Identifiers (UNII) for food allergens.24 The Centers for Disease Control and Prevention released a value set mapped to SNOMED-CT that includes food allergens.19 A recent study found that, while gaps still exist, SNOMED-CT satisfies most desirable terminology characteristics (eg, content coverage, concept orientation, formal definitions, multiple granularities, vocabulary structure, and subset capability) for encoding common food allergens.17 However, it is still unclear how well SNOMED-CT and UNII classify and encode a wide variety of free-text allergens documented in EHR systems.
The goals of this study were to: 1) determine what terms healthcare providers use to document adverse sensitivities related to food in a patient’s list of allergies, 2) encode these extracted terms using standard terminologies and assess the terminologies’ coverage, and 3) report, at a high level of abstraction, a population-level analysis of food concepts contained in a large allergy repository. In this study, we examined, encoded, and grouped foods that caused any adverse sensitivity in an allergy repository. We used a generic natural language processing (NLP) application, called the Medical Text Extraction, Reasoning, and Mapping System (MTERMS),25,26 to process and map local allergy entries to the standard terminologies. Our group previously demonstrated MTERMS’s ability to encode allergy information in clinical notes.26
MATERIALS AND METHODS
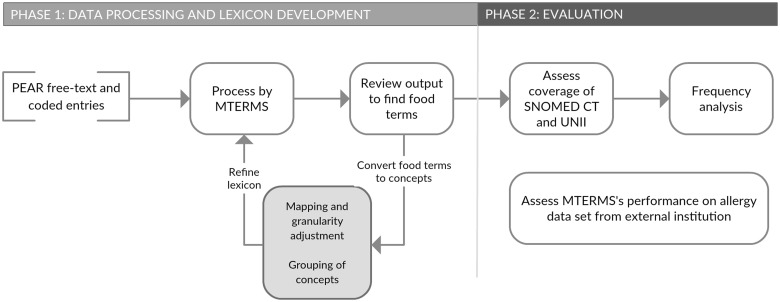
We used a two-phase approach to examine a large repository of food allergens (Figure 1).
Figure 1:
Overview of data processing, lexicon development, and evaluation.
Setting and Corpus
In the Partners Healthcare System, patients’ allergy information is entered by healthcare providers into the allergy module of the EHR systems used by each affiliated institution. Patients’ allergy data are integrated and stored in the Partners’ Enterprise-wide Allergy Repository (PEAR), so that each patient has a common allergy record that is shared within the federated provider/hospital network.27 As of October 26, 2014, PEAR consisted of 3 949 996 active records, including food, drug, and environmental allergies documented since the late 1980s. There are a number of ways to enter food allergens into our EHR systems’ allergy modules. One method is to use the “quick pick list,” which consists of a short list of commonly selected allergens. For example, the current food quick pick list includes 13 allergens: beef, caffeine, carbohydrate, chocolate, dairy products, eggs, fish, milk protein, peanuts, shellfish, soy, strawberries, and wheat. Clinicians can also utilize the “Drug Lookup” search box on the drug tab (unavailable on the food tab), which contains a dictionary of 335 food terms encoded using a commercial terminology. Alternatively, allergies can be entered as free text. Converting free-text entries to structured entries has historically relied on an existing string-match conversion tool and manual review, resulting in about 6% of total entries persisting as free text in the system.
For external validation, we collected 900 randomly selected allergy entries from the Emergency Department EHR at Tufts Medical Center between January and June 2012. Most of the allergy entries in the Tufts Medical Center EHR are coded to a commercial terminology; however, the system does include some local concepts for frequently seen allergens that are not included in the commercial terminology and some entries are free text.
This study was approved by both the Partners Healthcare System and Tufts Medical Center Institutional Review Boards.
Definitions
We define a “token” as a single word. We define a “term” as a collection of tokens (words) that are not normalized and may contain misspellings, acronyms, or plural versions. We define a “concept” as a representation of a collection of terms that represent the same allergy causative agent, with the term chosen to represent this collection of terms being a “normalized term.” We define a “group” as a collection of concepts that represent similar types of allergy causative agents. We define a “record” as an individual allergy. In contrast, an “entry” may contain multiple records or no records (ie, the entry does not represent an allergy). Cross-sensitivity is defined as “sensitivity to one substance that renders an individual sensitive to other substances of similar chemical structure.”28,29
Phase 1: Data Processing and Lexicon Refinement
In the first phase of this study, we iteratively developed an MTERMS lexicon by: 1) processing PEAR entries, then manually reviewing the output to find food terms; 2) adjusting the granularity of terms and mapping the resulting concepts to SNOMED-CT and UNII; and 3) grouping food terms at a high level of abstraction.
Processing PEAR Entries Using MTERMS
Free-text entries in PEAR contain diverse information relevant to drug, food, and environmental adverse sensitivities. At a more detailed level, this information includes allergies, intolerances, hypersensitivities, adverse reactions, criticality, severity, negations, and exceptions. MTERMS uses rules and a lexicon to: 1) tag individual allergens and reactions, 2) correct misspellings, and 3) handle contextual information (eg, negations, temporality, and exceptions) and other issues (eg, abbreviations and punctuations). MTERMS includes local and standard terminologies – SNOMED-CT (US version September 1, 2014),21 UNII (version November 20, 2014),24 and the Public Health Information Network Vocabulary Access and Distribution System (PHIN VADS) (version June 17, 2011),19 and other variations – in its lexicon and conducts a lexical look-up when processing free text. We enhanced the MTERMS lexicon by adding local interface terminology dictionaries, local allergy order sets (ie, serum allergen tests), a literature review,7,8,30–36 and a spelling checker.37 We did not consider vitamins, herbals, enteral nutrition, or parenteral nutrition as part of the food allergy list, because these substances are typically entered as drug allergies. We considered all color additives to be food additives, even though there could be other types of exposures from these substances.35 We recognize that insects (eg, grasshoppers and bees) are a food substance in some parts of the world; however, in our analysis we considered these to be environmental allergens rather than food allergens, because environmental exposure was the more common type of exposure for our patient population.
Manual Review of MTERMS Output to Find Food Terms
We manually reviewed the output from MTERMS to verify whether or not a term was a food term. First, we reviewed terms tagged by MTERMS as food terms, to validate that they represented foods and were not misclassified (eg, were actually drug allergens). Next, we reviewed terms tagged by MTERMS as another type of term (eg, drug allergens) to check for food terms that were misclassified. Then, we reviewed misspellings and acronyms that were corrected by the MTERMS spelling checker to an identified food concept, to determine whether these made sense to add as verified terms. We also reviewed cases in which negations and exceptions were present within the entry, to determine whether we were able to manually disambiguate whether a concept was negated, a true allergen, or resulted in an unidentifiable subgroup that did not represent an actual substance. For example, in the entry “all nuts except almonds,” we placed nuts into the “Exceptions” category, because the resulting subgroup is undefined, and placed almonds into the “Negated” category, because the patient in this example tolerates almonds. Finally, we reviewed untagged tokens to see whether they corresponded to an unidentified food term. We iteratively refined MTERMS’s lexicon and algorithms by performing a manual review and repeated the above steps until there were no detectable errors in the free-text PEAR entries.
Mapping of Concepts to SNOMED-CT and UNII and Adjusting Granularity of Food Terms to Concepts
Synonyms for some foods were not included in the standard terminologies (eg, SNOMED-CT includes “Isurus oxyrinchus,” but “mako shark” was not included in the terminology as a synonym for this term). In such cases, we manually mapped concepts without a synonym in the terminologies and added these synonyms to their corresponding concepts. In choosing between similar SNOMED-CT concepts in different hierarchies, we selected the substance level first. The terms “cream,” and “syrup” require semi-manual disambiguation due to multiple contexts of use in PEAR (ie, as a drug form, such as cough syrup, or as a food context, ie, “refers to cow milk fat”). We purposefully excluded the generic term “liver,” which does not (by itself) change clinical decision-making, because what animal the liver came from (eg, chicken or cow) is unknown; however, more specific concepts (eg, cod liver oil) were included.38
Adjustment of the terms’ granularity was done by merging terms that corresponded to the same substance into a single reference concept. If a term had an exact lexical match in SNOMED-CT or UNII, then we assigned it as a food allergen concept. We used the verified mappings for misspellings and acronyms, described above, to map the set of terms to their corresponding concepts. We mapped unmatched terms to existing concepts based on a semantic match (ie, the terms are synonyms at a level of granularity appropriate for clinical decision-making; eg, Macintosh apple = apple). Although there is little clinical evidence supporting allergies to flavor additives,39 this case presents a unique challenge to the task of granularity adjustment, due to its ambiguity. Several specific food flavoring agents exist in SNOMED-CT (eg, “apple flavor,” “mixed fruit flavor,” “orange flavor”); however, when there were gaps (eg, “banana flavor”), we mapped to a generic term (“food flavoring agent”), because we do not know how similar the protein structure of these natural and artificial flavors are to the causative agent (eg, “banana”).
Development of Food Allergen Groups
We further classified the individual allergen concepts into high-level food allergen groups, based on an extension and modification of FALCPA,5,6 cross-sensitivity findings,7,8,40,41 a review of SNOMED-CT’s structure and content regarding food allergen classes, and a literature review5–8,31–35 (Table 1).
Table 1:
Food Allergen Groups
| Group | Grouping rationale notes and examples |
|---|---|
| Shellfisha | The “Shellfish” group includes various species of mollusks, crustaceans, and echinoderms. Considerable cross-sensitivity exists between crustaceans (eg, shrimp, crab, and lobster), while a moderate risk of cross-sensitivity exists between crustaceans and mollusks (eg, clams and oysters).8 |
| Fruit or Vegetable | The “Fruit or Vegetable” group does not include grains (eg, corn) or legumes (eg, beans), but does includes tea, jasmine, and chamomile, which may cause similar allergic reactions. |
| Dairya | The “Dairy” group includes various dairy products, such as cow milk, lactose, milk protein, cheese, butter, etc. |
| Generic | We created a “Generic” umbrella group for ambiguous and multi-ingredient food allergens. The concept “nuts” is broad, because it may refer to peanuts and tree nuts, and, thus, belongs in the “Generic” group. “Seafood” is another umbrella term that includes fish and shellfish, and so we placed it in the “Generic” group. Other concepts that contain multiple ingredients or refer to a type of cuisine or food, such as Chinese food, baklava, pizza, ravioli, and chocolate, were classified in the “Generic” group. |
| Peanuta | The “Peanut” group includes peanuts, peanut butter, peanut oil, and grease. |
| Tree nutsa | Cross-sensitivity and co-allergy among tree nuts (eg, pistachios – cashew, and walnut – pecan) is common, as demonstrated through IgE binding to multiple tree nuts in serologic studies.7,8,40,41 |
| Egga | The “Egg” group includes egg components/products such as egg white, egg yolk, mayonnaise, eggnog, etc. |
| Grainb | While there is extensive in vitro cross-sensitivity between grains (eg, wheat and corn) and grass pollens,7,8 clinically, there is little cross-sensitivity; thus, we categorized grass pollens as environmental allergies. |
| Additive | The “Additive” group includes monosodium glutamate, dyes (eg, food coloring, Yellow Dye #5, and FD&C Blue No. 2), food preservative, sweeteners (eg, aspartame, sucrose, and artificial sweetener), caffeine, etc. |
| Fisha | The “Fish” group includes various species of fish, including salmon, tuna, swordfish, cod, anchovy, etc. |
| Soya | The “Soy” group includes various soy preparations, such as soy milk, soy sauce, tofu, soybean oil, frosting, soy protein, etc. |
| Seed | The “Seed” group includes various seeds, such as sesame seed, sunflower seed, cocoa, coffee, quinoa, etc. |
| Meat | The “Meat” group includes various preparations of meat, including beef, poultry, lamb, duck, ham, pepperoni, etc. |
| Spice | The “Spice” group includes garlic, black pepper, cinnamon, curry powder, sage, ginger, paprika, red pepper, etc. |
| Alcohol | The “Alcohol” group includes various ingestible alcohols, including fruit-based alcohols (eg, wine and red wine), grain-based alcohols (eg, beer and barley malt syrup), and liquors (eg, tequila). |
| Legume | The “Legume” group includes members of the legume family (eg, bean, chickpea, snow pea, red bean, and kidney bean), except peanut and soy, which are categorized into separate groups, based on previous cross-sensitivity studies. We placed the concept “legume” in the “Generic” group, because it is unclear whether the concept referred to peanuts, soy, or another member of the legume family. |
| Fungus | The “Fungus” group includes cultivated mushrooms, yeast, truffles, Portobello mushrooms, etc. |
| Extract | The “Extract” group includes types of edible cooking oils, such as olive oil, as well as other extracts, such as annatto, yeast extract, etc. |
| Infant formulas | The “Infant formula” includes infant formulas, such as Enfamil Prosobee Lipil, Similac, Elecare Powder, Nutramigen, Enfacare, as well as breast milk and baby food. |
| Nutritional supplement | The “Nutritional supplement” group includes nutritional or dietary supplements, such as Ensure, red yeast, fiber, etc. |
| No known food allergies | We captured “No known food allergy” as a category separate from food allergens, because this documentation is clinically useful. |
IgE, Immunoglobulin E.
aThis group is one of the eight major food allergens defined by the Food Allergen Labeling and Consumer Protection Act (FALCPA).
bA subgroup of “grain” corresponding to “wheat” is part of the eight major food allergens defined by FALCPA.
Studies have shown that there is a high rate of coexistence between peanut and tree nut allergies (∼25–50%) in some populations and homologous proteins between these foods by in vitro inhibition.7,8 Peanuts and soybeans are members of the legume family, but studies suggest that the in vitro cross-reacting antibodies are not clinically relevant, because patients with peanut allergy generally tolerate other legumes and soy.7,8 Although legumes are often considered to be vegetables, studies have found that legume-induced adverse reactions (eg, anaphylaxis) are often more severe than those caused by other vegetables.7,8 We therefore created a group called “Fruit or Vegetable,” because these foods often cause oral allergy syndrome or other mild adverse reactions.7,8
Because there is no evidence that shellfish cross-react with vertebrate fish,7,8 we categorized “Fish” and “Shellfish” as individual groups.
Phase 2: Evaluation Methods
Once our lexicon reached stability, we moved on to the evaluation phase and gleaned clinical insights from our corpus.
Assessment of Concept Mapping to SNOMED-CT and UNII
We assessed the concept mapping results and the coverage of SNOMED-CT and UNII for food terms in PEAR using the methods demonstrated in similar studies by Zhou.25,26,42 Mapping was assessed using four levels: exact match, narrower partial match, broader partial match, and no match. Percentages of concepts matched at each level were calculated.
Frequency Analysis
We calculated the frequency of food allergens within PEAR by group, as defined above. Free-text and structured entries were analyzed separately and were presented alongside the combined total for each group.
Assessing MTERMS Performance in Identifying Allergen Concepts
The allergy entries from the Emergency Department at Tufts Medical Center were used as external validation of MTERMS and our lexicon. The commonly used statistical measures of precision, recall, and F-measure were calculated.43
RESULTS
There were 2 730 250 patients with 3 949 996 active allergy records (including drug, food, and environmental allergies) in PEAR. Among these records, 93.7% (n = 3 701 332; unique = 5705) of the information on allergens was found to be in the form of structured entries, compared with 6.3% (n = 248 664; unique = 88 686) that was in the form of free-text entries. About 3.7% (100 194) of patients had at least one food allergy record. We identified 158 552 food allergy records (2140 unique allergen terms), accounting for 4.0% of the total records, of which 75 297 (47.5%) were stored in a free-text format.
Normalization of Terms to Food Allergen Concepts Results
We converted the 2140 unique allergen terms into 672 concepts. Of the 672 concepts in PEAR, 53 were only in the structured entries, 399 were only in the free-text entries, and 220 were found in both.
Assessment of Concept Mapping Between SNOMED-CT and UNII Results
We found that SNOMED-CT contained more food allergen concepts documented in PEAR than UNII (Table 2). Of the 672 total concepts, only 35.6% are included in the Centers for Disease Control and Prevention PHIN VADS subset. Investigation of the identified SNOMED-CT concepts found that 90.2% are under the SNOMED-CT substance hierarchy, and the rest are under the organism (6.7%), product (2.6%), qualifier value (0.3%; eg, “sprinkle” or “gum”), or finding (0.2%; eg, “poisoning by sea cucumber”) hierarchies.
Table 2:
Concept-Level Mapping for Food Allergens from PEAR to SNOMED-CT and from PEAR to UNII
| Matching | SNOMED-CT | UNII | Examples |
|---|---|---|---|
| (n = 672 total concepts) | n (%) | n (%) | |
| Exact | 549 (81.7) | 457 (68.0) | The fish “mahi mahi” is “mahi mahi – dietary” in SNOMED-CT and “mahi-mahi” in UNII. |
| Broader (Partial)a | 65 (9.7) | 27 (4.0) | In SNOMED-CT, “crab – dietary” is the closest concept to “Dungeness crab” that is found in UNII. |
| Narrow (Partial)b | 31 (4.6) | 38 (5.7) | The closest match to “crab” in UNII is “crab leg, unspecified,” as we could not find a “crab, unspecified” term. |
| Missing (No match) | 27 (4.0) | 150 (22.3) | “Gatorade,” “Mexican food,” “goose egg,” “Irish cream flavor,” “orange soda,” and “sprout” are not found in either terminology. Several exotic fruits or vegetables (eg, “Pitaya,” “lemongrass,” “logan fruit,” “mangosteen,” “mustard greens,” “wasabi,” “acai,” “Asian pear,” and “chayote”), a fish derivative (“caviar”), a shellfish (“langostino”), extracts (eg, “annatto extract”), additives (eg, “guarana,” “orange 5,” and “12-aminododecanoic acid”), a nutritional supplement (“red yeast”), a seed (“cocoa”), a grain (“corn meal”), and a flower used in tea (“Jasmine”) are not found in SNOMED-CT. UNII was the most deficient in multi-ingredient or ambiguous concepts. |
PEAR, Partners’ Enterprise-wide Allergy Repository; SNOMED-CT, Systematized Nomenclature of Medicine – Clinical Terms; UNII, Unique Ingredient Identifiers.
aThe “food allergen” concept in our lexicon has a less specific meaning than the corresponding term in SNOMED-CT or UNII.
bThe “food allergen” concept in our lexicon has a more specific meaning than the corresponding term in SNOMED-CT or UNII.
Frequency Analysis by Group Results
We analyzed the frequency of the PEAR food allergen records at the group level (Table 3). Our deep analysis on subgroups of grain and shellfish showed that 3111 (2.0% of total food terms) were wheat, 879 (0.6%) were corn, 5642 (3.6%) were crustaceans, and 2613 (1.7%) were mollusks. We found that the FDA FALCA’s eight food allergen groups (eggs, fish, milk, peanuts, shellfish, soy, tree nuts, and wheat) account for a total of 58.2% of the food terms in PEAR.
Table 3:
Frequency of Food Allergen Records by Group in PEAR
| Group | Total Records | Structured Records | Free-Text Records |
|---|---|---|---|
| n (%) | n (%) | n (%) | |
| Shellfisha | 30 522 (19.3) | 20 988 (25.2) | 9534 (12.7) |
| Fruit or Vegetableb | 29 137 (18.4) | 9875 (11.9) | 19 262 (25.6) |
| Dairya | 14 291 (9.0) | 11 030 (13.2) | 3261 (4.3) |
| Generic | 13 969 (8.8) | 1579 (1.9) | 12 390 (16.5) |
| Peanuta | 13 554 (8.5) | 12 524 (15.0) | 1030 (1.4) |
| Tree nutsa | 13 462 (8.5) | 668 (0.8) | 12 794 (17.0) |
| Egga | 9548 (6.0) | 8351 (10.0) | 1197 (1.6) |
| Grainc | 8157 (5.1) | 6006 (7.2) | 2151 (2.9) |
| Additive | 7377 (4.7) | 3987 (4.8) | 3390 (4.5) |
| Fisha | 4662 (2.9) | 2910 (3.5) | 1752 (2.3) |
| Soya | 3077 (1.9) | 2512 (3.0) | 565 (0.8) |
| Seed | 2462 (1.6) | 298 (0.4) | 2164 (2.9) |
| Meat | 2203 (1.4) | 450 (0.5) | 1753 (2.3) |
| Spice | 1470 (0.9) | 796 (1.0) | 674 (0.9) |
| Alcohol | 1428 (0.9) | 687 (0.8) | 741 (1.0) |
| Legumed | 1279 (0.8) | 67 (0.1) | 1212 (1.6) |
| Fungus | 1267 (0.8) | 201 (0.2) | 1066 (1.4) |
| Extract | 600 (0.4) | 319 (0.4) | 281 (0.4) |
| Infant formulas | 55 (0.0) | 4 (0.0) | 51 (0.1) |
| Nutritional supplement | 32 (0.0) | 3 (0.0) | 29 (0.0) |
| Total foods | 158 552 (100.0) | 83 255 (100.0) | 75 297 (100.0) |
| Exceptionse | – | 89 | 89 |
| Negatede | – | 169 | 169 |
| No known food allergy | – | 281 | 281 |
PEAR, Partners’ Enterprise-wide Allergy Repository.
aThis group is one of the eight major food allergens defined by the Food Allergen Labeling and Consumer Protection Act (FALCPA).
bThe “Fruit or Vegetable” group does not include grains (eg, corn) or legumes (eg, beans), but does include tea, jasmine, and chamomile, which may cause similar allergic reactions.
cA subgroup of “grain” corresponding to “wheat” is part of the eight major food allergens defined by the FALCPA. “Wheat” corresponds to 3111 (2.0%) of the total food terms.
dThe “Legume” group includes members of the legume family (eg, bean, chickpea, snow pea, red bean, and kidney bean), except peanut and soy, which are categorized as separate groups, based on previous cross-sensitivity studies. We placed the concept “legume” in the “Generic” group, because it is unclear whether the concept referred to peanut, soy, or another member of the legume family.
eWe investigated patterns for exceptions and negations. Looking at the example “all nuts except cashews,” we placed “nuts” into the “Exceptions” category, because the resulting subgroup is undefined, and placed “cashews” into the “Negated” category, because the patient in this example tolerates cashews.
Results of Validation on External Corpus of Allergy Entries
MTERMS achieved 100% precision and 97.0% recall, yielding an F1-measure of 98.5% on our external sample. We identified 169 food terms in the 900 Tufts Medical Center EHR allergy entries. In this external sample, our lexicon covered the majority of food allergens. The food allergen terms we manually identified as not being in PEAR were misspellings (eg, “lactose intolerent,” “califlower,” “broccli,” and “tomatoe soup”) and an abbreviation of “orange juice” (ie, “OJ”).
DISCUSSION
We developed a semi-automated approach to process and analyze the large quantity of food allergen entries in EHRs using NLP. We present an overview of food terms in the allergy repository and their respective coverage across standard terminologies. The proposed approach and the MTERMS NLP system achieved a high performance when identifying food allergies using comparable data from a different institution. Food allergies represented a substantial fraction of all the allergens recorded. Similar to previous studies,17 we found that the SNOMED-CT terminology had a higher coverage rate for food allergen concepts documented in patients’ allergy lists than the UNII terminology.
Food Allergens
To date, research on food allergies has largely been based on surveys, self-reported data, and billing codes.4,10,44 The prevalence of food allergies in our study was 3.7%, which is consistent with prevalence estimates made using clinically verified methods (eg, skin prick testing [4–6%]).2 Although prior research suggests that the FDA FALCPA subset accounts for 90% of food allergens,11,45 our results showed a lower rate, of 58.2%. One likely explanation for the above difference is that existing data reports are largely based on immune-mediated reactions to food.46 In practice, we found that patients tend to report all types of food adverse sensitivities (ie, any unpleasant reaction to food). Clinicians (excluding allergists) input such patient-reported data to PEAR via the EHRs allergy module. One unexpected finding was clinicians’ documentation of patients’ various dietary preferences related to food products (eg, vegetarian, gluten-free, Atkins diet, casein-free, and kosher) in the PEAR free-text allergen field.
Knowing the source and clinical certainty of an allergy is important in some clinical cases. One recommendation for EHR design to support such cases would be to highlight those allergies that have been verified by a sensitization test or a documented severe allergic reaction, in order to distinguish between true allergies and hypersensitivities, intolerances, or preferences. For example, in one of the commercial EHR systems currently in use at our organization, a provider can specify in a structured way whether the concept is an intolerance or an allergy; however, this system lacks similar capabilities for specifying whether a concept is a hypersensitivity or a preference.
SNOMED-CT and UNII for Representing Food Allergens
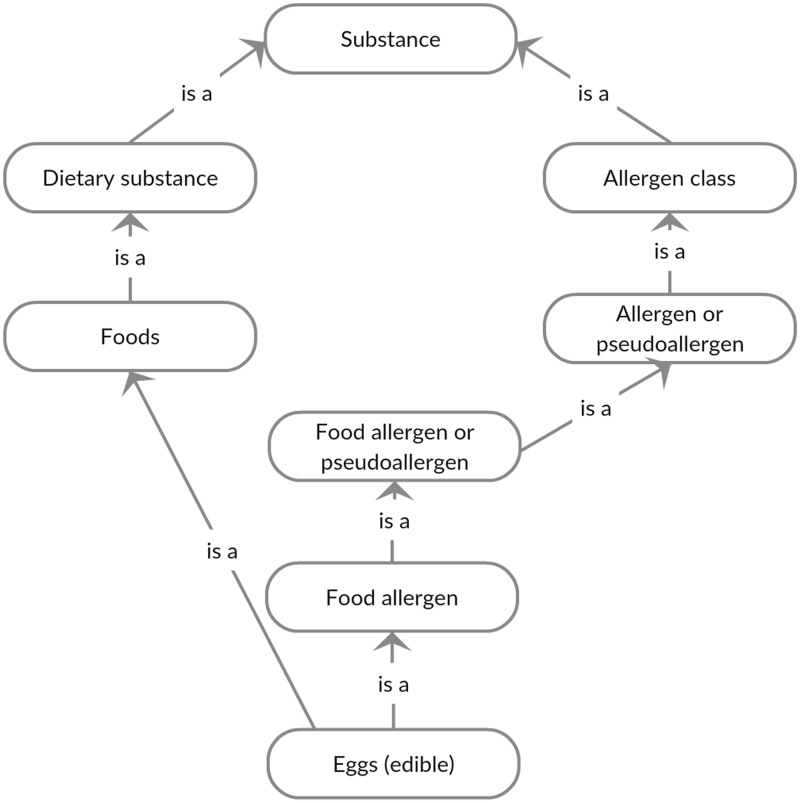
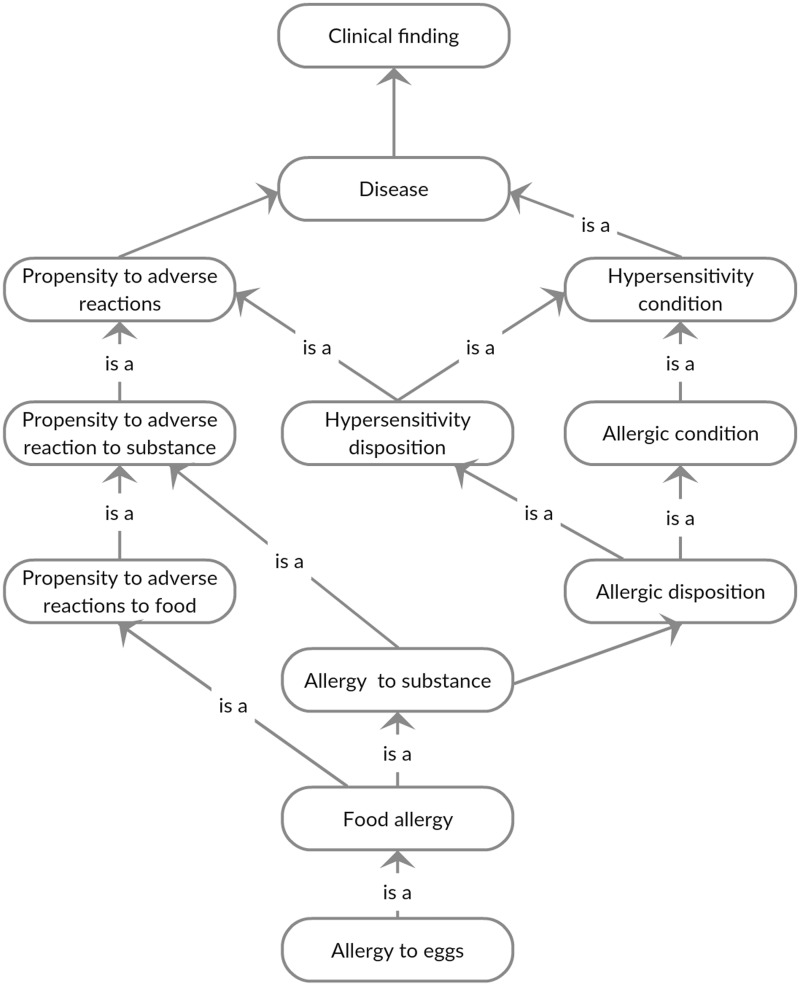
We examined the structure and content of SNOMED-CT (July 2014 release) to better understand how food allergies are documented. In terms of content, SNOMED-CT contains both specific ingredients and more generic multi-ingredient concepts from consumers’ vocabulary. The concepts in SNOMED-CT tend to be less specific than UNII (eg, “clam – dietary” vs “razor shell clam whole”), which may be sufficient for most clinical food allergy documentation use cases. We found that food allergy concepts are represented at both the substance (see Figure 2 in the Appendix) and disorder (see Figure 3 in the Appendix) levels.
At the substance level, food allergens are classified according to allergen class. Granularity at this level is markedly higher, and, when examining the SNOMED-CT hierarchy, we found approximately 900 food allergen terms (descendants of food allergen). Food concepts are also included under the dietary substance sub-tree. However, 258 of the 900 food allergens in SNOMED-CT are not descendants of the dietary substance sub-tree, indicating that future work is needed to check the consistency between these two classes. Guidelines are also needed for determining which concept (food allergen or dietary substance) should be used to encode food allergens documented in EHRs.
At the disorder level, allergens are described as a propensity to adverse reactions. The concepts under the disorder classification are typically precoordinated with the term “allergy to” and the causative agent (eg, “allergy to” + “cherry” = “allergy to cherry”). These concepts are fewer in number (ie, 47 concepts, compared with the 900 substance-level allergens). Interestingly, the substance used to define a food allergy does not always come from the descendant of disorder, and, 24 of the 47 food allergen concepts under the disorder classification belong to the dietary substance sub-tree (descendant of substance).
In UNII, food allergens are represented as a flat list, using the molecular structure of the substance, or, alternatively, defined by descriptive information (eg, “Chinese white shrimp”) in cases in which the structure of the substance is not available. Because concepts in UNII are represented more at the ingredient or substance level, the terminology is missing many of the multi-ingredient or ambiguous causative agents that are common in consumers’ vocabulary. Adding terms outside of the current substance-level scope of UNII to represent generic concepts would be necessary. One notable limitation of UNII is that it does not support hierarchical or semantic relationships between substance concepts.24
Exceptions and Negations
There is a pattern for exceptions within PEAR: a clinician documents a patient’s allergy at the class level but do not specify all of the allergens the patient is allergic to and, instead, specifies what the patient tolerates (eg, “all fruit except berries and oranges”). Similarly, there are patterns in PEAR free-text entries for tolerance (eg, “tolerates egg in food”) and diagnostic investigations (eg, “rule out food allergy”). Outside of “No known allergy,” negations within PEAR are very uncommon, because clinicians do not typically make lists of everything a patient is not allergic to, except when discussing results of sensitization tests (which are not documented in PEAR). There are cases in which a physician will write “No known drug allergies” and then list food allergens, which we can convert to food allergy concepts, because these terms are not a subset of drugs and are thus not negated. In the Tufts Medical Center EHR entries, the phrase “No identified allergies” shows up 42 times, often followed by a substance(s), which indicates that this concept was added to the allergy database within the EHR, allowing it to be selected. This suggests a need to reconcile or auto-remove this concept if any allergens are present.
Impact on EHR User Interface Design and Terminology Development
Compared to other types of allergens, food allergens are more likely to be documented in free-text entries, which is indicative of the lack of a comprehensive, controlled food allergen terminology. Several of the most common free-text food allergen entries in PEAR are not present in our EHR’s coded quick pick list (eg, “shrimp,” “walnuts,” and “lobster”). Granularity in this list should be at the concept level rather than the term level, and concepts in the “Generic” group should not be included on the list, to encourage more detailed documentation.
When entering a long list of patient-reported allergens, clinicians may find it easier to enter these as a free-text list rather than clicking through multiple tabs to document the allergens in a structured format. Implementing a more efficient user interface would reduce free-text entries, encouraging structured data entry using concepts on which clinical decision support rules can act.
SNOMED-CT could be improved by utilizing the disorder hierarchy for the food groups (e.g., grain) or subgroups (eg, wheat, corn, mollusks, and crustaceans) in which the children of these class-level groups are in the substance hierarchy. This would create greater cohesion between the hierarchies and enable group-level reporting using precoordinated terms. Additional concepts and synonyms should be added to both SNOMED-CT and UNII to fill the gaps we identified. It may also make sense for the International Health Terminology Standards Development Organisation to review SNOMED-CT’s food allergen categories to ensure that they are all at similar levels of granularity.
LIMITATIONS
From an allergist’s perspective, PEAR data represents unverified reported hypersensitivities/allergies to foods. PEAR does not document true food allergens, because allergists at our organization document verified food allergies in the problem list instead of in PEAR. Therefore, we could not differentiate between different types of adverse sensitivities to identify which are true allergens, as opposed to intolerances, hypersensitivities, or preferences. We used patient-reported data from a predominately adult population that was entered by a clinician at the time of reporting and may not capture reported allergies that occurred during childhood or infancy. We surmise that many of the food allergies documented in PEAR, especially those entered as free text, have not been clinically verified by, for example, sensitization tests. We assessed the coverage of UNII and SNOMED-CT using a set of terms collected from only one institution’s data repository. Prevalence estimates are likely conservative, as it is likely that many food allergens are not yet recorded.
There may be regional differences in the prevalence of specific types of allergies.45 Our population is largely based in New England; thus, the frequencies we report for various food allergens are influenced by regional plant life and foods and may not be generalizable to other regions of the world or other healthcare delivery systems.
FOOTNOTES
Contributions J.M.P. and F.R.G. contributed equally to this work. All authors provided a substantial contribution to the conception and design of this work, its data analysis and interpretation, and helped draft and revise the manuscript. All the authors are accountable for the integrity of this work.
CONCLUSION
We present a semi-automated approach to process, encode, and group food adverse sensitivities in the EHR. We found that food allergens are diverse, and there are knowledge gaps in existing documentation standards. Strategies for representing and standardizing food allergen concepts are needed to improve the documentation of this information in EHRs.
ACKNOWLEDGEMENTS
We would like to thank James Shalaby, Carol Broverman, Neil Dhopeshwarkar, Kin Wah Fung (NIH/NLM/LHC), George Robinson (FDB), Robert McClure, John Kilbourne (NIH/NLM), Shelly Spiro, Mina Kim, and Chunlei Tang for their assistance in this research.
APPENDIX
Figure 2:
Systematic Nomenclature of Medical Terms – Clinical Terms (SNOMED-CT) hierarchy for egg allergy (substance).
Figure 3:
Systematic Nomenclature of Medical Terms – Clinical Terms (SNOMED-CT) hierarchy for egg allergy (disorder).
FUNDING
This study was funded by Agency for HealthCare Research and Quality grant 1R01HS022728-01.
COMPETING INTERESTS
None.
REFERENCES
- 1.Chafen JJ, Newberry SJ, Riedl MA, et al. Diagnosing and managing common food allergies: a systematic review. JAMA. 2010;303(18):1848–1856. [DOI] [PubMed] [Google Scholar]
- 2.Rona RJ, Keil T, Summers C, et al. The prevalence of food allergy: a meta-analysis. J Allergy Clin Immunol. 2007;120(3):638–646. [DOI] [PubMed] [Google Scholar]
- 3.Clark S, Espinola J, Rudders SA, Banerji A, Camargo CA. Frequency of US emergency department visits for food-related acute allergic reactions. J Allergy Clin Immunol. 2011;127(3):682–683. [DOI] [PubMed] [Google Scholar]
- 4.Clark S, Espinola JA, Rudders SA, Banerji A, Camargo CA. Favorable trends in the frequency of U.S. emergency department visits for food allergy, 2001-2009. Allerg Asthma Proc. 2013;34(5):439–445. [DOI] [PubMed] [Google Scholar]
- 5.Randell AW, Whitehead AJ. Codex alimentarius: food quality and safety standards for international trade. Revue Scientifique et Technique. 1997;16(2):313–321. [DOI] [PubMed] [Google Scholar]
- 6.United States Food and Drug Administration. http://www.fda.gov/food/labelingnutrition/FoodAllergensLabeling/GuidanceComplianceRegulatoryInformation Food Allergen Labeling and Consumer Protection Act of 2004 (Public Law 108-282, Title II). [accessed on April 4, 2012]. [Google Scholar]
- 7.Sampson HA, Aceves S, Bock SA, et al. Food allergy: a practice parameter update – 2014. J Allergy Clin Immunol. 2014;134(5):1016–1025, e43. [DOI] [PubMed] [Google Scholar]
- 8.American College of Allergy, Asthma, and Immunology. Food allergy: a practice parameter. Ann Allergy, Asthma, Immunol. 2006;96(3 Suppl 2):S1–S68. [PubMed] [Google Scholar]
- 9.Nebraska-Lincoln Food Allergy Research and Resource Program. Food Allergens – International Regulatory Chart, http://farrp.unl.edu/c/document_library/get_file?uuid=f0c3a875-ce07-404f-b05f-8a7983e57daa&groupId=2103626&.pdf. Accessed July 27, 2014. [Google Scholar]
- 10.Chafen JJS, Newberry SJ, Riedl MA, et al. Prevalence, Natural History, Diagnosis, and Treatment of Food Allergy. RAND Health; 2010, WR757-1. http://www.rand.org/content/dam/rand/pubs/working_papers/2010/RAND_WR757-1.pdf Accessed July 27, 2014. [Google Scholar]
- 11.FARE – Food Allergy Research and Education. http://www.foodallergy.org/document.doc?id=194 Food Allergy Facts and Statistics for the U.S. [accessed on 29 July, 2014]. [Google Scholar]
- 12.Yunginger JW. Lethal Food Allergy in Children. New Engl J Med. 1992;327(6):421–422. [DOI] [PubMed] [Google Scholar]
- 13.An epidemiologic study of severe anaphylactic and anaphylactoid reactions among hospital patients: methods and overall risks. The International Collaborative Study of Severe Anaphylaxis. Epidemiology. 1998;9(2):141–146. [PubMed] [Google Scholar]
- 14.Harikumar S, Carpenter JE, Ledan L. Hospital-acquired anaphylaxis. US Pharm. 2013;38(7):HS10–HS14. [Google Scholar]
- 15.Kelso JM. Potential food allergens in medications. J Allergy Clin Immunol. 2014;133(6):1509–1518. [DOI] [PubMed] [Google Scholar]
- 16.Macy E, Poon KYT. Self-reported antibiotic allergy incidence and prevalence: age and sex effects. Am J Med. 2009;122(8):778 e1–7. [DOI] [PubMed] [Google Scholar]
- 17.Goss FR, Zhou L, Plasek JM, et al. Evaluating standard terminologies for encoding allergy information. JAMIA. 2013;20(5):969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Committee SaT. Allergy. In: Committee SaT, ed. House of Lords; 2006, http://www.bsaci.org/pdf/HoL_science_report_vol.1.pdf. [Google Scholar]
- 19.PHIN Vocabulary Access and Distribution System (VADS). Allergy/Adverse Event Type Value Set (Revised) ed: Centers for Disease Control; 2013, https://phinvads.cdc.gov/vads/ViewValueSet.action?id=7AFDBFB5-A277-DE11-9B52-0015173D1785# Accessed July 17, 2014. [Google Scholar]
- 20.Andre Boudreau ML. Pan-Canadian Approach to Allergy, Intolerance, and Adverse Reaction (Interoperable EHR). HL7 Patient Care Working Group 2011. http://wiki.hl7.org/images/d/d9/Canadian_Approach_Allergy-Intolerance-AR-20110719c-post_meetg.pdf. [Google Scholar]
- 21.SNOMED-CT (Systematized Nomenclature of Medicine – Clinical Terms). [accessed on September 1, 2014]. http://www.ihtsdo.org/snomed-ct. [Google Scholar]
- 22.D'Amore J., HL7 Task Force Examples. [Accessed on April 5, 2015] https://github.com/jddamore/HL7-Task-Force-Examples/blob/master/ALLERGY_Food_allergy.xml. [Google Scholar]
- 23.Value Sets Defined in FHIR. HL7. [Accessed on April 5, 2015]. http://www.hl7.org/implement/standards/fhir/2015May/terminologies-valuesets.html. [Google Scholar]
- 24.Substance Registration System-Unique Ingredient Identifier (UNII). http://www.fda.gov/ForIndustry/DataStandards/SubstanceRegistrationSystem-UniqueIngredientIdentifierUNII/default.htm [accessed on October 27, 2011]. [Google Scholar]
- 25.Zhou L, Plasek JM, Mahoney LM, et al. Using Medical Text Extraction, Reasoning and Mapping System (MTERMS) to process medication information in outpatient clinical notes. AMIA Ann Symp Proc. 2011;2011:1639–1648. [PMC free article] [PubMed] [Google Scholar]
- 26.Goss FR, Plasek JM, Lau JJ, Seger DL, Chang FY, Zhou L. An evaluation of a natural language processing tool for identifying and encoding allergy information in emergency department clinical notes. AMIA Annu Symp Proc. 2014; 2014: 580–588. [PMC free article] [PubMed] [Google Scholar]
- 27.Kuperman GJ, Marston E, Paterno M, et al. Creating an enterprise-wide allergy repository at Partners HealthCare System. AMIA Annu Symp Proc. 2003:376–380. [PMC free article] [PubMed] [Google Scholar]
- 28.Myers T. Mosby's Medical Dictionary. 7th ed. St. Louis, MO: Mosby/Elsevier; 2006. [Google Scholar]
- 29.Anovadiya AP, Barvaliya MJ, Patel TK, Tripathi CB. Cross sensitivity between ciprofloxacin and levofloxacin for an immediate hypersensitivity reaction. J Pharmacol Pharmacotherap. 2011;2(3):187–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epstein RH, St Jacques P, Stockin M, Rothman B, Ehrenfeld JM, Denny JC. Automated identification of drug and food allergies entered using non-standard terminology. JAMIA. 2013;20(5):962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Common Food Allergies. 2009 http://www.medicalnewstoday.com/articles/8624.php [accessed on 30 July, 2014]. [Google Scholar]
- 32.Gershman N. Food Allergy (including list of possible allergens). [accessed on 30 July, 2014]. http://www.allergyweb.com/article-list-of-food-allergies.html. [Google Scholar]
- 33.Food Allergens. 2013 http://www.fda.gov/food/ingredientspackaginglabeling/foodallergens/ucm2006768.htm [accessed on 30 July, 2014]. [Google Scholar]
- 34.The Food Allergy & Anaphylaxis Network. Food Allergy Training Guide for Hospital and Food Service Staff. 2006 [accessed on 30 July, 2014]. http://www.foodallergy.org/document.doc?id=149. [Google Scholar]
- 35.Summary of Color Additives for Use in United States in Foods, Drugs, Cosmetics, and Medical Devices. FDA. 2013. [accessed on 5/3/2015]; Available from: http://www.fda.gov/forindustry/coloradditives/coloradditiveinventories/ucm115641.htm. [Google Scholar]
- 36.InformAll Allergenic Food Database. iFAAM. [Accessed on 7/27/2014]. http://www.inflammation-repair.manchester.ac.uk/informall/allergenic-foods/. [Google Scholar]
- 37.Lai KH, Topaz M, Goss FR, Zhou L. Automated misspelling detection and correction in clinical free-text records. J Biomed Inform. 2015; 55:188-195. [DOI] [PubMed] [Google Scholar]
- 38.Ahn Y-Y, Ahnert SE, Bagrow JP, Barabási A-L. Flavor network and the principles of food pairing. Sci Rep. 2011;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matheu V, Zapatero L, Alcazar M, Martinez-Molero MI, Baeza ML. IgE-mediated reaction to a banana-flavored drug additive. J Allergy ClinI Immunol. 2000;106(6):1202–1203. [DOI] [PubMed] [Google Scholar]
- 40.Ewan PW. Clinical study of peanut and nut allergy in 62 consecutive patients: new features and associations. BMJ. 1996;312(7038):1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sicherer SH, Burks AW, Sampson HA. Clinical features of acute allergic reactions to peanut and tree nuts in children. Pediatrics. 1998;102(1):e6. [DOI] [PubMed] [Google Scholar]
- 42.Zhou L, Plasek JM, Mahoney LM, Chang FY, Dimaggio D, Rocha RA. Mapping Partners Master Drug Dictionary to RxNorm using an NLP-based approach. J Biomed Inform. 2012. 45(4):626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval. New York, Harlow, England: ACM Press; Addison-Wesley; 1999. [Google Scholar]
- 44.Wood RA, Camargo CA, Jr, Lieberman P, et al. Anaphylaxis in America: the prevalence and characteristics of anaphylaxis in the United States. J Allergy Clin Immunol. 2014;133(2):461–467. [DOI] [PubMed] [Google Scholar]
- 45.Salo PM, Arbes SJ, Jr, Jaramillo R, et al. Prevalence of allergic sensitization in the United States: results from the National Health and Nutrition Examination Survey (NHANES) 2005-2006. J Allergy Clin Immunol. 2014;134(2):350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gell PGH, Coombs RRA. Clinical Aspects of Immunology. Oxford: Blackwell; 1963. [Google Scholar]