Abstract
Objective To evaluate the phenotyping performance of three major electronic health record (EHR) components: International Classification of Disease (ICD) diagnosis codes, primary notes, and specific medications.
Materials and Methods We conducted the evaluation using de-identified Vanderbilt EHR data. We preselected ten diseases: atrial fibrillation, Alzheimer’s disease, breast cancer, gout, human immunodeficiency virus infection, multiple sclerosis, Parkinson’s disease, rheumatoid arthritis, and types 1 and 2 diabetes mellitus. For each disease, patients were classified into seven categories based on the presence of evidence in diagnosis codes, primary notes, and specific medications. Twenty-five patients per disease category (a total number of 175 patients for each disease, 1750 patients for all ten diseases) were randomly selected for manual chart review. Review results were used to estimate the positive predictive value (PPV), sensitivity, and F -score for each EHR component alone and in combination.
Results The PPVs of single components were inconsistent and inadequate for accurately phenotyping (0.06–0.71). Using two or more ICD codes improved the average PPV to 0.84. We observed a more stable and higher accuracy when using at least two components (mean ± standard deviation: 0.91 ± 0.08). Primary notes offered the best sensitivity (0.77). The sensitivity of ICD codes was 0.67. Again, two or more components provided a reasonably high and stable sensitivity (0.59 ± 0.16). Overall, the best performance ( F score: 0.70 ± 0.12) was achieved by using two or more components. Although the overall performance of using ICD codes (0.67 ± 0.14) was only slightly lower than using two or more components, its PPV (0.71 ± 0.13) is substantially worse (0.91 ± 0.08).
Conclusion Multiple EHR components provide a more consistent and higher performance than a single one for the selected phenotypes. We suggest considering multiple EHR components for future phenotyping design in order to obtain an ideal result.
Keywords: diagnosis codes, electronic health records, clinical notes, International Classification of Diseases, problem lists, medications, phenotype
INTRODUCTION
The dramatic increase in national and international adoption of electronic health record (EHR) systems is generating enormous amounts of computable clinical data. These data are emerging as a rich resource for a variety of secondary research uses, such as research into healthcare processes, comparative effectiveness, and basic biology; the latter is enabled by linkage of EHR data with bioreposititories. 1–3 However, since EHR data is collected primarily for clinical care, challenges exist in reusing these data for research, including inconsistent data quality, data fragmentation, missing data, and bias toward sick individuals. 4 To counter some of these challenges, investigators have deployed algorithms to find specific EHR phenotypes. The process of EHR phenotyping, or accurately identifying patients with a specific observable trait from large volumes of imperfect practice-based data, is one of the crucial challenges to efficient and effective use of EHRs for secondary analyses. 5–8 In this paper, we evaluated the phenotyping performance of three major EHR components often used in phenotyping—billing codes, medication exposures, and text diagnoses—over 10 common diseases. The goal was to provide insight into future design for phenotyping algorithms.
BACKGROUND
The adoption of EHR systems has not only improved patient care, but also enabled to conduct observational research on large, practice-based longitudinal data sets. 2 However, there are gaps between general practice and research settings that must be addressed. 9
EHR data are collected for patient care, to support the operations of healthcare, and to serve as a permanent legal record. Diagnosis, clinical testing, and treatment data are generated for the purpose of medical care and often represent an evolving understanding of the patient’s healthcare status, primary problems, and interactions with insurance. Inaccuracy or uncertainty remains an important nature of EHR data due to the fact that barely any medical observation can be accepted with absolute certainty. 4,10 There are many examples of uncertainty in clinical care, such as a patient with dementia who may not be able to provide an accurate history or similar initial patient presentations of different diagnoses; for example, Crohn’s disease vs ulcerative colitis.
Another significant barrier to using EHR data for clinical research arises from their incompleteness. 9 We have previously demonstrated that using a single center’s data for phenotyping leads to missed cases because of patient data fragmented across multiple sites. 11 We have also shown that patients with a longer history of interaction in the EHR have more accurate phenotyping results. 12 This incompleteness increases for patients who are seen at multiple healthcare centers that do not share patient data. 13
Phenotyping is neither easy nor perfect. Fortunately, EHRs contain sufficient information to accurately assign clinical phenotypes for many diseases. 4–6,14,15 Some EMR data are stored in structured components and can be effortlessly retrieved (e.g., diagnoses, procedures, and clinical laboratory results), while others are embedded in unstructured components and require additional tools (e.g., natural language processing [NLP] pipelines for extracting structured concepts from clinical notes). 9 International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9) diagnosis codes have been predominantly used in many EHR phenotyping exercises, 16–20 since most patients with the disease should be assigned a relevant code for billing purposes. However, due to their inaccuracy 21 or incompleteness, 11 using billing codes alone may result in low specificity or sensitivity. 22,23 Collaborative phenotyping groups, such as the Electronic Medical Records and Genomics Network, 24 demonstrated that combining billing codes with other EHR components; for example, medication and clinical notes, improves phenotyping performance for multiple diseases. 25–30 Other studies applied regression models and other machine learning approaches to identify disease and drug response phenotypes from EHR data. 31 These studies typically leveraged billing codes, medication exposures, laboratory or radiology data, and NLP features. Many have also shown that combining multiple classes of EHR data yields superior results as compared to using a single class of data.
These previous results from various diseases and study groups led to a reasonable assumption that data from multiple components of EHR may improve the positive predictive value (PPV), and possibly sensitivity, of phenotyping. However, to our knowledge, this hypothesis has not been systematically tested before. In this paper, we evaluated the phenotyping performance of three major EHR components across a broad spectrum of pre-selected diseases. We studied three EHR components: billing codes, clinical notes, and specific medications. We believe this study provides a deeper understanding of how leveraging different EHR components for phenotyping affects performance and present a useful guideline for future phenotyping design.
METHODS
Study Setting
This study was conducted at Vanderbilt University Medical Center (VUMC). VUMC is a comprehensive healthcare facility dedicated to patient care, research, and biomedical education. VUMC reflects the racial makeup of the surrounding community throughout Tennessee and the Southeast, and the majority of the records within this database (85%) are from subjects of European ancestry. 32
Data Set
VUMC had previously constructed a de-identified version of its integrated (combined inpatient-outpatient) EHR for epidemiological research in a practice-based setting. This practice-derived resource, called the synthetic derivative (SD), maintains a de-identified version of the entire VUMC EHR that contains the records of over two million unique individuals, including dense longitudinal clinical data for over one million individuals. 33 The SD incorporates clinical data from multiple components, including diagnostic and procedural codes, as well as provider inpatient and outpatient notes, laboratory data, and medication histories. The SD is scrubbed of all Health Insurance Portability Accountability Act identifiers; for example, the name “John Smith” in the original record is permanently replaced with a tag (e.g., [NAME AAA, BBB]). The scrub process maintains the semantic integrity of the text. The scrubbing process efficiency has been assessed, and our data de-identification process has an error rate of ∼0.01%. The SD contains only de-identified data, and all research using this resource has been determined by Vanderbilt’s Institutional Review Board to constitute non-human subjects research. This study was approved by Vanderbilt’s Institutional Review Board.
Data Extraction
We used all EHRs in the SD, which included clinical data for 2 326 150 unique patients. ICD-9 billing codes were retrieved from administrative claims data. We then extracted text diagnoses from “primary clinical notes,” defined as problem lists, admission notes, progress reports, consult notes, discharge summaries, or history and physical examinations. We ignored prescriptions, instructions, and communication letters. We searched primary notes for keywords to determine if a patient’s notes mentioned the disease. Simple negations were excluded using regular expressions (e.g., no diabetes). In addition, we ignored keywords found within family history sections by using a simplified version our prior published algorithm. 34
To obtain specific medications associated with diseases, we used the medications defined by MEDication Indication (MEDI). 35 MEDI is a freely-available, computable medication-indication resource that lists indications and the estimated prevalence for each based on evaluation in the SD. 36 For example, MEDI lists 37 indications for metformin, including type 2 diabetes mellitus (T2DM). T2DM is listed as the primary indication with a prevalence of 80%, which is significantly higher than for polycystic ovary syndrome (8%) and others. For each disease, we used the medications with a prevalence of at least 80% ( Supplementary Appendix A ). We set a high prevalence threshold to ensure selection of medications highly specific to our set of diseases. We hypothesized that a strict threshold would enable us to infer the presence of the disease solely from the presence of the medication.
Medication data in the SD are embedded in clinical narratives and were obtained with the MedEx NLP system in addition to electronic prescribing records from inpatient and outpatient order entry. MedEx extracts medication names and other signatures (dose, route, frequency) from clinical narratives. 37
Study Design
Ten diseases were selected for this evaluation study: atrial fibrillation, Alzheimer’s disease, breast cancer, gout, human immunodeficiency virus infection (HIV), multiple sclerosis, Parkinson’s disease, rheumatoid arthritis (RA), type 1 diabetes mellitus (T1DM), and T2DM. Each of these common diseases poses an enormous public health burden and several have been the focus of EHR-based research.
For each disease, patients were classified into one of the seven categories: 1) ICD-9 only (patients have corresponding ICD-9 s but no positive mention of the disease in primary notes and no specific medication prescribed), 2) primary clinical notes only (patients with positive mentions of the disease in their primary notes but no corresponding ICD-9 s or specific medication prescribed), 3) medications only (patients with specific medications prescribed but no corresponding ICD-9 s and no positive mention of the disease in primary notes), 4) ICD-9 and primary notes (patients have corresponding ICD-9 s and positive mentions of the disease in primary notes but no specific medication prescribed), 5) ICD-9 and medications (patients have corresponding ICD-9 s and specific medications prescribed but no positive mention of the disease in primary notes), 6) primary notes and medications (patients with positive mentions of the disease in their primary notes and corresponding medications prescribed but no ICD-9 s), and 7) ICD-9, primary notes, and medications (patients have corresponding ICD-9 s, medications, and positive mentions of the disease in primary notes).
A group of 25 patients per disease category (a total number of 175 for each disease, 1750 for the 10 diseases) were randomly selected. Each was reviewed by at least one of the five authors (P.L.T., H.M., R.M.C., J.W., and W.Q.W.), each of whom has a medical background. Reviewers went through EHR independently using their clinical knowledge to determine each as a true or not true case (i.e., do or do not have the given disease). Both negative and uncertain patients were classified as not true . More than 20% of were reviewed by two reviewers and the results were used to calculate a kappa score and estimate inter-rater agreement. Another board-certified internist (J.C.D.) adjudicated all labeling conflicts.
Review results were used to calculate the PPV of each category. We also estimated the sensitivity and F -score for each category by using stratified sampling over the categories. The sensitivity of category c was estimated using equation (1) , where C (c) is the set of categories for which the component c is positive and represents the number of found within the category i .
| (1) |
F -score is the harmonic mean of PPV and sensitivity, which is defined in equation (2) .
| (2) |
RESULTS
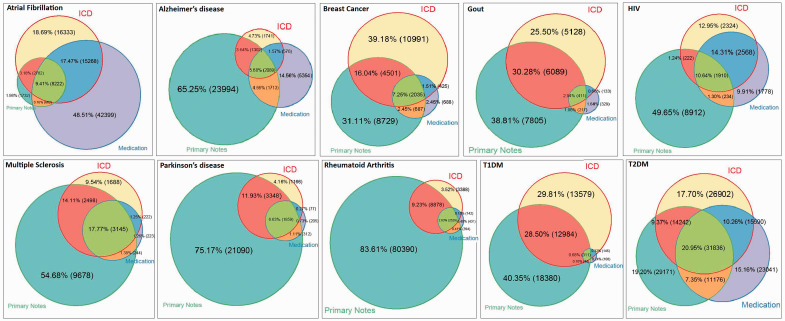
The distributions of patients with ICD-9, primary notes, and specific medications across the 10 diseases ( Figure 1 ) demonstrated that no single EHR component dominated others consistently across the different diseases studied. Diseases such as Alzheimer’s, Parkinson’s disease, and RA are mentioned in a substantial proportion of primary notes in records that do not contain the corresponding ICD-9 codes. For breast cancer, gout, and both types of diabetes, either ICD-9 or primary notes are included. For atrial fibrillation, a large number of possible cases came from specific medications mentions in absense of other evidence. This observation confirmed our hypothesis that additional sources beyond diagnosis codes are worth considering for improving both sensitivity and PPV when phenotyping from EHR data.
Figure 1:
Weighted Venn diagrams of the distributions of patients with ICD-9, primary notes, and specific medications. Each color represents a resource. Different area colors represent the number of patients that were found within intersecting resources.
A total of 1750 (175 per disease, 25 per disease category) were randomly selected and reviewed by at least one author with a clinical background. Over 20% of patients were reviewed by two. The kappa scores suggested substantial agreement between reviewers: P.L.T. and H.M., R.M.C and H.M., R.M.C. and P.L.T., and W.Q.W. and P.L.T. were 0.68 (95% confidence interval, 0.48–0.89), 0.74 (0.56–0.92), 0.83 (0.67–0.99), and 0.90 (0.85–0.95), respectively. The majority of the discrepancies between reviewers fell between the true and uncertain cases. For example, should an obese teenager with an insulin-dependent diabetes be classified as T1DM (when not clearly specified by the treating physicians)? Or, should a patient with multiple HIV codes but no definitive medications and labs be considered as a true case? These differences were reviewed and resolved by a third physician blinded to the original determinations and their raters.
Based on the manual chart review results, the PPVs using single components without corroborating evidence from another data type (e.g., ICD-9 without primary notes and medications) were poor: 0.06–0.37 ( Table 1 ). Mediocre performances (0.55–0.71) were obtained when using single components regardless of other components; for example, ICD-9 with or without primary notes and medications. In patients with two or more corresponding ICD-9 codes regardless of medications or text mentions, the average PPV went up to 0.84 with a standard deviation 0.12. However, we observed a more stable and higher accuracy when using at least two components (mean ± standard deviation: 0.91 ± 0.08). Primary notes offered the best estimated sensitivity (0.77, Table 2 ) of the categories. The sensitivity for ICD-9 was 0.67, which dropped to 0.50 when requiring at least 2 ICD-9 codes. Requiring two or more components provided a reasonably high and stable sensitivity (0.59 ± 0.16).
Table 1:
Positive prediction values of various categories based on chart review results
| Disease | ICD-9 Only | PN Only | Meds Only | ICD-9+Meds | ICD-9+PN | Meds+PN | ICD-9+both | ICD-9 | Meds | PN | ≥2 ICD-9 s | ≥2 Components |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFIB | 0.52 | 0.72 | 0.08 | 0.72 | 1.00 | 1.00 | 1.00 | 0.72 | 0.35 | 0.96 | 0.88 | 0.84 |
| Alzheimer's | 0.28 | 0.20 | 0.00 | 0.80 | 0.88 | 0.92 | 0.88 | 0.69 | 0.40 | 0.32 | 0.74 | 0.88 |
| Breast CA | 0.12 | 0.72 | 0.04 | 0.88 | 0.96 | 1.00 | 1.00 | 0.45 | 0.81 | 0.84 | 1.00 | 0.97 |
| Gout | 0.56 | 0.84 | 0.00 | 0.92 | 1.00 | 1.00 | 1.00 | 0.81 | 0.69 | 0.91 | 0.93 | 1.00 |
| HIV | 0.52 | 0.00 | 0.00 | 0.92 | 0.84 | 0.88 | 1.00 | 0.81 | 0.69 | 0.20 | 0.89 | 0.95 |
| MS | 0.20 | 0.08 | 0.12 | 0.88 | 0.88 | 0.88 | 1.00 | 0.78 | 0.93 | 0.41 | 0.86 | 0.94 |
| Parkinson | 0.48 | 0.16 | 0.04 | 0.84 | 1.00 | 0.88 | 0.96 | 0.89 | 0.87 | 0.33 | 0.94 | 0.98 |
| RA | 0.36 | 0.20 | 0.00 | 0.64 | 0.76 | 0.88 | 0.84 | 0.68 | 0.73 | 0.27 | 0.77 | 0.78 |
| T1DM | 0.28 | 0.12 | 0.04 | 0.16 | 0.92 | 0.84 | 0.76 | 0.59 | 0.49 | 0.45 | 0.62 | 0.91 |
| T2DM | 0.36 | 0.68 | 0.24 | 0.60 | 0.80 | 1.00 | 0.84 | 0.65 | 0.65 | 0.80 | 0.73 | 0.81 |
| Average | 0.37 | 0.37 | 0.06 | 0.74 | 0.90 | 0.93 | 0.93 | 0.71 | 0.66 | 0.55 | 0.84 | 0.91 |
| Standard Deviation | 0.15 | 0.32 | 0.08 | 0.23 | 0.09 | 0.06 | 0.09 | 0.13 | 0.20 | 0.29 | 0.12 | 0.08 |
PN, Primary Notes.
AFIB, atrial fibrillation; HIV, human immunodeficiency virus infection; MS, multiple sclerosis; RA, rheumatoid arthritis; T1DM, type 1 diabetes mellitus.
Table 2:
Sensitivities of various categories based on chart review results
| Disease | ICD-9 Only | PN Only | Meds Only | ICD-9+Meds | ICD-9+PN | Meds+PN | ICD-9+both | ICD-9 | Meds | PN | ≥2 ICD-9 s | ≥2 Components |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFIB | 0.24 | 0.03 | 0.09 | 0.31 | 0.08 | 0.02 | 0.23 | 0.85 | 0.65 | 0.36 | 0.64 | 0.63 |
| Alzheimer's | 0.05 | 0.47 | 0.00 | 0.04 | 0.11 | 0.15 | 0.18 | 0.38 | 0.38 | 0.91 | 0.24 | 0.49 |
| Breast CA | 0.09 | 0.42 | 0.00 | 0.02 | 0.29 | 0.05 | 0.14 | 0.53 | 0.21 | 0.89 | 0.55 | 0.49 |
| Gout | 0.18 | 0.40 | 0.00 | 0.01 | 0.37 | 0.01 | 0.03 | 0.58 | 0.05 | 0.82 | 0.34 | 0.42 |
| HIV | 0.21 | 0.00 | 0.00 | 0.40 | 0.03 | 0.04 | 0.33 | 0.96 | 0.76 | 0.39 | 0.82 | 0.79 |
| MS | 0.05 | 0.11 | 0.00 | 0.03 | 0.32 | 0.03 | 0.46 | 0.85 | 0.52 | 0.92 | 0.63 | 0.83 |
| Parkinson | 0.06 | 0.36 | 0.00 | 0.01 | 0.36 | 0.03 | 0.19 | 0.61 | 0.23 | 0.93 | 0.42 | 0.58 |
| RA | 0.05 | 0.60 | 0.00 | 0.00 | 0.25 | 0.01 | 0.08 | 0.38 | 0.10 | 0.95 | 0.31 | 0.35 |
| T1DM | 0.21 | 0.12 | 0.00 | 0.00 | 0.65 | 0.00 | 0.01 | 0.88 | 0.02 | 0.79 | 0.60 | 0.67 |
| T2DM | 0.10 | 0.21 | 0.06 | 0.10 | 0.12 | 0.12 | 0.29 | 0.61 | 0.56 | 0.74 | 0.42 | 0.63 |
| Average | 0.12 | 0.27 | 0.02 | 0.09 | 0.26 | 0.05 | 0.19 | 0.67 | 0.35 | 0.77 | 0.50 | 0.59 |
| Standard deviation | 0.08 | 0.20 | 0.03 | 0.14 | 0.19 | 0.05 | 0.14 | 0.21 | 0.27 | 0.22 | 0.18 | 0.16 |
PN, primary notes.
AFIB, atrial fibrillation; HIV, human immunodeficiency virus infection; MS, multiple sclerosis; RA, rheumatoid arthritis; T1DM, type 1 diabetes mellitus.
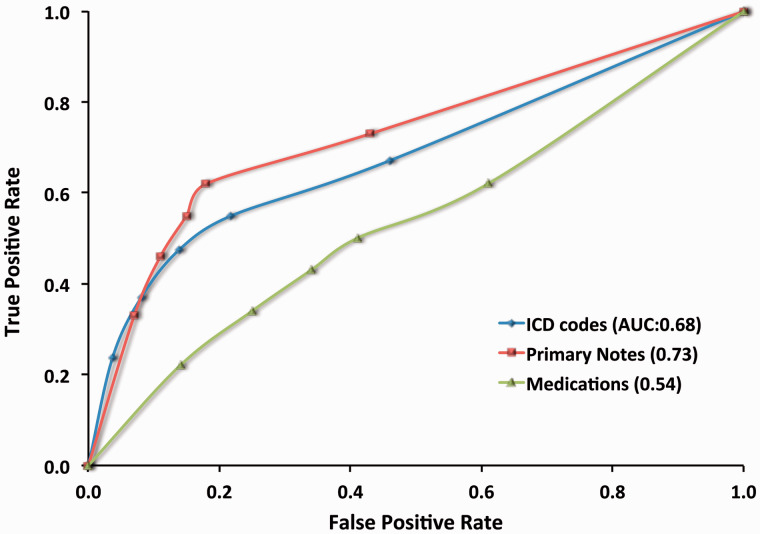
Among the three components, primary notes had the best performance with the area under curve (AUC) score 0.73 ( Figure 2 ). ICD-9 was similar but slightly less, with an AUC of 0.68. Medications underperformed both with an AUC of 0.54, lower than either primary notes or ICD-9. Primary notes and ICD-9 showed the similar performance of positive prediction values, which was significantly higher than medications ( Table 4 ). Overall, the best phenotyping performance ( F score: 0.70 ± 0.12) was achieved by using two or more components ( Table 3 ). Although the F score of using ICD-9 (0.67 ± 0.14) was only slightly lower than using two or more components, its PPV (0.71 ± 0.13) is substantially lower than when using at least two components (0.91 ± 0.08).
Figure 2:
Receiver operating characteristic (ROC) curve for ICD-9, primary notes, and specific medications. ROC was performed using data of 1750 reviewed cases across 10 diseases. AUC: Area under the curve.
Table 4:
Positive prediction values and sensitivities of requiring 1 to 10 ICD9 codes, primary notes, and medication mentions
|
Positive prediction values
|
Sensitivities
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| ICD codes | AFIB | 0.72 | 0.88 | 0.96 | 0.98 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.85 | 0.64 | 0.46 | 0.43 | 0.39 | 0.37 | 0.31 | 0.27 | 0.25 | 0.24 |
| Alzheimer | 0.69 | 0.74 | 0.78 | 0.77 | 0.76 | 0.74 | 0.74 | 0.71 | 0.69 | 0.67 | 0.38 | 0.24 | 0.24 | 0.24 | 0.20 | 0.18 | 0.15 | 0.11 | 0.09 | 0.08 | |
| BRCA | 0.45 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.53 | 0.55 | 0.45 | 0.43 | 0.40 | 0.39 | 0.34 | 0.33 | 0.31 | 0.29 | |
| Gout | 0.81 | 0.93 | 0.92 | 0.90 | 0.94 | 0.93 | 0.92 | 0.95 | 1.00 | 1.00 | 0.58 | 0.34 | 0.33 | 0.27 | 0.25 | 0.21 | 0.17 | 0.13 | 0.10 | 0.09 | |
| HIV | 0.81 | 0.89 | 0.91 | 0.93 | 0.96 | 0.95 | 0.98 | 0.97 | 0.97 | 0.97 | 0.96 | 0.82 | 0.61 | 0.55 | 0.46 | 0.43 | 0.41 | 0.35 | 0.33 | 0.30 | |
| MS | 0.78 | 0.86 | 0.89 | 0.92 | 0.91 | 0.93 | 0.95 | 0.97 | 1.00 | 1.00 | 0.85 | 0.63 | 0.50 | 0.48 | 0.43 | 0.42 | 0.38 | 0.33 | 0.31 | 0.30 | |
| Parkinson | 0.89 | 0.94 | 0.94 | 0.98 | 0.97 | 0.97 | 0.97 | 0.96 | 0.96 | 0.96 | 0.61 | 0.42 | 0.42 | 0.39 | 0.34 | 0.32 | 0.28 | 0.26 | 0.26 | 0.26 | |
| RA | 0.68 | 0.77 | 0.83 | 0.85 | 0.87 | 0.90 | 0.92 | 0.91 | 0.90 | 0.92 | 0.38 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | |
| T1DM | 0.59 | 0.62 | 0.71 | 0.75 | 0.77 | 0.79 | 0.79 | 0.81 | 0.79 | 0.79 | 0.88 | 0.60 | 0.60 | 0.60 | 0.55 | 0.51 | 0.45 | 0.39 | 0.36 | 0.35 | |
| T2DM | 0.65 | 0.73 | 0.81 | 0.77 | 0.81 | 0.81 | 0.81 | 0.80 | 0.84 | 0.81 | 0.61 | 0.42 | 0.39 | 0.31 | 0.28 | 0.23 | 0.20 | 0.18 | 0.15 | 0.12 | |
| Average | 0.71 | 0.84 | 0.88 | 0.89 | 0.90 | 0.90 | 0.91 | 0.91 | 0.92 | 0.91 | 0.66 | 0.50 | 0.43 | 0.40 | 0.36 | 0.34 | 0.30 | 0.27 | 0.25 | 0.24 | |
| SD | 0.13 | 0.12 | 0.09 | 0.09 | 0.09 | 0.09 | 0.09 | 0.10 | 0.11 | 0.12 | 0.21 | 0.18 | 0.12 | 0.12 | 0.11 | 0.11 | 0.10 | 0.10 | 0.10 | 0.10 | |
| Primary notes | AFIB | 0.96 | 0.98 | 0.97 | 0.97 | 0.96 | 0.96 | 0.96 | 0.96 | 0.98 | 0.98 | 0.36 | 0.36 | 0.30 | 0.30 | 0.30 | 0.30 | 0.30 | 0.30 | 0.30 | 0.35 |
| Alzheimer | 0.32 | 0.78 | 0.79 | 0.84 | 0.85 | 0.82 | 0.85 | 0.83 | 0.82 | 0.84 | 0.91 | 0.59 | 0.53 | 0.52 | 0.43 | 0.33 | 0.31 | 0.26 | 0.24 | 0.22 | |
| BRCA | 0.84 | 0.92 | 0.93 | 0.93 | 0.93 | 0.94 | 0.95 | 0.95 | 0.94 | 0.94 | 0.89 | 0.72 | 0.65 | 0.59 | 0.57 | 0.52 | 0.50 | 0.47 | 0.42 | 0.42 | |
| Gout | 0.91 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.99 | 0.99 | 0.99 | 0.99 | 0.82 | 0.71 | 0.69 | 0.68 | 0.66 | 0.63 | 0.61 | 0.57 | 0.55 | 0.52 | |
| HIV | 0.20 | 0.72 | 0.72 | 0.69 | 0.70 | 0.69 | 0.68 | 0.69 | 0.70 | 0.68 | 0.39 | 0.35 | 0.53 | 0.45 | 0.40 | 0.36 | 0.35 | 0.34 | 0.32 | 0.29 | |
| MS | 0.41 | 0.93 | 0.95 | 0.98 | 0.97 | 0.97 | 1.00 | 1.00 | 1.00 | 1.00 | 0.92 | 0.62 | 0.51 | 0.44 | 0.37 | 0.37 | 0.35 | 0.32 | 0.29 | 0.28 | |
| Parkinson | 0.33 | 0.88 | 0.89 | 0.90 | 0.91 | 0.91 | 0.89 | 0.88 | 0.88 | 0.89 | 0.93 | 0.64 | 0.56 | 0.52 | 0.48 | 0.47 | 0.41 | 0.35 | 0.34 | 0.33 | |
| RA | 0.27 | 0.82 | 0.81 | 0.82 | 0.83 | 0.86 | 0.85 | 0.85 | 0.84 | 0.82 | 0.95 | 0.56 | 0.50 | 0.45 | 0.43 | 0.41 | 0.40 | 0.38 | 0.35 | 0.32 | |
| T1DM | 0.45 | 0.78 | 0.84 | 0.84 | 0.87 | 0.85 | 0.87 | 0.88 | 0.88 | 0.90 | 0.79 | 0.73 | 0.69 | 0.64 | 0.62 | 0.53 | 0.53 | 0.51 | 0.50 | 0.49 | |
| T2DM | 0.80 | 0.89 | 0.90 | 0.88 | 0.86 | 0.84 | 0.80 | 0.83 | 0.83 | 0.82 | 0.74 | 0.31 | 0.26 | 0.19 | 0.17 | 0.15 | 0.11 | 0.09 | 0.09 | 0.08 | |
| Average | 0.55 | 0.87 | 0.88 | 0.88 | 0.89 | 0.88 | 0.88 | 0.89 | 0.89 | 0.89 | 0.77 | 0.56 | 0.52 | 0.48 | 0.44 | 0.41 | 0.39 | 0.36 | 0.34 | 0.33 | |
| Standard deviation | 0.29 | 0.09 | 0.09 | 0.09 | 0.08 | 0.09 | 0.10 | 0.09 | 0.09 | 0.10 | 0.22 | 0.16 | 0.15 | 0.15 | 0.15 | 0.14 | 0.14 | 0.14 | 0.13 | 0.13 | |
| Medication mentions | AFIB | 0.35 | 0.73 | 0.72 | 0.73 | 0.75 | 0.72 | 0.73 | 0.78 | 0.79 | 0.82 | 0.65 | 0.51 | 0.46 | 0.44 | 0.39 | 0.33 | 0.30 | 0.29 | 0.29 | 0.26 |
| Alzheimer | 0.40 | 0.66 | 0.68 | 0.65 | 0.64 | 0.66 | 0.65 | 0.65 | 0.66 | 0.68 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.35 | 0.35 | 0.33 | 0.29 | |
| BRCA | 0.81 | 0.74 | 0.74 | 0.78 | 0.80 | 0.87 | 0.86 | 0.88 | 0.86 | 0.88 | 0.21 | 0.21 | 0.21 | 0.21 | 0.21 | 0.21 | 0.21 | 0.21 | 0.21 | 0.18 | |
| Gout | 0.69 | 0.75 | 0.76 | 0.77 | 0.80 | 0.84 | 0.86 | 0.85 | 0.83 | 0.88 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | |
| HIV | 0.69 | 0.82 | 0.83 | 0.85 | 0.91 | 0.90 | 0.90 | 0.96 | 0.96 | 0.96 | 0.76 | 0.51 | 0.42 | 0.36 | 0.31 | 0.29 | 0.27 | 0.24 | 0.23 | 0.23 | |
| MS | 0.93 | 0.88 | 0.90 | 0.94 | 0.94 | 0.96 | 0.95 | 0.97 | 0.97 | 0.96 | 0.52 | 0.62 | 0.55 | 0.50 | 0.46 | 0.43 | 0.40 | 0.32 | 0.29 | 0.27 | |
| Parkinson | 0.87 | 0.73 | 0.76 | 0.78 | 0.75 | 0.74 | 0.76 | 0.75 | 0.75 | 0.72 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.23 | 0.21 | |
| RA | 0.73 | 0.60 | 0.62 | 0.65 | 0.65 | 0.72 | 0.74 | 0.71 | 0.71 | 0.71 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | |
| T1DM | 0.49 | 0.43 | 0.34 | 0.31 | 0.29 | 0.30 | 0.26 | 0.33 | 0.33 | 0.29 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | |
| T2DM | 0.65 | 0.73 | 0.73 | 0.71 | 0.70 | 0.67 | 0.65 | 0.63 | 0.63 | 0.62 | 0.56 | 0.56 | 0.50 | 0.45 | 0.42 | 0.37 | 0.33 | 0.29 | 0.28 | 0.27 | |
| Average | 0.66 | 0.71 | 0.71 | 0.72 | 0.72 | 0.74 | 0.74 | 0.75 | 0.75 | 0.75 | 0.35 | 0.32 | 0.29 | 0.27 | 0.26 | 0.24 | 0.23 | 0.21 | 0.20 | 0.19 | |
| Standard deviation | 0.19 | 0.12 | 0.15 | 0.17 | 0.18 | 0.18 | 0.20 | 0.19 | 0.19 | 0.20 | 0.26 | 0.23 | 0.20 | 0.18 | 0.16 | 0.14 | 0.13 | 0.12 | 0.11 | 0.10 | |
AFIB, atrial fibrillation; BRCA, breast cancer; HIV, human immunodeficiency virus infection; MS, multiple sclerosis; RA, rheumatoid arthritis; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus.
Table 3:
F -scores of various categories based on chart review results
| Disease | ICD-9 Only | PN Only | Meds Only | ICD-9+Meds | ICD-9+PN | Meds+PN | ICD-9+both | ICD-9 | Meds | PN | ≥2 ICD-9 s | ≥2 Components |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFIB | 0.33 | 0.07 | 0.09 | 0.43 | 0.14 | 0.04 | 0.37 | 0.78 | 0.45 | 0.53 | 0.74 | 0.72 |
| Alzheimer's | 0.08 | 0.28 | 0.00 | 0.08 | 0.20 | 0.26 | 0.30 | 0.49 | 0.39 | 0.47 | 0.36 | 0.63 |
| Breast CA | 0.10 | 0.53 | 0.00 | 0.05 | 0.44 | 0.09 | 0.24 | 0.49 | 0.33 | 0.86 | 0.71 | 0.65 |
| Gout | 0.27 | 0.54 | 0.00 | 0.01 | 0.54 | 0.03 | 0.05 | 0.68 | 0.09 | 0.86 | 0.49 | 0.59 |
| HIV | 0.29 | 0.00 | 0.00 | 0.56 | 0.06 | 0.07 | 0.49 | 0.88 | 0.72 | 0.27 | 0.85 | 0.86 |
| MS | 0.08 | 0.09 | 0.01 | 0.05 | 0.47 | 0.06 | 0.63 | 0.81 | 0.67 | 0.56 | 0.73 | 0.89 |
| Parkinson | 0.11 | 0.22 | 0.00 | 0.01 | 0.52 | 0.06 | 0.32 | 0.73 | 0.36 | 0.49 | 0.58 | 0.73 |
| RA | 0.08 | 0.30 | 0.00 | 0.01 | 0.38 | 0.03 | 0.15 | 0.49 | 0.17 | 0.43 | 0.44 | 0.48 |
| T1DM | 0.24 | 0.12 | 0.00 | 0.00 | 0.76 | 0.00 | 0.03 | 0.71 | 0.03 | 0.58 | 0.61 | 0.77 |
| T2DM | 0.16 | 0.32 | 0.09 | 0.17 | 0.21 | 0.21 | 0.43 | 0.63 | 0.60 | 0.77 | 0.53 | 0.70 |
| Average | 0.17 | 0.25 | 0.02 | 0.14 | 0.37 | 0.08 | 0.30 | 0.67 | 0.38 | 0.58 | 0.60 | 0.70 |
| Standard deviation | 0.10 | 0.19 | 0.04 | 0.20 | 0.22 | 0.09 | 0.19 | 0.14 | 0.24 | 0.19 | 0.15 | 0.12 |
PN, primary note.
AFIB, atrial fibrillation; HIV, human immunodeficiency virus infection; MS, multiple sclerosis; RA, rheumatoid arthritis; T1DM, type 1 diabetes mellitus.
DISCUSSION
The lack of automated methods to convert imperfect EHR data into quality phenotypes has become a fundamental impediment to leveraging the huge volumes of EHR data now available for clinical and genomic research. 4,7 Much of historical EHR research has relied largely upon administrative codes, but much research has demonstrated the benefit of additional information to phenotyping sensitivity and PPV. 5,24 Our results validate these findings, demonstrating that the use of multiple components of EHR data significantly improves PPV and F -score. Taken as a single class of data, ICD-9 had the best PPV and F -score, and PN mentions had the best sensitivity and slightly higher AUC than ICD-9 codes. However, no single component of EHR data, when further support from other data was absent, was adequate for an accurate identification task in the ten diseases we studied (average PPV < 0.37).
Overall, the best performing single class of data was arguably ICD-9 codes, when taken regardless of the presence of other evidence, delivering a decent performance (PPV 0.71 ± 0.13, sensitivity 0.67 ± 0.21, F -score 0.67 ± 0.14). However, we observed poor PPVs on breast cancer, Alzheimer's disease, and both types of diabetes. The identification of patients with Alzheimer's disease 38 or diabetes 11,12,28 is especially challenging. Further analysis indicates that a considerable number of patients with breast cancer ICD-9 s have only pathology requests from outside facilities but no further information in our EHR to confirm the presence of disease. Our results confirmed that using ICD-9 codes without other supportive evidence does not work well since the ICD-9 codes are often miscoded or used when a diagnosis was suspected but not actually confirmed.
As seen in prior studies, requiring two or more ICD-9 codes significantly improved the PPV (0.71 vs 0.84, P < .02). PPV continued to improve by requiring more codes but with a corresponding decrease in sensitivity. Particularly, using two or more ICD-9 codes reduces the false positives caused by outsourcing labs or after a diagnosis is ruled out. Given their overall strong performance, using multiple ICD-9 codes may be an efficient phenotyping strategy when NLP tools or other EHR components are not available. Such a strategy is typically employed in phenome-wide association studies using EHR data to improve PPV, which typically require multiple codes to qualify a case. 39 It was also notable in this study that we saw similar improvements in PPV by requiring multiple instances of medication mentions or PN text mentions.
The potential use of clinical notes to improve phenotyping sensitivity and granularity has been addressed by numerous studies. 31,40–42 Therefore, it is not surprising that primary clinic notes provide the best overall sensitivity. However, many challenges remain to precisely retrieve relevant information from EHRs. In our study, NLP-induced errors were largely caused by word sense disambiguation failures (e.g., does “RA” represent rheumatoid arthritis , right atrium , or room air ). Although we excluded the family history sections, some illness histories of family members are found within other part of primary notes; for example, in social history and in the history of present illness. Use of more advanced section tagging applications, 34 word sense disambiguation methods, 43,44 and algorithms such as ConTEXT 45 might improve the PPV of NLP-derived phenotype mentions.
In summary, the best performance is achieved when evidence can be found within two or more EHR components. This observation confirms our hypothesis that multiple components improve phenotyping performance. Moreover, our results across various diseases suggest that the phenotyping performance of using two or more components is significantly more consistent than simply using ICD-9 codes, or using multiple ICD-9 codes. This consistent higher performance reveals the potential value of multiple components for future phenotyping design.
This study has several limitations. First, this study evaluates only three major EHR components: diagnosis codes, medications, and primary notes. Laboratory tests, as another important part of EHR, have not been included into this evaluation because we are not able to find a knowledge base that identifies specific laboratory tests for diseases that easily incorporates into our approach. Some diseases have highly specific laboratory tests; for example, low-density lipoprotein for hyperlipidemia and troponin for myocardial infarction, while others do not. The results of our study and our previous work imply that additional sources such as specific laboratory tests may be beneficial. 46 Computational approaches to statistically associate laboratory data with diagnoses, or parsing of diagnoses from expert systems such as Quick Medical Reference 47 or DXplain, 48 could accelerate creation of such a resource.
Secondly, the medication prevalence threshold (which operates as a PPV of the medicine for the disease) we chose was so rigorous (0.8) that some commonly prescribed medications may be neglected; for example, allopurinol for gout (prevalence: 0.5, which may be artificially low due to the automated method by which prevalence was calculated 36 ). We chose this high threshold under the hypothesis that perhaps highly specific medications would provide a sufficient PPV that they may be able to qualify an individual as a case without other substantiating evidence. However, even for HIV infections, in which specific medication prevalence were >0.90, the PPV of the medication for the disease in the absence of other confirming data sources was low (0%). Thus, our data suggest that medication exposures alone are not sufficient for inferring the presence of a diagnosis in most cases, even with highly specific medications. The performance of medications may improve if more sensitive medications or combinations of medications are involved. Thirdly, this study is also limited by the selection of ICD-9 codes. We only consider the most commonly used ones; for example, 250.00 for T2DM. An inclusion of more focused codes (e.g., 250.62) may improve our results. For example, more specific codes for types 1 and 2 diabetes (e.g., T2DM with complications) demonstrate stronger odds ratios for known genetic associations than more general codes, suggesting a higher PPV for the more specific codes (e.g., rs2647044 with type 1 diabetes with complications had odds ratios >2.2 while the generic type 1 diabetes code had an odds ratio of 1.42; similar results are found for type 2 diabetes and TCF7L2 variants). 39
Finally, this evaluation is conducted on 10 preselected chronic diseases using EHR data at a single medical center. For a more thorough evaluation, this study needs to be repeated at different locations on more phenotypes. Acute diseases may also perform differently, as sensitivity may fall quickly with requirements for multiple codes.
CONCLUSION
This study, to our knowledge, is one of the first attempt to systematically evaluate the phenotyping performance of major EHR components used in phenotyping. Our results demonstrated that multiple EHR components provide more consistent and higher performance than single elements. We suggest that multiple EHR components should be considered in future phenotyping design for the best performance.
FUNDING
The project was supported by the following grants: National Library of Medicine R01 LM010685, National Institute of General Medical Sciences P50 GM115305 and R01 GM103859, American Heart Association 13POST16470018, and National Human Genome Research Institute U01 HG006378.
CONFLICT OF INTEREST
All authors have declared that no competing interest exists.
AUTHORS’ CONTRIBUTIONS
Study initialization: W-Q.W. and J.C.D.
Study design: W.-Q.W. and J.C.D.
Acquisition of data: W.-Q.W.
Analysis and interpretation of data: W.-Q.W., P.L.T., H.M., R.M.C., J.L.W., J.C.D.
Drafting of the manuscript: W.-Q.W., P.L.T., H.M., R.M.C., J.L.W., J.C.D.; all authors contributed to refinement of the manuscript and approved the final manuscript.
Grant holder: J.C.D. and W.-Q.W.
Supplementary Material
REFERENCES
- 1. Shea S, Hripcsak G . Accelerating the use of electronic health records in physician practices . New Engl J Med. 2010. ; 362 ( 3 ): 192 – 195 . [DOI] [PubMed] [Google Scholar]
- 2. Wilke RA, Xu H, Denny JC, et al. . The emerging role of electronic medical records in pharmacogenomics . Clin Pharmacol Therap. 2011. ; 89 ( 3 ): 379 – 386 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roden DM, Xu H, Denny JC, Wilke RA . Electronic medical records as a tool in clinical pharmacology: opportunities and challenges . Clin Pharmacol Therap. 2012. ; 91 ( 6 ): 1083 – 1086 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hripcsak G, Albers DJ . Next-generation phenotyping of electronic health records . JAMIA. 2013. ; 20 ( 1 ): 117 – 121 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kho AN, Pacheco JA, Peissig PL, et al. . Electronic medical records for genetic research: results of the eMERGE consortium . Sci Trans Med. 2011. ; 3 ( 79 ): 79re71 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Newton KM, Peissig PL, Kho AN, et al. . Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network . JAMIA. 2013. ; 20 ( e1 ): e147 – e154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robinson PN . Deep phenotyping for precision medicine . Hum Mutat. 2012. ; 33 ( 5 ): 777 – 780 . [DOI] [PubMed] [Google Scholar]
- 8. Tracy RP . ‘Deep phenotyping': characterizing populations in the era of genomics and systems biology . Curr Opin Lipidol. 2008. ; 19 ( 2 ): 151 – 157 . [DOI] [PubMed] [Google Scholar]
- 9. Wei WQ, Denny JC . Extracting research-quality phenotypes from electronic health records to support precision medicine . Genome Med . 2015. ; 7 ( 1 ): 41 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shortliffe EH, Cimino JJ . Biomedical Informatics: Computer Applications in Health Care and Biomedicine . 3rd ed . New York: : Springer; ; 2006. . [Google Scholar]
- 11. Wei WQ, Leibson CL, Ransom JE, et al. . Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus . JAMIA. 2012. ; 19 ( 2 ): 219 – 224 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei WQ, Leibson CL, Ransom JE, Kho AN, Chute CG . The absence of longitudinal data limits the accuracy of high-throughput clinical phenotyping for identifying type 2 diabetes mellitus subjects . Int J Med Inform. 2013. ; 82 ( 4 ): 239 – 247 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bourgeois FC, Olson KL, Mandl KD . Patients treated at multiple acute health care facilities: quantifying information fragmentation . Arch Intern Med. 2010. ; 170 ( 22 ): 1989 – 1995 . [DOI] [PubMed] [Google Scholar]
- 14. Shivade C, Raghavan P, Fosler-Lussier E, et al. . A review of approaches to identifying patient phenotype cohorts using electronic health records . JAMIA. 2014. ; 21 ( 2 ): 221 – 230 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richesson RL, Horvath MM, Rusincovitch SA . Clinical research informatics and electronic health record data . Yearbook Med Inform. 2014. ; 9 ( 1 ): 215 – 223 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldberg DS, Lewis JD, Halpern SD, Weiner MG, Lo Re V, 3rd . Validation of a coding algorithm to identify patients with hepatocellular carcinoma in an administrative database . Pharmacoepidemiol Drug Safety. 2013. ; 22 ( 1 ): 103 – 107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldberg D, Lewis J, Halpern S, Weiner M, Lo Re V, 3rd . Validation of three coding algorithms to identify patients with end-stage liver disease in an administrative database . Pharmacoepidemiol Drug Safety. 2012. ; 21 ( 7 ): 765 – 769 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tu K, Mitiku T, Guo H, Lee DS, Tu JV . Myocardial infarction and the validation of physician billing and hospitalization data using electronic medical records . Chronic Dis Can. 2010. ; 30 ( 4 ): 141 – 146 . [PubMed] [Google Scholar]
- 19. Tu K, Mitiku T, Lee DS, Guo H, Tu JV . Validation of physician billing and hospitalization data to identify patients with ischemic heart disease using data from the Electronic Medical Record Administrative data Linked Database (EMRALD) . Can J Cardiol. 2010. ; 26 ( 7 ): e225 – e228 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tu K, Wang M, Jaakkimainen RL, et al. . Assessing the validity of using administrative data to identify patients with epilepsy . Epilepsia. 2014. ; 55 ( 2 ): 335 – 343 . [DOI] [PubMed] [Google Scholar]
- 21. O'Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM . Measuring diagnoses: ICD code accuracy . Health Services Res. 2005. ; 40 ( 5 Pt 2 ): 1620 – 1639 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kern EF, Maney M, Miller DR, et al. . Failure of ICD-9-CM codes to identify patients with comorbid chronic kidney disease in diabetes . Health Services Res. 2006. ; 41 ( 2 ): 564 – 580 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grams ME, Waikar SS, Macmahon B, Whelton S, Ballew SH, Coresh J . Performance and limitations of administrative data in the identification of AKI . CJASN. 2014. ; 9 ( 4 ): 682 – 689 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. eMERGE . The Electronic Medical Records and Genomics (eMERGE) Network . 2014. . http://www.gwas.net . Accessed 3 March 2014 . [Google Scholar]
- 25. Cooke CR, Joo MJ, Anderson SM, et al. . The validity of using ICD-9 codes and pharmacy records to identify patients with chronic obstructive pulmonary disease . BMC Health Services Res. 2011. ; 11 : 37 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian TY, Zlateva I, Anderson DR . Using electronic health records data to identify patients with chronic pain in a primary care setting . JAMIA. 2013. ; 20 ( e2 ): e275 – e280 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goetz MB, Hoang T, Kan V, Rimland D, Rodriguez-Barradas M . Development and validation of an algorithm to identify patients newly diagnosed with HIV infection from electronic health records . AIDS Res Hum Retroviruses. 2014. ; 30 ( 7 ): 626 – 633 . [DOI] [PubMed] [Google Scholar]
- 28. Kho AN, Hayes MG, Rasmussen-Torvik L, et al. . Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study . JAMIA. 2012. ; 19 ( 2 ): 212 – 218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carroll RJ, Thompson WK, Eyler AE, et al. . Portability of an algorithm to identify rheumatoid arthritis in electronic health records . JAMIA. 2012. ; 19 ( e1 ): e162 – e169 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei WQ, Feng Q, Weeke P, et al. . Creation and validation of an EMR-based algorithm for identifying major adverse cardiac events while on statins . Joint Summits on Translational Science, AMIA. San Francisco; 2014 . [PMC free article] [PubMed] [Google Scholar]
- 31. Wei WQ, Tao C, Jiang G, Chute CG . A high throughput semantic concept frequency based approach for patient identification: a case study using type 2 diabetes mellitus clinical notes . AMIA… Annual Symposium Proceedings/AMIA Symposium. AMIA Symposium. 2010. ; 2010 : 857 – 861 . [PMC free article] [PubMed] [Google Scholar]
- 32. Dumitrescu L, Ritchie MD, Brown-Gentry K, et al. . Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records . Genetics Med. 2010. ; 12 ( 10 ): 648 – 650 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roden DM, Pulley JM, Basford MA, et al. . Development of a large-scale de-identified DNA biobank to enable personalized medicine . Clin Pharmacol Therap. 2008. ; 84 ( 3 ): 362 – 369 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Denny JC, Spickard A, 3rd, Johnson KB, Peterson NB, Peterson JF, Miller RA . Evaluation of a method to identify and categorize section headers in clinical documents . JAMIA. 2009. ; 16 ( 6 ): 806 – 815 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei WQ, Cronin RM, Xu H, Lasko TA, Bastarache L, Denny JC . Development of an ensemble resource linking MEDications to their Indications (MEDI) . AMIA Summits Transl Sci. 2013. ; 2013 : 172 . [PubMed] [Google Scholar]
- 36. Wei WQ, Mosley JD, Bastarache L, Denny JC . Validation and Enhancement of a Computable Medication Indication Resource (MEDI) Using a Large Practice-based Dataset . AMIA… Annual Symposium Proceedings/AMIA Symposium. 2013. : 1448 – 1456 . [PMC free article] [PubMed] [Google Scholar]
- 37. Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC . MedEx: a medication information extraction system for clinical narratives . JAMIA. 2010. ; 17 ( 1 ): 19 – 24 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pippenger M, Holloway RG, Vickrey BG . Neurologists' use of ICD-9CM codes for dementia . Neurology. 2001. ; 56 ( 9 ): 1206 – 1209 . [DOI] [PubMed] [Google Scholar]
- 39. Denny JC, Bastarache L, Ritchie MD, et al. . Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data . Nat Biotechnol. 2013. ; 31 ( 12 ): 1102 – 1110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB . Data from clinical notes: a perspective on the tension between structure and flexible documentation . JAMIA . 2011. ; 18 ( 2 ): 181 – 186 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tange HJ, Schouten HC, Kester AD, Hasman A . The granularity of medical narratives and its effect on the speed and completeness of information retrieval . JAMIA. 1998. ; 5 ( 6 ): 571 – 582 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei WQ, Feng Q, Jiang L, et al. . Characterization of statin dose response in electronic medical records . Clin Pharmacol Therap . 2014. ; 95 ( 3 ): 331 – 338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andreopoulos B, Alexopoulou D, Schroeder M . Word Sense Disambiguation in biomedical ontologies with term co-occurrence analysis and document clustering . Int J Data Min Bioinform. 2008. ; 2 ( 3 ): 193 – 215 . [DOI] [PubMed] [Google Scholar]
- 44. Xu H, Markatou M, Dimova R, Liu H, Friedman C . Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues . BMC Bioinformatics. 2006. ; 7 : 334 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chapman BE, Lee S, Kang HP, Chapman WW . Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm . J BiomedI Inform . 2011. ; 44 ( 5 ): 728 – 737 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Warner JL, Alterovitz G . Phenome based analysis as a means for discovering context dependent clinical reference ranges . AMIA… Annual Symposium Proceedings/AMIA Symposium. AMIA Symposium. 2012. ; 2012 : 1441 – 1449 . [PMC free article] [PubMed] [Google Scholar]
- 47. Quick Medical Reference . http://www.openclinical.org/aisp_qmr.html , 2014 Accessed March 3, 2015 . [Google Scholar]
- 48. Barnett GO, Cimino JJ, Hupp JA, Hoffer EP . DXplain. An evolving diagnostic decision-support system . JAMA. 1987. ; 258 ( 1 ): 67 – 74 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.