Abstract
Neolamarckia cadamba is a fast-growing tropical hardwood tree that is used extensively for plywood and pulp production, light furniture fabrication, building materials, and as a raw material for the preparation of certain indigenous medicines. Lack of genomic resources hampers progress in the molecular breeding and genetic improvement of this multipurpose tree species. In this study, transcriptome profiling of differentiating stems was performed to understand N. cadamba xylogenesis. The N. cadamba transcriptome was sequenced using Illumina paired-end sequencing technology. This generated 42.49 G of raw data that was then de novo assembled into 55,432 UniGenes with a mean length of 803.2bp. Approximately 47.8% of the UniGenes (26,487) were annotated against publically available protein databases, among which 21,699 and 7,754 UniGenes were assigned to Gene Ontology categories (GO) and Clusters of Orthologous Groups (COG), respectively. 5,589 UniGenes could be mapped onto 116 pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. Among 6,202 UniGenes exhibiting differential expression during xylogenesis, 1,634 showed significantly higher levels of expression in the basal and middle stem segments compared to the apical stem segment. These genes included NAC and MYB transcription factors related to secondary cell wall biosynthesis, genes related to most metabolic steps of lignin biosynthesis, and CesA genes involved in cellulose biosynthesis. This study lays the foundation for further screening of key genes associated with xylogenesis in N. cadamba as well as enhancing our understanding of the mechanism of xylogenesis in fast-growing trees.
Introduction
Wood is the most abundant biological resource on earth. It is an important raw material for lumber and paper manufacturing, and is increasingly being exploited as an environmentally cost-effective, renewable source for energy production [1]. Wood formation is an important plant developmental event resulting from the accumulation of secondary cell walls in the xylem. Wood formation or xylogenesis involves a sequence of developmental events at the cellular level including cell division, cell expansion, deposition of secondary cell walls, lignification and programmed cell death [2]. Secondary cell wall formation and lignification are critical steps in the maturation of xylem tracheary elements and fibre cells. The secondary cell wall is mainly composed of cellulose (40–50%), hemicellulose (~25%), and lignin (25–35%) as well as small amounts of pectin and protein [2].
Biofuel production has recently stimulated interest in understanding the biosynthesis of secondary cell walls, including its transcriptional regulation. For cellulose biosynthesis, both direct (sucrose synthase, SuSY) and indirect (invertase, INV) pathways produce UDP-glucose, which is the direct substrate for cellulose biosynthesis [3]. In Arabidopsis thaliana, cellulose synthases (CesA) form a complex in which AtCesA4, AtCesA7 and AtCesA8 are essential for secondary cell wall formation [4], whereas AtCesA1, AtCesA3 and AtCesA6 are involved in primary cell wall synthesis [5]. The biosynthesis of xylan, a major hemicellulose in the secondary cell wall, involves numerous glycosyltransferases (GTs). GT43 family members IRX9, IRX9L, IRX14 and IRX14L, GT47 family members IRX10, IRX10L, FRA8/IRX7 and F8H, and GT8 family members IRX8, PARVUS, GUX1, GUX2 and GUX4, are all involved in xylan backbone elongation, synthesis of the reducing end tetrasaccharide sequence and addition of glucuronic acid or a 4-O-methylglucuronic acid branch to the xylan backbone [6–9]. Lignin is synthesized from the cinnamyl alcohol monomers p-coumaryl, coniferyl, and sinapyl, and these three monolignols are synthesized in the cytoplasm from phenylalanine using different biosynthetic enzymes [10, 11]. Additionally, caffeoyl shikimate esterase (CSE) hydrolyzes caffeoyl shikimate into caffeate and together with 4-coumarate: CoA ligase (4CL) in the lignin biosynthetic pathway, bypasses the second hydroxycinnamoyl-CoA: shikimate/quinate hydroxycinnamoyltransferase (HCT) reaction [12]. Also, there are several transcription factors that have been shown to regulate secondary cell wall biosynthesis in A. thaliana [13]. Some transcription factors, for example MYB103, are not only able to activate genes involved in cellulose biosynthesis, such as CesA8, but also lignin or xylan biosynthesis genes, such as F5H, especially in S lignin biosynthesis [14].
Neolamarckia cadamba (syn. Anthocephalus chinensis), a member of the Rubiaceae family, is widely distributed in South Asia and South China due to its high economic value [15]. To date, N. cadamba research has mainly focused on its medicinal value in the treatment of various ailments and extraction of bioactive compounds [16]. Among these, triterpenoid saponins are documented as important active components and their biosynthesis requires the mevalonate (MVA) pathway in the cytosol and the non-mevalonate or 2-C-methyl-D-erythritol4-phosphate/1-deoxy-D-xylulose 5-phosphate (MEP/DOXP) plastid pathway for backbone formation [17]. Subsequently, the backbone undergoes various oxidation, substitution, and glycosylation steps mediated by a series of enzymes including geranyl diphosphate synthase(GPPS), farnesyl diphosphate synthase (FPS), squalene synthase (SQS), squalene epoxidase (SQE), β-amyrin synthase (β-AS), cytochrome P450-dependent monooxygenases (CYP 450s) and glycosyltransferases (UGTs) [18]. However, there have been no studies to date that dissect the molecular mechanisms underlying the biosynthesis of these bioactive compounds in N. cadamba. Apart from medicinal utilization, N. cadamba wood is also a suitable alternative material for building, furniture, pulp production and biomass utilization in tropical and subtropical regions [19]. We recently studied N. cadamba as a model system to analyze heteroxylan formation at the biochemical and molecular level during wood formation. Microsomes isolated from the middle and basal stem region exhibited higher UDP-Xyl synthase and xylosyltransferase enzyme activity and higher gene expression related to heteroxylan biosynthesis compared to the apical part of the stem [20]. Despite this investigation, little is known about cell wall biosynthesis pathways and transcriptional regulatory networks involved in N. cadamba xylogenesis.
RNA sequencing (RNA-Seq) is a high throughput technology based on next-generation sequencing (NGS) that enables genetic studies of species without the requirement of corresponding sequenced reference genome information [21]. RNA-Seq has evolved in to a powerful method for transcriptome profiling due to its accuracy, high throughput nature and reproducibility [22]. Subsequently, it has dramatically improved the efficiency and cost effectiveness of gene discovery [23]. To better understand the molecular basis of wood formation in N. cadamba, we initiated a RNA-seq project (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA232616) to analyze and characterize the transcriptomes of several classes of genes involved in wood formation. We hope this profiling study will help initiate a systematic breeding program geared towards genetically improving the quality and productivity of this multipurpose timber-yielding tree.
Materials and Methods
Plant materials
N.cadamba was grown as described previously [20]. Clones were proliferated and rooted as described previously [24] and grown in a greenhouse at 28/24°C, 14/10 h(day/night) and 330 μmol m2 s1 light. Peeled apical (just under apical bud, A), middle (M) and basal (just on soil surface, B) stem segments (1 cm in length) were sampled from a one-year-old plant, representing three different stages from primary to secondary growth, respectively (Fig 1). Each tissue was collected from two individual plants representing two biological replicates. All samples were quickly cut into pieces and immediately frozen in liquid nitrogen for RNA extraction.
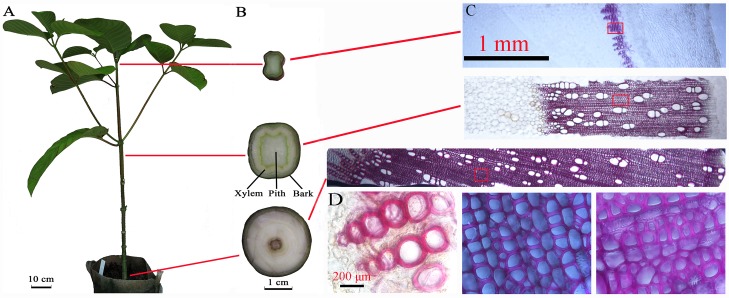
Fig 1. Tissues of N. cadamba used in deep sequencing.
(A) The whole plant of greenhouse-grown N. cadamba. (B) The apical, middle and basal stem segments with bark from N. cadamba. (C) Phloroglucinol staining of transverse stem section. (D) Magnified view of red box region in C. Scale bar: A = 10 cm, B = 1 cm, C = 1 mm and D = 200 μm.
Sectioning of stems
Tissues from the three stem regions were fixed in FAA [5% (v/v) formalin, 5% (v/v) glacial acetic acid, 65% (v/v) ethylalcohol]. Next, tissues were embedded in 3% (w/v) agarose and sections of 40μm thickness cut using a Leica VT1000S vibratome fitted with a razor blade. Histochemical analysis using Wiesner and Maűle staining in Phloroglucinol-HCl was performed as previously described [25]. Stained stem sections were observed under an Olympus BX43F light microscope.
RNA extraction, library construction and RNA-Seq
Total RNA from each sample was isolated using CTAB plus the OMEGA Plant RNA isolation kit as described previously [26]. Three libraries labeled A, M and B were constructed from RNA extracted from the apical, middle and basal stem segments, respectively. The RNA samples were checked for integrity on a 1.2% agarose gel and quantified using a Nanodrop 1000 spectrophotometer. RIN (RNA integrity number) values (> 8.0) of the samples were evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Construction of the six libraries and RNA-Seq analysis were performed by Biomarker Biotechnology Corporation (Beijing, China). The mRNA enrichment and library construction were carried out according to protocol of NEB kit (E7490, E6110, E7500). Finally, the six libraries were sequenced using Illumina HiSeq™ 2000.
De novo assembly
Clean reads were filtered from the raw reads by removing the adapter sequences and low quality sequences including reads with unknown nucleotides > 5% and more than 10% bases with a quality score (Q value) of < 20. The clean reads were then de novo assembled using the Trinity platform (http://trinityrnaseq.sourceforge.net/) with the parameters ‘K-mer = 25, group pairs distance = 300’ [21]. The reads obtained for the three stem samples and their two biological replicates were assembled together. Short reads were first assembled into longer contigs and then joined into transcripts based on the paired-end information and similarity between contigs. Finally, the longest transcript was taken as the sample UniGene. To facilitate access and utilization of the N. cadamba transcriptome sequencing data, all UniGene sequences have been uploaded to the Transcriptome Shotgun Assembly Sequence Database (TSA) at NCBI with accession numbers GASC01000001 to GASC01055370.
Functional annotation
Putative functions of the UniGenes were annotated by carrying out a BLASTx analysis [27] with an E-value threshold of 10−5 against protein databases including the NCBI non-redundant (nr) database [27], the Swiss-Prot protein database [28], the Gene Ontology (GO) database [29], the Clusters of Orthologous Groups of Proteins (COG) database [30], and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database [31].
Expression annotation
For each sample, the SOAPaligner (http://soap.genomics.org.cn/soapaligner.html) platform was used to map back reads to each UniGene. The number of mapped clean reads for each UniGene was counted and normalized into an RPKM value (reads per kb per million reads) [32]. The mean RPKM value for each UniGene from the biological replicates for each tissue was applied for downstream analysis. After that, differentially expressed genes (DEG) between samples were detected using DESeq software with a general chi-square test based on RPKM values. FDR (false discovery rate) was used to identify the P-value threshold in multiple tests in order to compute the significance of differences in each UniGene expression between two samples [33]. Genes were regarded as differentially expressed by the FDR < 0.01 and the absolute value of log2Ratio ≥ 1. In our study, DEGs between two samples were screened and used to compare GO classifications. Then, detailed expression profiles, I (up-regulation), II (irregularly regulated), III (irregularly regulated), IV (down-regulation) were distinguished for DEGs by using log10 RPKMA, log10 RPKMM and log10 RPKMB values, where I = (log10 RPKMA ≤ log10 RPKMM ≤ log10 RPKMB), II = (log10 RPKMA ≤ log10 RPKMM ≥ log10 RPKMB), III = (log10 RPKMA ≥ log10 RPKMM ≤ log10 RPKMB), and IV = (log10 RPKMA ≥ log10 RPKMM ≥ log10 RPKMB). UniGenes that were more abundantly expressed in libraries M and B compared to A were identified.
Identification and regulation pathways analysis of cell wall-related transcription factors
A total of 82 transcription factors and transcriptional regulatory families of A. thaliana were downloaded from the PlnTFDB database [34]. UniGenes were searched against this database using the local NCBI-2.2.30+ BLASTx algorithm (E-value≤1E-10). Thetranscription factor KNAT7 was searched using local TBLASTN with the A. thaliana KNAT7 amino acid sequence (E-value≤1E-10) against the transcriptome library. UniGene sequences were double-checked by BLASTx searches against protein databases including the NCBI non-redundant (nr) database and the A. thaliana TAIR10 database.
According to the cell wall-related transcription factor regulatory network described by Schuetz et al [13] and Ohman et al [14], transcription factors involved in regulating lignin, cellulose and hemicellulose biosynthesis were identified in N. cadamba as described above. Relative (yellow—blue scale) and absolute (white—red scale) expression profiles of these genes were implicated in three different lignification processes represented by the A, M and B stem segments.
Discovery of cellulose, mannan and monolignol biosyntheticgenes
Sequences of cellulose biosynthesis-related, mannan biosynthesis-related and monolignol biosynthesis-related proteins from A. thaliana were downloaded from the TAIR database (www.arabidopsis.org; shown in S1 File). The cellulose, mannan and monolignol biosynthesis-related structural genes were searched using TBLASTN with the amino acid sequences of the proteins (E-value ≤1E-10) against the transcriptomic library [27]. Then, these enzymes were identified in N. cadamba as previously described for KNAT7. According to the known cellulose [3], mannan [35, 36] and lignin [10–12] biosynthesis pathways, the UniGenes were ascribed to metabolic pathways.
Discovery of cadambine, triterpenoidsaponin and phytosterol biosynthesis-related genes
Cadambine, triterpenoid saponin and phytosterol biosynthesis-related protein tryptophan synthase (TSA1-2: At3g54640, At4g02610; TSB1-2: At5g54810, At4g27070), tryptophan decarboxylase (TDC: X67662.1), geranyl diphosphate synthase (GPPS: AT2G34630), geraniol synthase (GES: JN882024.1), geraniol10-hydroxylase (G10H: KF561461.1), secologanin synthase (SLS: KF415117.1), strictosidine synthase (STR1-3: At1g74020, AT1G74020, At1g74000), farnesyl diphosphate synthase (FPS1-2: AT5G47770, AT4G17190), squalene synthase (SQS1-2:AT4G34640, AT4G34650), squalene epoxidase (SQE1-3: AT1G58440, AT2G22830, AT4G37760), β-amyrin synthase (β-AS: AT1G78950), and cycloartenolcyclase (CAS1: AT2G07050) from A. thaliana or Catharanthus roseus were downloaded from the TAIR database or Uniprot database [28]. The cadambine, triterpenoid saponin and phytosterol biosynthesis-related structural genes in N. cadamba were subsequently identified, allowing them to be assigned to metabolic pathways [18, 37].
Real-time quantitative PCR analysis (RT-qPCR)
For RT-qPCR analysis, RNA samples were reverse transcribed into first-strand cDNA using PrimeScript® RT Master Mix (Takara, China) according to the manufacturer’s protocol. The cDNA was diluted fifteen-fold and used as the template for RT-qPCR. Amplifications were carried out in triplicate in a total volume of 20μL containing 10μL of 2×SYBR® Premix Ex Taq™ II (Takara, China), 2 μl of each primer (5 μM), 2 μl of cDNA, and 4 μl of ddH2O. Thermocycling conditions were as follows: an initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 58°C annealing for 30 s and 72°C extension for 15 s, and an infinite hold at 10°C. The specificity of the PCR amplicon was checked using a heat dissociation protocol (from 65 to 95°C) after the final PCR cycle. The primers used in the RT-qPCR are shown in S2 File and the cyclophilin (JX902587) gene was used as the internal reference.
Results
Determination of cell wall components during xylogenesis
Previously, we analyzed N. cadamba cell wall composition during xylogenesis [20] and found that the levels of both lignin and non-cellulosic polysaccharides increased with stem maturity, exhibiting highest levels in the basal stem segment and lowest in the apical stem segment. The major non-cellulosic polysaccharide in the stem segments was heteroxylan with a substantially lower level of heteromannan. We also found that the proportion of cellulose decreased with stem maturity, with the highest level observed in the apical stem segment and the lowest in the middle segment, increasing in the basal stem segment. However, the proportion of lignin in both the middle and basal segments was significantly higher than in the apical stem segment [20]. As shown in histochemical analysis, lignin deposition was found only in vessel elements of the apical stem segments in contrast to the middle and basal stem segments that show lignin deposition in tracheary elements and fibers (Fig 1C and 1D).
RNA-Seq and de novo assembly
To obtain a global overview of the N. cadamba transcriptome and gene expression during progression of xylogenesis in the stem, RNA was extracted from 1-year-old greenhouse grown plants initially raised from tissue culture. Three libraries (A, M, B) were constructed and RNA-Seq was performed using two biological replicates. RNA-Seq analysis generated 42.49G of raw data, and Q30 percentages (percentage of sequences with sequencing error rates < 0.1%) were found to be over 80% (Table 1). Based on the de novo assembly using Trinity [32], all clean short reads from the six libraries were assembled together into 5,870,723 contigs based on their overlap regions. The contigs were joined into 111,864 transcripts, and finally, a total of 55,432 UniGenes were identified with a mean length of 803.2bp and an N50 length of 1,501bp, among which 13,280 UniGenes (23.96%) were greater than 1 kb. The length distributions of contigs, transcripts and UniGenes are shown in Table 2. The data shows that the throughput and sequencing quality was high enough for further downstream analyses.
Table 1. RNA-Seq data summary for all samples.
| Samples | BMK-ID | Total reads | Total bases (nt) | GC% | Q30% |
|---|---|---|---|---|---|
| Apical stem | A1a | 25,085,358 | 5,066,725,234 | 46.03% | 81.51% |
| segment | A2 | 32,126,330 | 6,489,002,687 | 44.08% | 81.48% |
| Middle stem | M1 | 38,555,036 | 7,787,264,195 | 44.64% | 82.13% |
| segment | M2 | 31,421,956 | 6,346,729,891 | 44.06% | 81.26% |
| Basal stem | B1 | 47,999,394 | 9,695,200,117 | 44.29% | 82.53% |
| segment | B2 | 35,170,009 | 7,103,808,681 | 43.95% | 82.14% |
| Total | 210.39M | 42.49G |
a biological replicates
Table 2. Length distribution of assembled contigs, transcripts, UniGenes and predicted ORFs of UniGenes.
| Length range | Contigs | Transcripts | UniGenes | ORF |
|---|---|---|---|---|
| 200–300 | 5,820,454(99.14%) | 21,414(19.14%) | 17,894(32.28%) | 38,179(69.20%)a |
| 300–500 | 21,669(0.37%) | 19,663(17.58%) | 14,433(26.04%) | 4,556(8.26%) |
| 500–1000 | 14,456(0.25%) | 21,046(18.81%) | 9,825(17.72%) | 5,443(9.87%) |
| 1000–2000 | 9,122(0.16%) | 27,098(24.22%) | 7,816(14.10%) | 5,177(9.38%) |
| 2000+ | 5,022(0.09%) | 22,643(20.24%) | 5,464(9.86%) | 1,816(3.29%) |
| Total number | 5,870,723 | 111,864 | 55,432 | 55,171 |
| Total length | 269,782,291 | 137,157,312 | 44,522,744 | 23,683,593 |
| N50 length | 44 | 2,023 | 1,501 | 1,086 |
| Mean length | 45.95 | 1226.11 | 803.20 | 429.28 |
a The length range is <300 bp
Functional annotation
Approximately 47.8% of the UniGenes (26,487) were annotated by BLASTx, with a threshold of 10−5, using five public databases (NCBI non-redundant (nr) database, Swiss-Prot protein database, Gene Ontology (GO) database, Clusters of Orthologous Groups of Proteins (COG) database and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database). One half (12,293) of the UniGenes had a length of > 1000 bp. The functional annotation distributions of UniGenes are shown in Table 3.
Table 3. Summary for UniGene functional annotation.
| Annotated databases | All sequence | > = 300 bp | > = 1000 bp |
|---|---|---|---|
| COG | 7,754 | 7,093 | 5,255 |
| GO | 21,699 | 18,499 | 11,022 |
| KEGG | 5,589 | 4,780 | 3,042 |
| Swissprot | 19,590 | 16,967 | 10,559 |
| nr | 26,404 | 22,213 | 12,290 |
| All | 26,487 | 22,267 | 12,293 |
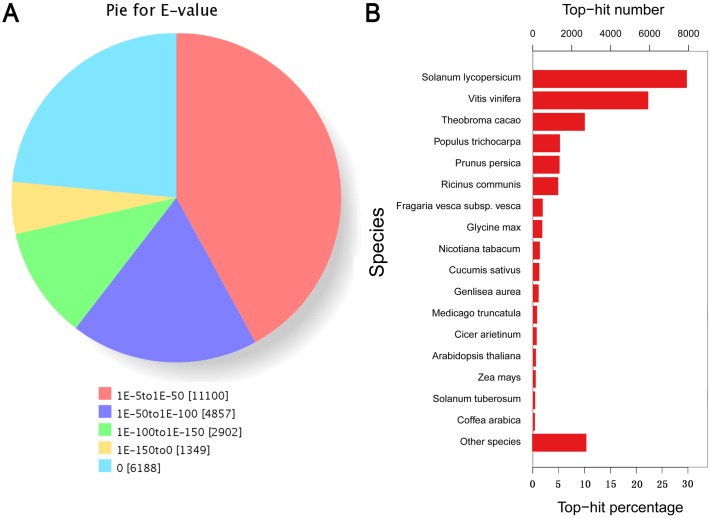
Based on nr annotation and the E-value distribution, 57.9% of UniGenes showed a very strong homology (E-value < 10−50) to available plant sequences (Fig 2A). The 17 top-hit species based on nr annotation are shown in Fig 2B. Nearly 73% of UniGenes could be annotated with sequences from the 5 top-hit species (Fig 2B, S3 File), which were Solanum lycopersicum, Vitis vinifera, Theobroma cacao, Populus trichocarpa and Prunus persica.
Fig 2. Characteristics of the homology search of N. cadamba UniGenes in the nr database.
(A) E-value distribution of the top BLASTx hits against the nr database for each UniGene. (B) Number and percentage of UniGenes matching the 17 top species using BLASTx in the nr database.
Generally, Gene Ontology (GO) analysis was carried out to classify the functions of the assembled UniGenes in terms of their associated biological processes, cellular components, and molecular functions [29]. To better understand functional categories, GO analysis was employed to annotate UniGenes by known proteins using the Blast2GO program [38], after which the GO functional classifications of these UniGenes were performed using WEGO software [39]. A total of 21,699 UniGenes were classified according to the three GO categories. Under the biological process category, large numbers of UniGenes were categorized as cellular process (17,099, 78.8%) and metabolic process (16,414, 75.6%). Within the cellular component category, large numbers of UniGenes were categorized as cell part, cell and organelle component categories. As for the molecular function category, binding (13,353, 61.5%) and catalytic activity (11,293, 52.0%) were the most highly represented categories (S1 Fig, S4 File). Furthermore, a total of 205, 165, 337, 61and 19 UniGenes were annotated within the cellulose biosynthetic process (GO:0030244), lignin biosynthetic process (GO:0009809), xylan biosynthetic process (GO:0045492), glucuronoxylan biosynthetic process (GO:0010417) and mannan synthase activity (GO:0051753), respectively (S5 File).
The COG protein database is an attempt on phylogenetic classification of the proteins encoded in complete genomes of species [40]. All UniGenes were subjected to a search against the COG database for functional prediction and classification resulting in the assignment of 7,754 UniGenes. The COG database represented major phylogenetic lineages of N. cadamba, as shown in S2 Fig and S6 File. According to the COG annotation, these UniGenes were classified into 25 different functional classes, with the largest cluster being general function prediction only (group R, 2,177, 28.1%) followed by replication, recombination and repair (group L, 1,154, 14.9%). However, no UniGene was assigned to extracellular structures (group W). It was noteworthy that there were 618, 375 and 263 UniGenes sharing homology with carbohydrate transport and metabolism (group G), secondary metabolites biosynthesis, transport and catabolism (group Q) and cell wall/membrane/envelope biogenesis (group M), respectively, including UniGenes related to cell wall biosynthesis. This data will be useful in exploring protein classification and evolution rates [41].
By mapping to the KEGG reference pathways, 5,589 UniGenes were assigned to116 pathways (S7 File) in the KEGG database [31]. Ribosome pathways (Ko03010) were the most enriched (201), followed by plant hormone signal transduction (Ko04075, 190). Moreover, 79 UniGenes were mapped to phenylpropanoid biosynthesis pathways (Ko00940), with a majority of genes participating in lignin biosynthesis, and 146 UniGenes were mapped to starch and sucrose metabolism (ko00500), with a majority of genes participating in cellulose, mannan and heteroxylan biosynthesis (S8 File).
Differentially expressed genes in different stem segments
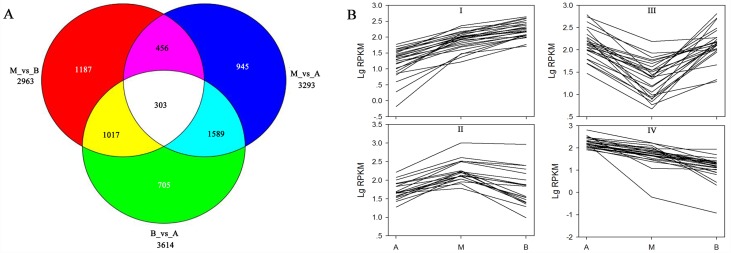
Differentially expressed genes (DEGs) among the three different stem segments were identified by DESeq software [33]. Based on this analysis, a total of 6,202 UniGenes were identified as DEGs in at least two libraries (Fig 3A, S9 File). Among these UniGenes, 3,293, 3,614 and 2,963 showed differential expression between A and M, A and B, M and B, respectively (Fig 3A, S1 Table). GO and COG classification is shown in S1 and S3 Figs respectively. Amongst the DEGs, 4 groups were defined according to their variations in expression profiles, containing 1,119, 1,436, 1,588 and 2,059 UniGenes, respectively. Group I was defined as being up-regulated, group IV as down-regulated, and groups II and III had irregular expression patterns (Fig 3B). Each of these four groups is shown in S10 File. Moreover, there were 1,634 UniGenes with a higher expression profile in the M and B libraries compared to the A library, and these UniGenes are shown in S9 File.
Fig 3. DEGs Venn diagram and expression profile.
(A) Venn diagram of DEGs. A, apical stem segment; M, middle stem segment; B, basal stem segment. (B) Four expression profiles of DEGs. I and IV indicate UniGenes which are up-regulated and down-regulated, respectively, and II and III indicate those with irregular expression. Twenty-five UniGenes were selected for the diagram for each expression profile.
Among the DEGs and 1,634 UniGenes, there were 25 and 10 UniGenes annotated in the cellulose biosynthetic process (GO:0030244) category, 42 and 23 in the lignin biosynthetic process (GO:0009809), 74 and 31 in the xylan biosynthetic process (GO:0045492), 11 and 5 in the glucuronoxylan biosynthetic process (GO:0010417), and 10 and 0 in the mannan synthase activity (GO:0051753) category, respectively (S11 File).
Transcription factors related to cell wall component biosynthesis
A total of 1,782 N. cadamba UniGenes with high sequence identity (E-value ≤ 1E-10) corresponding to 80 out of 82 A. thaliana transcription factor families downloaded from PlnTFDB were found in the stem libraries. The five most abundant transcription factor gene families were bHLH, C3H, C2H2, MYB and the HB group (S12 File). These are mainly associated with plant growth, development, stress responses, cell differentiation, morphogenesis, RNA metabolism, secondary cell wall formation and secondary metabolism [42–46].
Secondary cell wall formation is a critical step in the maturation of tracheary elements and fibre cells in the xylem [6, 47] and regulated by MYB [14, 48–54] and NAC [55–64] transcription factor families. In this study, 77 and 45 UniGenes were found with high sequence identity (E-value ≤ 1E-10) corresponding to the A.thaliana MYB and NAC transcription factor families, respectively (S12 File). Of 77 MYB transcription factor UniGenes, 28 UniGenes belonged to DEGs among the A, M and B libraries, including 12 UniGenes with an expression profile where the abundance in the M and B libraries was higher than in the A library. Of the 45 NAC transcription factor UniGenes, 19 UniGenes, including 12 UniGenes with the same expression profile as above, were identified as DEGs.
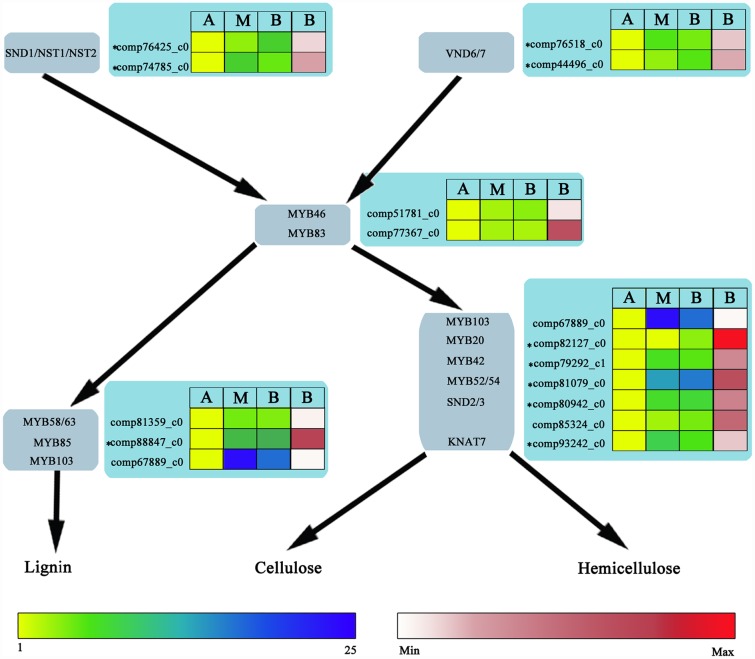
A network of transcription factors involved in the biosynthesis of the secondary cell wall has been identified in A. thaliana [13, 14, 47, 52, 57, 58]. Based on this network, UniGene transcription factor sequences identified in this study were double-checked by BLAST searches against protein databases including the NCBI nr database and the A. thaliana TAIR10 database. Candidate genes corresponding to most of the known transcription factors involved in regulatory networks in A. thaliana have been found in N. cadamba (Fig 4). Moreover, most of them were identified as DEGs and had an expression profile with higher expression abundance in the M and B libraries than in the A library (Fig 4, S12 File), including UniGenes comp51781_c0 (MYB46), comp77367_c0 (MYB83), comp67889_c0 (MYB103), comp81359_c0 (MYB56/63) and comp85324_c0 (SND2/3), despite no significant differences observed.
Fig 4. N. cadamba UniGenes in the transcriptional network regulating secondary cell wall (SCW) biosynthesis according to A. thaliana.
Light grey boxes indicate major transcription factors of the Arabidopsis transcriptional network leading to biosynthesis of the three major SCW constituents. Light blue boxes indicate N. cadamba putative UniGenes encoding orthologues. * indicates the DEGs. Yellow-blue scale and white-red scale indicate relative and absolute expression profiles, respectively. A, apical stem segment; M, middle stem segment; B, basal stem segment. Absolute expression level (RPKM) is only shown for the basal stem segment.
Genes involved in the cellulose and mannan biosynthetic pathways
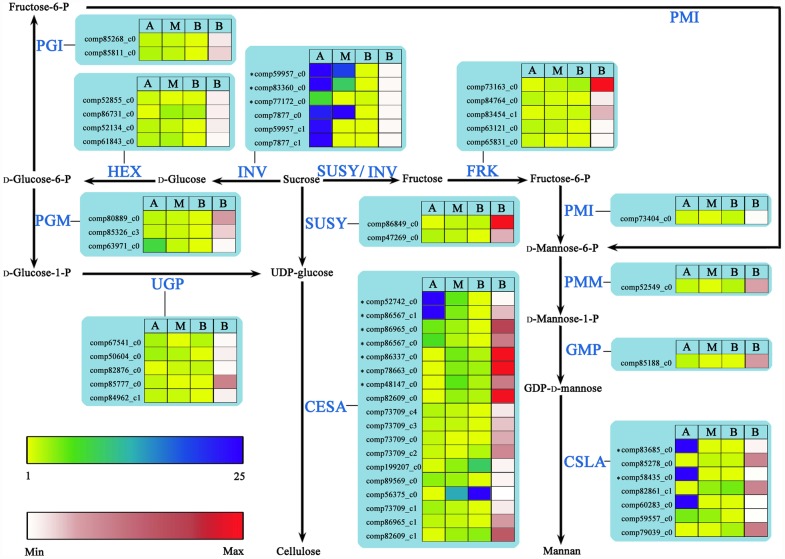
Sucrose catabolism produces both direct and indirect substrates for cellulose biosynthesis, and indirect substrates for mannan biosynthesis in planta, which means that cellulose and mannan biosynthetic pathways converge on sucrose [3, 36]. Most of the UniGenes encoding enzymes participating in cellulose or mannan biosynthetic pathways were not identified as DEGs. However, the majority of these UniGenes had a higher expression abundance in apical stem segments compared to the middle and basal stem segments, including UniGenes comp86965_c0 showing 82% identity with AtCesA1, and comp52742_c0, comp86567_c0 and comp86567_c1 showing high identity with AtCesA2/6/9 by BLASTx. In contrast, the UniGenes comp86337_c0 (82% identity with AtCesA7 by BLASTx) and comp78663_c0 (73% identity with AtCesA4 by BLASTx) exhibited opposite expression pattern (Fig 5, S13 File).
Fig 5. UniGenes involved in cellulose and mannan biosynthesis in wood-forming tissues of N. cadamba.
Sugar and polymer intermediates are shown in black, whereas the proteins (enzymes) involved in each step are shown in blue. Detailed protein names, annotation and RNA-Seq expression data are provided in S12 File. * indicates the DEGs. Yellow-blue scale and white-red scale indicate relative and absolute expression profiles, respectively. A, apical stem segment; M, middle stem segment; B, basal stem segment. Absolute expression level (RPKM) is only shown for basal stem segment. CESA, cellulose synthase; CSLA, cellulose synthase like A; FRK, fructokinase; GMP, GDP-D-mannose pyrophosphorylase; HEX, hexokinase; INV, invertase; PGI, phosphoglucose isomerase; PGM, phosphoglucomutase; PMI, phosphomannose isomerase; PMM, phosphomannomutase; SUSY, sucrose synthase; UGP, UDP-glucosepyrophosphorylase.
Genes involved in the monolignol biosynthetic pathway
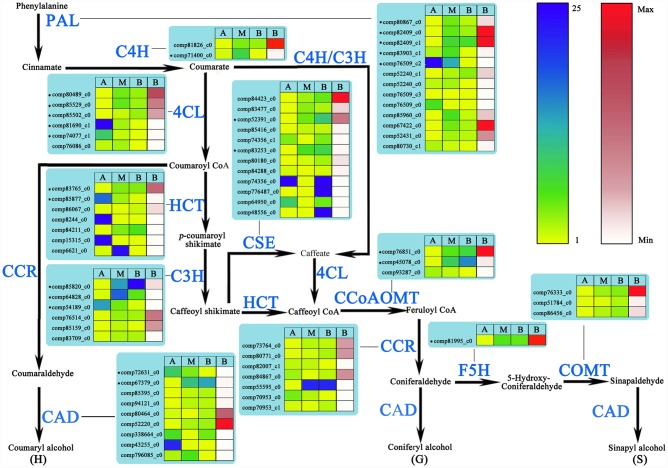
The main monolignols of lignin are coumaryl, coniferyl and sinapyl alcohols, which are finally incorporated into the lignin polymer as p-hydroxyphenyl (H), guaiacyl (G) andsyringyl (S) units, respectively [11]. In this study, a total of 69 UniGenes were found in N. cadamba transcriptomes with an E-value ≤ 1E-10 corresponding to all eleven monolignol biosynthesis enzymes in A. thaliana (Fig 6). The expression of 1–3 members of each gene family (PAL, 4CL, HCT, C3H, CSE, CCoAOMT, F5H and CAD) increased in both M and B libraries as compared to the A library (Fig 6, S14 File). Additionally, compared with the number of genes that encode each one of the eleven key enzymes involved in the lignin biosynthesis pathway in the A. thaliana TAIR database, there are more UniGenes in N. cadamba and genes in the Populus trichocarpa [65] and Eucalyptus grandis [3] genomes (Table 4).
Fig 6. UniGenes involved in lignin biosynthesis in wood forming tissues of N. cadamba.
Phenylalanine, intermediates and monolignols are shown in black, while the proteins (enzymes) involved in each step are shown in blue. Detailed protein names, annotation and RNA-Seq expression data are provided in S13 File. * indicates the DEGs. Yellow-blue scale and white-red scale indicate relative and absolute expression profiles, respectively. A, apical stem segment; M, middle stem segment; B, basal stem segment. Absolute expression level (RPKM) is only shown for basal stem segment. 4CL, 4-coumarate: CoA ligase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; COMT, caffeic acid O-methyltransferase; CSE, Caffeoylshikimate esterase; F5H, ferulate 5-hydroxylase; HCT, p-hydroxycinnamoyl-CoA: quinate shikimate p-hydroxycinnamoyltransferase; PAL, phenylalanine ammonia-lyase.
Table 4. Comparison in number of genes that encode eleven key enzymes in the monolignol biosynthesis pathway among A. thaliana, N. cadamba, P. trichocarpa and E. grandis.
| Enzymes | A. thaliana | N. cadamba | P. trichocarpa | E. grandis |
|---|---|---|---|---|
| PAL | 4 | 13 | 5 | 9 |
| C4H | 1 | 2 | 3 | 2 |
| 4CL | 4 | 6 | 17 | 13 |
| CCR | 2 | 7 | 9 | 9 |
| CAD | 9 | 9 | 16 | 46 |
| HCT | 1 | 7 | 7 | 5 |
| C3H | 1 | 6 | 4 | 4 |
| CSE | 1 | 12 | 2 | 4 |
| CCoAOMT | 1 | 3 | 6 | 17 |
| COMT | 1 | 3 | 25 | 67 |
| F5H | 1 | 1 | 3 | 2 |
Genes involved in the cadambine, triterpenoid saponin and phytosterol biosynthetic pathways
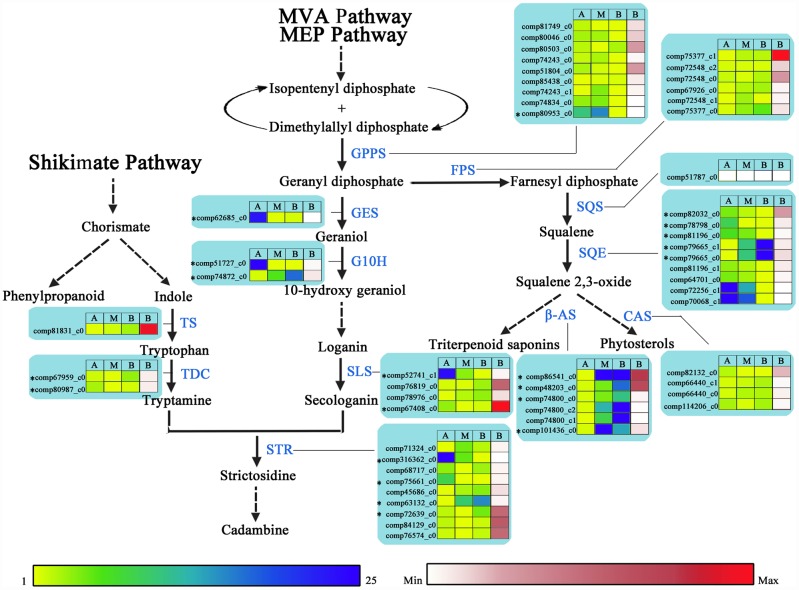
Medicinal ingredients isolated from N. cadamba plants have traditionally been used in the treatment of various human ailments, such as diabetes mellitus, wounds and fever, as well as for their antimicrobial activity and antitumor properties [16]. Medicinal properties of N. cadamaba might be due to the presence of bioactive compounds such as alkaloids, triterpenoids and iridoids in plant tissues [66–68]. In the present study, a total of 54 UniGenes were found in the N. cadamba transcriptomes with E-values≤ 1E-10 corresponding to all eleven enzymes in A. thaliana or C. roseus (Materials and methods), except for the UniGene comp51787_c0, which was found in the transcriptomes from the other tissues sample (Fig 7, S15 File).
Fig 7. UniGenes involved incadambine, triterpenoid saponin and phytosterol biosynthesis.
Intermediates, cadambine, triterpenoid saponins and phytosterols are shown in black, whereas the proteins (enzymes) involved in each step are shown in blue. Detailed protein names, annotation and RNA-Seq expression data are provided in S15 File. * indicates the DEGs. Yellow-blue scale and white-red scale indicate relative and absolute expression profiles, respectively. A, apical stem segment; M, middle stem segment; B, basal stem segment. Absolute expression level (RPKM) is only shown for basal stem segment. CAS, Cycloartenol cyclase; FPS, farnesyl diphosphate synthase; G10H, geraniol 10-hydroxylase; GES, geraniol synthase; GPPS, geranyl diphosphate synthase; SLS, secologanin synthase; SQE, squalene epoxidase; SQS, Squalene synthase; STR, strictosidinesynthase; TDC, tryptophan decarboxylase; TS, tryptophan synthase; β-AS, β-amyrin synthase.
Expression analysis by RT-qPCR
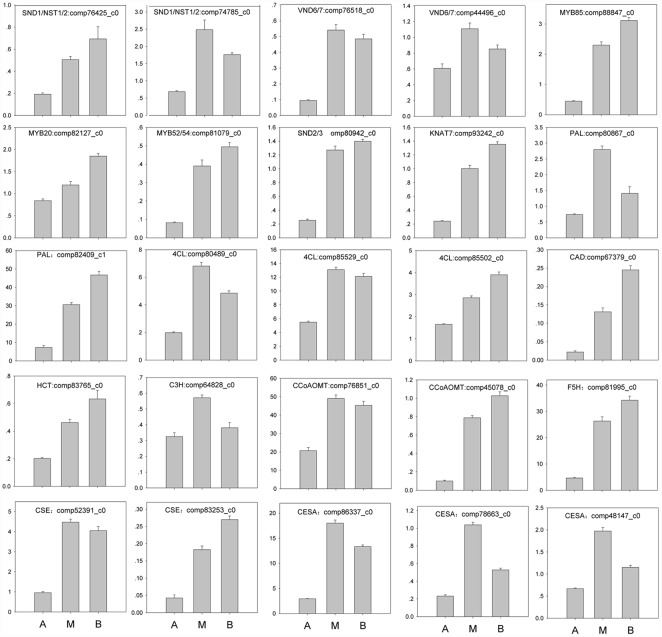
DEGs related to cell wall biosynthesis, including transcription factors and structural genes with expression levels higher in both basal and middle stem segments compared to apical stem segments, were examined for their expression patterns in the three stem segments representing different stages of xylogenesis. Genes for which no primers could be designed were excluded (Fig 8). Additionally, genes involved in cadambine, triterpenoid saponin or phytosterol biosynthetic pathways were also examined for their expression in roots, middle stem segments, young leaves, mature leaves, flowers, bark and cambium, with the exception of a few genes for which no primers could be selected (S4 Fig). As shown in Fig 8, all genes were expressed at a higher level in both middle and basal stem regions compared to the apical part of the stem, demonstrating a high correlation between RNA-seq and RT-qPCR data.
Fig 8. Expression of UniGenes related to transcription factors regulating SCW biosynthesis, lignin, or cellulose biosynthesis.
Error bars on each column indicate SEs from three replicates. A, apical stem segment; M, middle stem segment; B, basal stem segment.
Discussion
N. cadamba transcriptome database by Illumina HiSeqTM 2000 sequence
It is easier to clone and characterize target genes in model plants such as Arabidopsis, rice, and poplar that have a fully sequenced genome and other available tools for genetic manipulation. However, to carry out similar work or any genetic improvement research, it is difficult to clone genes of interest in plants such as N. cadamba for which no genomic information is available. Despite the fact that the N.cadamba genome is not sequenced, RNA-Seq has been used to provide the sequences of expressed genes. In this report, 55,171 UniGenes with intact ORFs were predicted using the Getorf software (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html, Table 2). Thus, genes of interest from N. cadamba can directly be cloned through the database and/or together with 5’ or 3’RACE.
Among the 55,432 unique sequences, 28,945 UniGenes (52.2%) had no functional annotation in the present study (Table 2, S9 File). Thus, half of the UniGenes did not match known protein families in the five public protein databases. Therefore, we consider them to represent unknown protein families, indicating that novel information was discovered in our Illumina data sets, in particular the 1,649 UniGenes without functional annotation among the DEGs. There were 49,230 UniGenes (88.8%) that comprise a group that did not show differential expression between the three stem segments (S9 File). The large number of genes with common expression levels in the three stem region transcriptomes suggests that the majority of the transcripts are involved in basal cellular metabolism processes and that the distinctive characteristics of each xylem region are derived from differences in the expression levels of a relatively small number of genes.
Regulation of secondary metabolism during xylogenesis
The major goal of the present study was to carry out a preliminary screen for key genes involved in N. cadamba xylogenesis. Several studies in Arabidopsis have identified a network of transcription factors regulating the expression of numerous genes directly involved in the biosynthesis of secondary cell walls [13, 14]. The master switches for fibre (SND1, NST1, NST2) [47, 57, 69], protoxylem (VND7) and metaxylem (VND6) [58] differentiation are thought to initiate the transcriptional network for secondary cell wall formation by binding to SNBE (Secondary wall NAC-Binding Element) regulatory regions in the promoters of target genes. These include the two core transcription factors, MYB46 and MYB83, whose promoters each contain several SNBE promoter elements and are thought to be direct targets of secondary cell wall NAC genes [48, 49, 53, 64]. The MYB46/83 node activates the expression of numerous other transcription factors, whose activity amplifies the transcriptional network and thereby promotes lignin, cellulose and/or hemicelluloses biosynthesis [48, 49, 53, 54]. In this study, most of these transcription factor genes were up-regulated during xylogenesis in N. cadamba (Figs 4 and 8). These data indicate that these transcription factors are involved in secondary cell wall biosynthesis during xylogenesis in N. cadamba.
UniGenes in several metabolic pathways involved in secondary wall formation and/or maintenance showed differential expression between different regions of the stem (Figs 5 and 6). Transcription factors MYB58, MYB63, or MYB85 specifically regulate lignin biosynthesis/deposition, with the MYB58 and MYB63 directly activating expression of nearly all the genes involved in the lignin biosynthetic pathway. Both MYBs are thought to bind at conserved AC regulatory elements found upstream of the majority of lignin biosynthetic genes [50, 52, 54], with the exception that MYB103 is required for F5H expression and syringyl lignin biosynthesis [14]. Transcription factors SND2, SND3, and MYB103 are able to activate the promoters of genes involved in cellulose biosynthesis, such as CesA8, while MYB52 andMYB54 activate gene promoters involved in cellulose and xylan biosynthesis. In this study, the expression levels of these downstream transcription factors and some UniGenes in most metabolic steps in lignin and cellulose biosynthesis were similar to those of the upstream transcription factors. Expression levels of these transcription factors were significantly higher in both middle and basal stem segments of N. cadamba, compared to the apical stem segment (Figs 4–6, S12–S14 Files). Our previous study has found that microsomes isolated from the middle and basal stem segments exhibit the highest activity of UDP-xylsynthase and xylosyltransferase, and higher expression of genes related to heteroxylan biosynthesis compared to the apical segment of N. cadamba stem [20]. Overall, these data will be beneficial to further understanding the regulatory networks involved in secondary cell wall formation.
Genes involved in the cellulose/monolignol biosynthetic pathway
The UniGenes comp86965_c0 (82% identity with AtCesA1 by BLASTx), and comp52742_c0, comp86567_c0 and comp86567_c1 (high identity with AtCesA2/6/9 by BLASTx) exhibited higher expression levels in apical stem segments compared to the middle and basal stem segments (Fig 5, S13 File). Vascular bundles in the apical stem segment are formed from procambial cells and consist of primary xylem tissues [20], consistent with expression of these CesAs, which are proposed to be involved in primary cell wall synthesis based on their homologies with A. thaliana CesAs [5]. However, the differentially expressed UniGenes, comp86337_c0 (82% identity with AtCesA7 by BLASTx) and comp78663_c0 (73% identity with AtCesA4 by BLASTx) exhibited higher expression in both middle and basal stem segments compared to apical stem segments (Fig 5, S13 File). Furthermore, the amount of secondary xylem increased in both middle and basal stem segments [20]. This was primarily due to the activity of CesAs involved in secondary cell wall biosynthesis as reported in the case of A. thaliana, where CesA complex enzymes CesA4, CesA7 and CesA8 are required for cellulose synthesis [4]. All these observations indicate that the differentially expressed UniGenes are required for primary or second cell wall cellulose biosynthesis during stem development in N. cadamba.
In the monolignol biosynthetic pathway, the expression of 1–3 members of gene families (PAL, 4CL, HCT, C3H, CSE, CCoAOMT, F5H and CAD) increased in both the M and B libraries as compared to the A library (Fig 6, S14 File). The expression of these genes corresponded with our earlier cell wall component analysis of apical, middle and basal stem segments in N. cadamba showing that lignin levels were higher in the middle and basal stem segments compared to the apical segment [20]. These observations indicate that these genes are the main transcripts and strongest candidates for involvement in lignin biosynthesis in N. cadamaba. Furthermore, the number of N. cadamba UniGenes and the number of genes found in the P. trichocarpa [65] and E. grandis [3] genomes that encode each one of the eleven key enzymes involved in the lignin biosynthesis pathway were more than the number found in the A. thaliana TAIR database (Table 4). These comparisons suggest that lignin biosynthesis in trees is more complex and requires more genes.
The new genetic model tree for xylogenesis
Unlike the model plant Arabidopsis, tree species such as N. cadamaba accumulate higher amounts of secondary xylem. There are many more genes related to lignin biosynthesis in trees, compared to Arabidopsis (Table 4), which forms relatively little lignin rich in G units [70]. In contrast, trees typically have roughly equal amounts of G and S units comprising their lignin. This suggests that although Arabidopsis has been considered an excellent genetic model for the study of lignin biosynthesis in trees [71], there are a number of disadvantages. These include fewer xylem cell types, small plant size, and an annual growth habit, which means that studies into seasonal variation of xylem differentiation, dormancy, and cambial aging process cannot be carried out [72]. N. cadamba is a fast-growing tree for which a highly efficient in vitro regeneration system has been successfully established [24]. Furthermore, the CRISPR-Cas9 system for genome engineering has been established and applied widely to elucidate the functional organization of the genome at the systems level, and establish causal linkages between genetic variations and biological phenotypes [73]. These factors suggest that N. Cadamba might be established as a model plant for cell wall biosynthesis and wood development studies in the future by adopting new genetic technologies.
Conclusions
In this study, we have conducted the first large-scale analysis of the N. cadamba transcriptome and identified several genes responsible for N. cadamba xylogenesis using Illumina paired-end sequencing technology. With DEG profiling, our results have provided a vast amount of information about genes that are differentially expressed during xylogenesis. 1,634 UniGenes exhibited significantly higher expression levels in the basal and middle stem segments compared to the apical stem segment. They included NAC and MYB transcription factors related to secondary cell wall biosynthesis, genes related to most metabolic steps of lignin biosynthesis and CesA genes involved in cellulose biosynthesis. Further analysis of the generated transcriptome dataset will provide new insights into molecular mechanisms of wood formation in fast-growing trees.
Supporting Information
(TIF)
(TIF)
(TIF)
Error bars on each column indicate SEs from three replicates. R, root; S, middle stem segment; L, young leaf; ML, mature leaf; F, flower; B, bark; C, cambium.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLS)
(XLSX)
(XLS)
(XLSX)
The color red, green, blue and violet indicate UniGenes participating in lignin, cellulose, mannan and heteroxylan biosynthesis process, respectively.
(XLSX)
(XLSX)
(XLSX)
The color red and green indicate the UniGenes from DEGs and UniGenes with the expression profile showing that the expression abundance in the M and B libraries was higher than in the A library in DEGs, respectively.
(XLSX)
The color red and blue indicate the UniGenes identified as DEGs and UniGenes with expression profiles where the expression abundance in the M and B libraries was higher than in the A library in DEGs, respectively.
(XLSX)
(XLSX)
The color green and red indicate the UniGenes from DEGs and UniGenes with an expression profile showing that abundance in the M and B libraries was higher than in the A library in DEGs, respectively.
(XLSX)
(XLSX)
* indicates M as control sample and A as test sample.
(DOCX)
Acknowledgments
This work was supported by the National Science Foundation of China (Grant Number 31470681), the special funds project of Forestry Public Welfare Industry Research of China (Grant Number 201004020) and State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources in China.
Data Availability
All UniGenes are available from the NCBI Transcriptome Shotgun Assembly Sequence Database (TSA) (accession number) from GASC01000001 to GASC01055370.
Funding Statement
This work was supported by the National Science Foundation of China (Grant Number 31470681), the special funds project of Forestry Public Welfare Industry Research of China (Grant Number 201004020). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Paux E, Tamasloukht M, Ladouce N, Sivadon P, Grima-Pettenati J. Identification of genes preferentially expressed during wood formation in Eucalyptus. Plant Mol Biol. 2004; 55(2): 263–280. [DOI] [PubMed] [Google Scholar]
- 2.Plomion C, Leprovost G, Stokes A. Wood formation in trees. Plant Physiol. 2001; 127(4): 1513–1523. [PMC free article] [PubMed] [Google Scholar]
- 3.Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, et al. The genome of Eucalyptus grandis. Nature. 2014; 510(7505): 356–362. 10.1038/nature13308 [DOI] [PubMed] [Google Scholar]
- 4.Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci U S A. 2003; 100(3): 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, et al. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci U S A. 2007; 104(39): 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu AM, Hornblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, et al. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 2010; 153(2): 542–554. 10.1104/pp.110.154971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pena MJ, Zhong R, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, et al. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007; 19(2): 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromley JR, Busse-Wicher M, Tryfona T, Mortimer JC, Zhang Z, Brown DM, et al. GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 2013; 74(3): 423–434. 10.1111/tpj.12135 [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Teng Q, Huang W, Zhong R, Ye ZH. The F8H glycosyltransferase is a functional paralog of FRA8 involved in glucuronoxylan biosynthesis in Arabidopsis. Plant Cell Physiol. 2009; 50(4): 812–827. 10.1093/pcp/pcp025 [DOI] [PubMed] [Google Scholar]
- 10.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003; 54: 519–546. [DOI] [PubMed] [Google Scholar]
- 11.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010; 153(3): 895–905. 10.1104/pp.110.155119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science. 2013; 341(6150): 1103–1106. 10.1126/science.1241602 [DOI] [PubMed] [Google Scholar]
- 13.Schuetz M, Smith R, Ellis B. Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot. 2013; 64(1): 11–31. 10.1093/jxb/ers287 [DOI] [PubMed] [Google Scholar]
- 14.Ohman D, Demedts B, Kumar M, Gerber L, Gorzsas A, Goeminne G, et al. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 2012; 73(1): 63–76. 10.1111/tpj.12018 [DOI] [PubMed] [Google Scholar]
- 15.Ouyang KX, Liu MQ, Pian RQ, Liu SS, Chen XY. Isolation and analysis of alpha-expansin genes in the tree Anthocephalus chinensis (Rubiaceae). Genet Mol Res. 2013; 12(2): 1061–1073. 10.4238/2013.April.10.2 [DOI] [PubMed] [Google Scholar]
- 16.Dubey A, Nayak S, Goupale DC. Anthocephalus cadamba: A review. Pharmacogn J. 2011; 2(18): 71–76. [Google Scholar]
- 17.Lichtenthaler HK. The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1999; 50: 47–65. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F, Li X, Li Z, Xu X, Peng B, Qin X, et al. UPLC/Q-TOF MS-based metabolomics and qRT-PCR in enzyme gene screening with key role in triterpenoid saponin biosynthesis of Polygala tenuifolia. PLoS One. 2014; 9(8): e105765 10.1371/journal.pone.0105765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lal M, Dutt D, Tyagi CH, Upadhyay JS, Upadhyay S. Characterization of Anthocephalus cadamba and its delignification by kraft pulping. Tappi J. 2010; 9(3): 30–37. [Google Scholar]
- 20.Zhao X, Ouyang K, Gan S, Zeng W, Song L, Zhao S, et al. Biochemical and molecular changes associated with heteroxylan biosynthesis in Neolamarckia cadamba (Rubiaceae) during xylogenesis. Front Plant Sci. 2014; 5: 602 10.3389/fpls.2014.00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011; 29(7): 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijay N, Poelstra JW, Kunstner A, Wolf JB. Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol Ecol. 2013; 22(3): 620–634. 10.1111/mec.12014 [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009; 10(1): 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H, Li J, OuYang K, Zhao X, Li P, Liao B, et al. Direct adventitious shoot organogenesis and plant regeneration from cotyledon explants in Neolamarckia cadamba. Plant Biotechnol. 2014; 31(2): 115–121. [Google Scholar]
- 25.Atanassova R, Favet N, Martz F, Chabbert B, Tollier M, Monties B, et al. Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 1995; 8(4): 465–477. [Google Scholar]
- 26.Ouyang K, Li J, Huang H, Que Q, Li Pei And Chen X. A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. BiotechnolBiotechnol Equip. 2014; 28(6): 1008–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997; 25(17): 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004; 32(Database issue): D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botstein D, Cherry JM, Ashburner M, Ball CA, Blake JA, Butler H, et al. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000; 25(1): 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000; 28(1): 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004; 32(Database issue): D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008; 5(7): 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 33.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010; 11(10): R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010; 38(Database issue): D822–D827. 10.1093/nar/gkp805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki S, Li L, Sun YH, Chiang VL. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006; 142(3): 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert GJ. Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr Opin Plant Biol. 2004; 7(3): 277–284. [DOI] [PubMed] [Google Scholar]
- 37.Pan Q, Wang Q, Yuan F, Xing S, Zhao J, Choi YH, et al. Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-metabolomics. PLoS One. 2012; 7(8): e43038 10.1371/journal.pone.0043038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005; 21(18): 3674–3676. [DOI] [PubMed] [Google Scholar]
- 39.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006; 34(Web Server): W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li D, Deng Z, Qin B, Liu X, Men Z. De novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensisMuell. Arg.). BMC Genomics. 2012; 13: 192 10.1186/1471-2164-13-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng C, Chen M, Xu CJ, Bai L, Yin XR, Li X, et al. Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genomics. 2012; 13: 19 10.1186/1471-2164-13-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003; 20(5): 735–747. [DOI] [PubMed] [Google Scholar]
- 43.Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, et al. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013; 161(3): 1202–1216. 10.1104/pp.112.205385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci. 2008; 65(7–8): 1150–1160. 10.1007/s00018-007-7473-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010; 15(10): 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 46.Chan RL, Gago GM, Palena CM, Gonzalez DH. Homeoboxes in plant development. Biochim Biophys Acta. 1998; 1442(1): 1–19. [DOI] [PubMed] [Google Scholar]
- 47.Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006; 18(11): 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009; 50(11): 1950–1964. 10.1093/pcp/pcp139 [DOI] [PubMed] [Google Scholar]
- 49.Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007; 19(9): 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009; 21(1): 248–266. 10.1105/tpc.108.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. Transcriptional and post transcriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci U S A. 2006; 103(15): 6055–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008; 20(10): 2763–2782. 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko JH, Kim WC, Han KH. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 2009; 60(4): 649–665. 10.1111/j.1365-313X.2009.03989.x [DOI] [PubMed] [Google Scholar]
- 54.Zhong R, Ye ZH. Transcriptional regulation of lignin biosynthesis. Plant Signal Behav. 2009; 4(11): 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, et al. Contribution of NAC transcription factors to plant adaptation to land. Science. 2014; 343(6178): 1505–1508. 10.1126/science.1248417 [DOI] [PubMed] [Google Scholar]
- 56.Lin YC, Li W, Sun YH, Kumari S, Wei H, Li Q, et al. SND1 transcription factor-directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. Plant Cell. 2013; 25(11): 4324–4341. 10.1105/tpc.113.117697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005; 17(11): 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005; 19(16): 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi M, Goue N, Igarashi H, Ohtani M, Nakano Y, Mortimer JC, et al. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010; 153(3): 906–914. 10.1104/pp.110.154013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi M, Kubo M, Fukuda H, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008; 55(4): 652–664. 10.1111/j.1365-313X.2008.03533.x [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011; 66(4): 579–590. 10.1111/j.1365-313X.2011.04514.x [DOI] [PubMed] [Google Scholar]
- 62.Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell. 2010; 22(10): 3461–3473. 10.1105/tpc.110.075036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007; 19(1): 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong R, Lee C, Ye ZH. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant. 2010; 3(6): 1087–1103. 10.1093/mp/ssq062 [DOI] [PubMed] [Google Scholar]
- 65.Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010; 51(1): 144–163. 10.1093/pcp/pcp175 [DOI] [PubMed] [Google Scholar]
- 66.Brown RT, Chappel CL. Anthocephalus alkaloids: Cadamine and isocadamine. Tetrahedron Lett. 1976; 17(19): 1629–1630. [Google Scholar]
- 67.Handa SS, Borris RP, Cordell GA, Phillipson JD. NMR spectral analysis of cadambine from Anthocephalus chinensis. J Nat Prod. 2004; 46 (3): 325–330. [Google Scholar]
- 68.Xu XY, Yang XH, Li SZ, Song QS. Two new triterpenoid glycosides from the leaves of Anthocephalus chinensis. J Asian Nat Prod Res. 2011; 13(11): 1008–1013. 10.1080/10286020.2011.606812 [DOI] [PubMed] [Google Scholar]
- 69.Zhong R, Richardson EA, Ye ZH. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007; 225(6): 1603–1611. [DOI] [PubMed] [Google Scholar]
- 70.Do CT, Pollet B, Thevenin J, Sibout R, Denoue D, Barriere Y, et al. Both caffeoyl Coenzyme a 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007; 226(5): 1117–1129. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J, Elo A, Helariutta Y. Arabidopsis as a model for wood formation. Curr Opin Biotechnol. 2011; 22(2): 293–299. 10.1016/j.copbio.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 72.Carvalho A, Paiva J, Louzada J, Lima-Brito J. The transcriptomics of secondary growth and wood formation in conifers. Mol Biol Int. 2013; 2013: 974324 10.1155/2013/974324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157(6): 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
Error bars on each column indicate SEs from three replicates. R, root; S, middle stem segment; L, young leaf; ML, mature leaf; F, flower; B, bark; C, cambium.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLS)
(XLSX)
(XLS)
(XLSX)
The color red, green, blue and violet indicate UniGenes participating in lignin, cellulose, mannan and heteroxylan biosynthesis process, respectively.
(XLSX)
(XLSX)
(XLSX)
The color red and green indicate the UniGenes from DEGs and UniGenes with the expression profile showing that the expression abundance in the M and B libraries was higher than in the A library in DEGs, respectively.
(XLSX)
The color red and blue indicate the UniGenes identified as DEGs and UniGenes with expression profiles where the expression abundance in the M and B libraries was higher than in the A library in DEGs, respectively.
(XLSX)
(XLSX)
The color green and red indicate the UniGenes from DEGs and UniGenes with an expression profile showing that abundance in the M and B libraries was higher than in the A library in DEGs, respectively.
(XLSX)
(XLSX)
* indicates M as control sample and A as test sample.
(DOCX)
Data Availability Statement
All UniGenes are available from the NCBI Transcriptome Shotgun Assembly Sequence Database (TSA) (accession number) from GASC01000001 to GASC01055370.