Abstract
Aim:
Recent evidence shows that localization of mRNAs and their protein products at cellular protrusions plays a decisive function in the metastasis of cancer cells. The aim of this study was to identify the variety of proteins encoded by protrusion-localized mRNAs and their roles in the metastasis and invasion of liver cancer cells.
Methods:
Highly metastatic hepatocellular carcinoma cell line HCCLM3 and non-metastatic hepatocellular carcinoma cell line SMMC-7721 were examined. Cell protrusions (Ps) were separated from cell bodies (CB) using a Boyden chamber assay; total mRNA population in CB and Ps fractions was analyzed using high-throughput direct RNA sequencing. The localization of STAT3 mRNA and protein at Ps was confirmed using RT-qPCR, RNA FISH, and immunofluorescence assays. Cell migration capacity and invasiveness of HCCLM3 cells were evaluated using MTT, wound healing migration and in vitro invasion assays. The interaction between Stat3 and growth factor receptors was explored with co-immunoprecipitation assays.
Results:
In HCCLM3 cells, 793 mRNAs were identified as being localized in the Ps fraction according to a cut-off value (Ps/CB ratio) >1.6. The Ps-localized mRNAs could be divided into 4 functional groups, and were all closely related to the invasive and metastatic properties. STAT3 mRNA accumulated in the Ps of HCCLM3 cells compared with non-metastatic SMMC-7721 cells. Treatment of HCCLM3 cells with siRNAs against STAT3 mRNA drastically decreased the cell migration and invasion. Moreover, Ps-localized Stat3 was found to interact with pseudopod-enriched platelet-derived growth factor receptor tyrosine kinase (PDGFRTK) in a growth factor-dependent manner.
Conclusion:
This study reveals STAT3 mRNA localization at the Ps of metastatic hepatocellular carcinoma HCCLM3 cells by combining application of genome-wide and gene specific description and functional analysis.
Keywords: hepatocellular carcinoma, metastasis, mRNA localization, STAT3, platelet-derived growth factor receptor, HCCLM3 cells, SMMC-7721 cells, Boyden chamber assay
Introduction
Hepatocellular carcinoma (HCC) is the fourth most common malignant cancer in the world. Approximately 250 000 people worldwide die of HCC each year1,2. More than 60% of patients who suffer from HCC are not diagnosed at an early stage because of the unclear pathogenesis of the disease, a hallmark of HCC. A delayed diagnosis of HCC results in only a 5% survival rate after 5 years3. Most cancer-associated mortality is related to the metastasis of malignant cancer cells. There are several discrete steps involved in the process of cancer metastasis: loss of cellular adhesion and acquisition of the ability to escape from the tumor tissue; increased invasiveness and motility driving malignant cancer cells to migrate to blood vessels; entrance into the blood vessels and survival in the circulation; exit into new tissue and, eventually, growth in the new environment4.
The formation and stabilization of cellular protrusions play a decisive role in cell migration5. The polymerization of actin within lamellipodia and filopodia drives the protrusion of the cell leading edge, a process related to metastatic and invasive properties. Actin filaments of filopodia and lamellipodia are organized into different types of structures at the ventral surface of the cell by specific actin-organizing proteins. The dendritic unbundled filaments in a lamellipodium generate force to drive membrane protrusion and cell motility. The tight actin bundle in a filopodium forms a projection to allow the cell to infiltrate into small gaps6. Proteomic analyses of filopodia and lamellipodia have revealed the extent of the proteome of these cellular protrusions7. However, it is still unclear how proteins are specifically targeted to or maintained in these subcellular domains.
The localization of specific mRNAs has emerged as a novel mechanism behind the targeting and local expression of proteins at cellular protrusions8. Indeed, recent evidence shows that local mRNA translation plays a key role in protein localization to protrusions9. The localization of mRNA relies on either active transport along actin filaments or microtubules or via diffusion and capture of transcripts at specific locations10. Several human cancer cell lines have been studied, using microarrays and proteomic analyses, to identify the specific proteins and transcripts present in the pseudopodia and involved in pseudopod protrusion, tumor cell migration and invasion11. However, a comprehensive analysis of the mRNAs and proteins localized in the protrusions of metastatic HCC cells and their function in the invasion and metastasis of these cells is still lacking12,13,14.
In this study, we combined cellular fractionation with direct RNA sequencing (DRS) to identify and quantify with high sensitivity and low bias, the total mRNA population in the cell body and protrusions of the highly metastatic HCC cell line, HCCLM3, and the non-metastatic HCC cell line, SMMC-772115,16. A Boyden chamber assay was used to separate the cell body fraction from the cellular protrusion fraction and to isolate mRNA molecules enriched in both fractions of these cell lines17. STAT3 mRNA was found to be enriched in pseudopodia, and its role in the process of cell migration and invasiveness was elucidated. In addition, we found that the protrusion-localized protein Stat3 interacts with the platelet-derived growth factor receptor (PDGFR), a cell surface tyrosine kinase. These results contribute to a broader understanding of the mechanisms and signaling regulations involved in pseudopodia formation and cancer cell metastasis.
Materials and methods
Boyden chamber assay for cell protrusion isolation and direct RNA sequencing
Isolation of cellular protrusions was performed according to previously published methods with slight modifications17,18. We used 9.6-cm2 (six-well format) cell culture inserts (Millipore, MA, USA) with a 1-μm pore size, uncoated polystyrene membrane for the Boyden chamber assay. The membranes were coated with a 10 μg/mL final concentration of collagen type-I (Sigma, St Louis, MO, USA) and incubated at 37 °C for 2 h. The membranes were then placed into a six-well cell culture plate without collagen type-I and incubated overnight in a 5% CO2 humidified incubator. Afterwards, we transferred the membranes to another six-well tissue culture dish containing serum free DMEM (Corning, NY, USA). Cells were serum starved overnight and detached using 0.5% trypsin-EDTA, and the trypsin was inactivated by suspending cells in DMEM containing 10% FCS. Cells were pelleted and re-suspended in serum free medium, and 1–2×106 cells were transferred to one membrane insert. Cell protrusions were allowed to grow through the membrane for 24–30 h before protein and RNA purification.
To isolate cell bodies and cell protrusions, the cell culture insert membranes were washed once with PBS, and then the cell body and cell protrusions were harvested using a sharp cell scraper (Costar, NY, USA). First, we thoroughly washed the lower side of the membrane with 1 mL TRI-reagent (Sigma, St Louis, MO, USA), and then used a cell scraper to scrape off the cell protrusion. The cell scraper was used only once on the membrane to avoid contaminating the protrusion fraction of lysed cells. Cells on the upper side of the membrane were subsequently scraped and collected with TRI-reagent. Approximately 1 μg of glycogen was used to facilitate the precipitation of RNA. Single RNA molecules were sequenced by direct RNA sequencing (DRS) using the HiSeq 2000 platform. DRS was performed once with pools of RNA representing the protrusion fraction (PF) or cell body fraction (CF) from four independent chamber insert membranes. The raw bioinformatics data analysis and sequence alignment was completed using the HiSeq 2000 platform.
Co-immunoprecipitation and Western blot analysis
Cells were lysed in RIPA buffer containing: 50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L PMSF, 1 mmol/L EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Sigma, St Louis, MO, USA). The supernatant was used for immunoprecipitation and Western blot assays. For the immunoprecipitation of phosphorylated Stat3/Stat3 with vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR), nerve growth factor receptor (NGFR) and hepatocyte growth factor receptor (HGFR), 10 μg of the specific antibody was added to 500 μL of supernatant and incubated at 4 °C with agitation for 1 h after which 50 μL of protein A-Sepharose beads was added, and the incubation at 4 °C was continued for 2 h. The beads were then washed four times for 3 min at 4 °C with a wash buffer (25 mmol/L HEPES-KOH, pH 7.5, 150 mmol/L KCl, and 2 mmol/L MgCl2). The same protocol was performed for reverse Co-IP experiments. The protein was eluted from the beads with 200 μL of 50 mmol/L Tris-HCl, pH 8.0, 100 mmol/L NaCl, 10 mmol/L EDTA, and 1% SDS by incubating for 10 min at 65 °C. The precipitated protein was then analyzed using a Western blot assay.
For Western blot assays, cells were lysed in RIPA buffer containing: 50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L PMSF, 1 mmol/L EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Sigma, St Louis, MO, USA). Then, 50 μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in an 8% gradient polyacrylamide gel (Bio-Rad). Proteins were transferred to a nitrocellulose membrane, probed with specific antibodies and detected by incubation with enhanced chemiluminescence (ECL) substrate followed by exposure of the membrane to autoradiography film. β-Actin was used as loading control. The following antibodies and dilutions were used: mouse anti-actin (Beyotime, Haimen, China), rabbit anti-STAT3 (Abcam, Cambridge, UK), rabbit anti-TP53 (Abcam, Cambridge, UK), rabbit anti-EEF2 (Abcam, Cambridge, UK), mouse anti-PDGFR beta (Abcam, Cambridge, UK), mouse anti-VEGFR1+VEGFR2 (Abcam, Cambridge, UK), mouse anti-VEGFR2 (Abcam, Cambridge, UK), rabbit anti-Met (c-Met) (Abcam, Cambridge, UK), mouse anti-p75 NGFR (Abcam, Cambridge, UK), rabbit anti-TrkA (Abcam, Cambridge, UK), and rabbit anti-histone H3 antibody (Beyotime, Haimen, China). The secondary antibodies used were: HRP-labeled goat anti-rabbit IgG (H+L) 1:1000 (Beyotime, Haimen, China) and HRP-labeled goat anti-mouse IgG (H+L) 1:1000 (Beyotime, Haimen, China).
Fluorescence in situ hybridization and immunofluorescence
Cells were processed for fluorescence in situ hybridization (FISH) and immunofluorescence according to the protocols described in a previous paper19. For in situ hybridization, cells were hybridized with a pool of FAM-conjugated STAT3 DNA oligonucleotide probes. For immunofluorescence, a 1:50 dilution of a mouse anti-Stat3 antibody (Oncogene Science, Cambridge, MA, USA) was used as a primary antibody. For the secondary antibody, a 1:1000 dilution of an anti-mouse Cy3-conjugated antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) was used. In addition, the following primary and secondary antibodies were also used for immunofluorescence: mouse anti-tubulin 1:500 (Beyotime, Haimen, China); secondary antibody Alexa Fluor 488-labeled goat anti-mouse IgG (H+L) 1:500 (Beyotime, Haimen, China), Alexa Fluor 555-labeled donkey anti-mouse IgG (H+L) 1:500 (Beyotime, Haimen, China). All immunofluorescence images were taken with a resolution ratio of 100 μm and 0.2-s exposure time using a CX41-32RFL fluorescence microscope (Olympus, Japan).
Statistical analysis
All experiments were carried out in triplicate unless otherwise stated in the Results section. Data are expressed as the mean±standard deviation (SD) of three independent experiments and were analyzed with SPSS software using Student's t test with significance defined as P<0.05.
Results
Isolation of mRNA from the cell bodies and protrusions of HCCLM3 cells
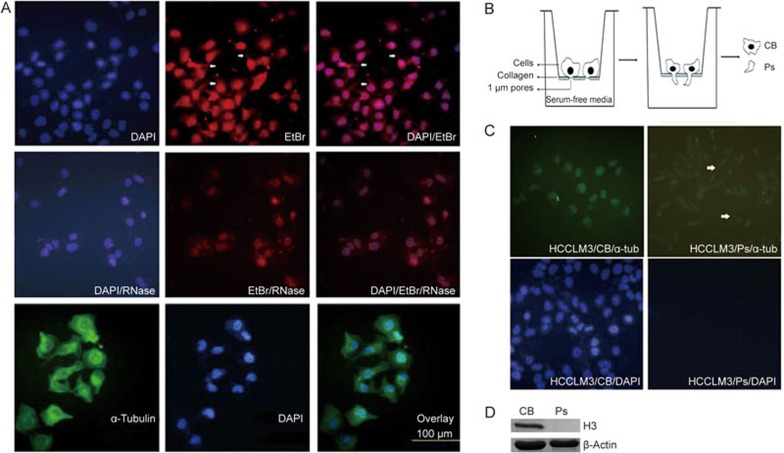
To identify and characterize the transcripts localized at the protrusions of metastatic HCC cells, we used the human hepatocellular carcinoma cell line HCCLM3, a well-characterized HCC cell line with high metastatic properties20. The total RNA and DNA from HCCLM3 cells grown on coverslips was visualized as cytoplasmic and nuclear EtBr staining (Figure 1A, upper panel). As expected, most cytoplasmic RNA signal vanished after RNase-treatment (Figure 1A, middle panel). The omnipresent cytoplasmic protein α-tubulin was used as a cytoplasmic staining control in the fluorescence assay. To isolate RNA from cell bodies and protrusions, a slightly modified Boyden chamber assay was used17. Because the average diameter of the cell body is approximately 10–20 μm and the size of the cell protrusion is less than 1 μm, the cell protrusion was separated by a Boyden chamber with a microporous membrane, through which only the thin cell protrusion could migrate (Figure 1B). The nuclei of the cell bodies were stained with DAPI to validate the migration and presence of cell protrusions (Ps) through the porous membrane of the Boyden chamber and the absence of migration of the cell bodies (CB) on the lower-side of the porous membrane (Figure 1C). Figure 1C shows that both the pseudopod and cell body fractions were stained for α-tubulin (green) but only the cell body fraction was stained for nuclei (blue). Western blotting confirmed that the nuclear marker histone H3 was absent from the protrusion fraction (Ps) (Figure 1D). These results show that we successfully isolated the cell protrusion fraction from the cell body fraction using a modified Boyden chamber assay.
Figure 1.
Trans-migration of HCCLM3 protrusions in a modified Boyden chamber assay. (A) Immunofluorescence image showing the typical morphology of human hepatic cancer HCCLM3 cells. The cellular cytoplasm was stained for α-tubulin (α-tub, green) and nuclei by DAPI (blue) (bottom). The cellular cytoplasm was stained with ethidium bromide (EtBr), with (+) or without (−) RNase-treatment (top and middle). Scale bar: 100 μm. (B) Schematic drawing showing the principle of the Boyden chamber set-up for the isolation and purification of migrating protrusions. Cells were grown on a 1-μm pore size microporous membrane, allowing trans-migration of only the cell protrusions. The lower side of the membrane is coated with collagen I protein. (C) Protrusions of HCCLM3 cells migrated through a 1-μm microporous membrane after 24 h assay time. α-Tubulin (green) and DAPI staining (blue) is shown without and after wipe (wipe) to remove cellular material present on the upper side of membranes. Arrowheads indicate protrusions present on the lower side of the membrane. (D) Histone H3 is absent from the Boyden chamber lower side protrusion fraction. Western blotting was performed with histone H3 antibody on cell body (CB) fraction and protrusions (Ps) protein fractions originating from pooled material from three independent Boyden chambers.
Direct RNA sequencing (DRS) of total RNA from the Ps and CB fractions of HCCLM3 cells
After purification, total RNA from both the Ps and CB fractions was submitted to high-throughput direct RNA sequencing (DRS). We were able to map 11 807 299 reads from the Ps mRNA fraction and 12 355 884 reads from the CB mRNA fraction. The relative similarity in the number of mapped reads suggests a comparable amount of starting RNA material used for DRS. The number of reads for each mRNA identified by DRS was normalized to represent the number of mRNA Reads Per Kb per Million reads (RPKM)21. The calculated RPKM values thereby represent the relative abundance of a given mRNA in the total number of mapped sequences. We selected mRNAs with RPKM values larger than 5 in both Ps and CB for downstream analysis, to eliminate insignificant results in the calculation of ratios22. The total number of mRNAs with RPKM values larger than 5 in both Ps and CB was 7591 (Supplementary Table S1). The relative mRNA localization in Ps compared to CB was calculated using the ratio Ps/CB. While the experimental conditions and the cell morphology may affect the absolute value of the mRNA localization ratio, the ratio calculated by this formula is relatively independent on these variables.
We previously established that a Ps/CB ratio larger than 1.6 represents a protrusion-enriched localized mRNA. We used this cut-off value to select the enriched localized mRNA and arranged them in descending order. There were 793 mRNAs with a localization ration >1.6 that were identified as being localized in the Ps fraction (Supplementary Table S2). The top 50 mRNAs localized at protrusions are shown in Table 1. RPKM represents the expression value of the gene, indicating the relative value of specific localized mRNA from the CB and Ps fractions. There was enough total mRNA from the Ps fraction to acquire a specific mRNA with a localization ratio >1.6 because the localization ratio reflects the relative localization in protrusions compared with cell bodies, not the absolute quantity for a given mRNA in the protrusions.
Table 1. HCCLM3-protrusion localized RNA.
| Gene ID | Name | HB-RPKM | HP-RPKM | Ratio |
|---|---|---|---|---|
| 001382 | CRABP2 | 30.55 | 76.80 | 2.513 |
| 008237 | USP11 | 8.71 | 21.27 | 2.442 |
| 003508 | JUP | 29.10 | 68.21 | 2.344 |
| 010381 | T UBB 3 | 47.43 | 106.90 | 2.254 |
| 009022 | CLIC3 | 8.59 | 19.28 | 2.244 |
| 051545 | ZNF 581 | 15.23 | 34.04 | 2.235 |
| 007001 | P RDX 2 | 97.80 | 216.74 | 2.216 |
| 005499 | PPP1CA | 61.79 | 136.84 | 2.215 |
| 010226 | PLIN 3 | 48.06 | 106.00 | 2.206 |
| 008175 | SF3A2 | 19.95 | 43.91 | 2.201 |
| 030851 | TAX1BP 3 | 5.32 | 11.71 | 2.201 |
| 000226 | ALDOA | 884.37 | 1933.11 | 2.186 |
| 009572 | NR1D 1 | 5.81 | 12.69 | 2.184 |
| 030846 | EHD 2 | 38.90 | 84.53 | 2.173 |
| 009149 | DYRK 1B | 7.28 | 15.80 | 2.171 |
| 005296 | PIK3R2 | 7.27 | 15.68 | 2.157 |
| 054345 | SOX 18 | 18.04 | 38.87 | 2.155 |
| 006237 | RRAS | 27.81 | 59.43 | 2.137 |
| 007917 | BAG6 | 32.20 | 68.65 | 2.132 |
| 004007 | PRICKLE3 | 11.10 | 23.63 | 2.129 |
| 003217 | HOXB 7 | 5.97 | 12.61 | 2.112 |
| 023144 | ZC3H3 | 10.27 | 21.65 | 2.107 |
| 007157 | TP 53 | 8.65 | 18.18 | 2.102 |
| 009524 | TECR | 39.92 | 83.39 | 2.089 |
| 006774 | STAT 3 | 23.05 | 47.91 | 2.079 |
| 000832 | CAPZB | 29.28 | 60.83 | 2.077 |
| 001819 | DRG2 | 14.40 | 29.90 | 2.076 |
| 000618 | BCYRN 1 | 27.05 | 55.91 | 2.067 |
| 006813 | STXBP2 | 18.29 | 37.80 | 2.067 |
| 029108 | PYCARD | 33.87 | 69.96 | 2.066 |
| 009114 | ATP6V0D1 | 30.18 | 62.34 | 2.066 |
| 002879 | GPX4 | 229.43 | 472.63 | 2.060 |
| 064122 | FN3K | 6.37 | 13.10 | 2.056 |
| 057146 | TMEM159 | 18.59 | 38.13 | 2.051 |
| 001938 | EEF2 | 353.43 | 723.94 | 2.048 |
| 055690 | PACS 1 | 12.12 | 24.81 | 2.047 |
| 401115 | C4orf48 | 10.50 | 21.50 | 2.047 |
| 002316 | FLNA | 151.19 | 309.33 | 2.046 |
| 010318 | TNIP1 | 34.23 | 70.02 | 2.045 |
| 004782 | NFIC | 7.78 | 15.89 | 2.043 |
| 140606 | SELM | 35.84 | 73.08 | 2.039 |
| 004784 | NFIX | 5.54 | 11.30 | 2.039 |
| 064763 | ZNF574 | 7.83 | 15.95 | 2.037 |
| 003142 | HLX | 8.84 | 17.99 | 2.036 |
| 007040 | TGFB1 | 55.71 | 113.39 | 2.035 |
| 150274 | HSCB | 5.30 | 10.74 | 2.027 |
| 003136 | HLA-H | 7.72 | 15.63 | 2.025 |
| 079877 | DCAKD | 7.29 | 14.74 | 2.022 |
| 000071 | ACTG1 | 708.54 | 1426.47 | 2.013 |
| 029995 | LMCD 1 | 13.94 | 28.06 | 2.012 |
To identify the protrusion-localized mRNAs that are most relevant to the metastasis of HCCLM3 cells, cellular fractionation with the Boyden chamber and DRS was repeated with the non-metastatic HCC cell line, SMMC-772120. Using the same cut-off value and localization ratio as described above, we found that the population of protrusion-localized mRNAs in SMMC-7721 cells is very different from HCCLM3 cells, as shown by the top 50 protrusion-localized mRNAs described in Supplementary Table S3. These results suggest that a specific population of transcripts are enriched in the protrusions of highly metastatic hepatocellular carcinoma cells.
Analysis of HCCLM3 cell protrusion-localized mRNAs
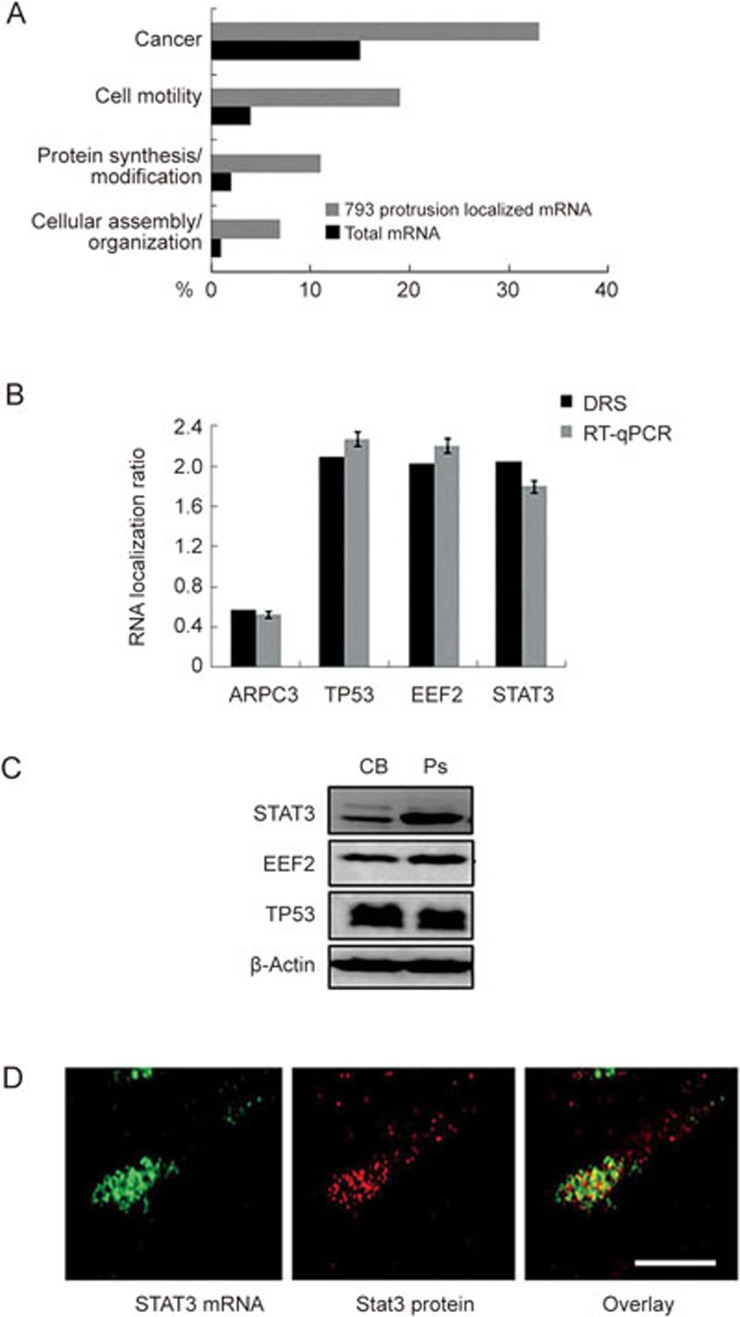
The localized mRNAs in the protrusions of HCCLM3 cells can be divided into four groups according to their major functions: cancer, cell motility, protein synthesis/modification and cellular assembly/organization (Figure 2A). Next, we validated the mRNA localization results for three tumor-related genes: TP53, STAT3 and EEF2 from the DRS assay using quantitative RT-PCR (RT-qPCR). Following normalization with ARPC3 mRNA, which is present in both protrusions and cell body, we obtained concordant results for TP53, STAT3 and EEF2 mRNA localization by the two methods (Figure 2B). Western blot analysis using CB and Ps protein extracts normalized for β-actin showed that, in the protrusions, Stat3 levels were much higher than in the cell body (Figure 2C). However, TP53 and EEF2 did not show any difference in expression between the two fractions (Figure 2C). Stat3 is a member of the STAT protein family, which is involved in cell invasion and metastasis in cancer23. To investigate the distribution of STAT3 mRNA and its production in cell protrusions, we fixed the cells and performed FISH and immunofluorescence (IF) on STAT3 mRNA and proteins in these protrusions. Our results confirmed both the localization of STAT3 mRNA and accumulation of Stat3 protein in cell protrusions (Figure 2D, cell bodies have very weak signal, data not shown).
Figure 2.
Transcriptome analysis of purified mRNA from HCCLM3 protrusions. (A) Annotation analysis of the 793 HCCLM3 protrusion localized mRNAs compared to the total cellular mRNA detected at ≥5 RPKM. Annotations were made using the IPA Ingenuity platform. (B) RT-qPCR confirmation analysis of selected HCCLM3 protrusion localized RNA identified by DRS. RNA localization ratios are normalized to ARPC3 mRNA. (C) Western blot analysis of putative protrusion-localized proteins. Western blotting was performed on pooled material from three independent Boyden chamber experiments representing cell body fraction (CB) and protrusions (Ps). Subsequent Western blot analyses were performed with antibodies against Stat3, EEF2 and TP53. β-Actin was used as loading control. (D) Fluorescent in situ hybridization on STAT3 mRNA (left panel) and immunofluorescence (IF) on Stat3 protein (middle panel) in HCCLM3 cells. Scale bar: 2 μm.
Knockdown of Stat3 decreases the metastatic and invasive capacity of HCCLM3 cells
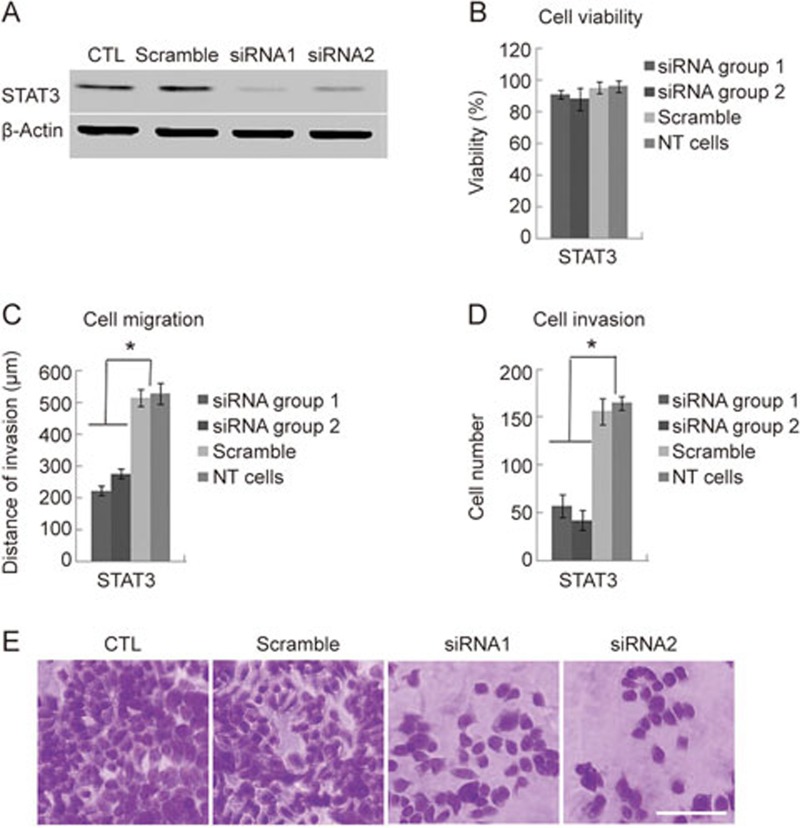
Signal transducer and activator of transcription 3 (STAT3) activation has been linked to the EMT program in hepatocellular carcinoma24, and we found that STAT3 mRNA is localized to the protrusions of HCCLM3 cells. To explore the role of protrusion-localized mRNA in tumor cell invasiveness, siRNA sequences targeting knockdown of STAT3 were used. HCCLM3 cells were incubated with control or with target-specific siRNA for 48 h. As anticipated, there was more than 80% knockdown in the protein level for Stat3 after transfection compared with the non-silencing control siRNA (Figure 3A). Following the confirmation of knockdown, the MTT assay was used to test the proliferative capacity of the cells. We found no significant differences in proliferation ability between HCCLM3 cells transfected with target-specific siRNAs and control cells transfected with scrambled siRNA (Figure 3B). Next, we used the wound healing migration assay to compare the migration capacity of scrambled siRNA-transfected cells with target-specific siRNA-transfected cells. A higher level of cell migration was observed in the control group compared to the STAT3-depleted group (Figure 3C). After STAT3 depletion, the invasive ability of the cells was measured by a Transwell Matrigel invasion assay. After 24 h, STAT3-depleted cells were significantly less invasive compared with control cells (Figure 3D and 3E). Altogether, these results show that the knockdown of STAT3 decreases the motility and invasive capacity of HCCLM3 cells.
Figure 3.
Stat3 protein knockdown affects migration and invasion of metastatic HCCLM3 cells. (A) Western blot against Stat3 protein from HCCLM3 cancer cells transfected with scrambled or specific siRNAs against STAT3 mRNA. (B) The effect of specific pseudopod-enriched Stat3 protein knockdown on cell viability was evaluated using the MTT assay. NT cells: non transformed cells. (C) Cell migration was assessed using the wound healing scratch assay. The average migration distances at 48 h from triplicate experiments are shown. NT cells: non transformed cells. (D) The invasiveness of HCCLM3 cells was assessed using a matrigel invasion approach. The number of transwelled cells was counted and expressed as mean±SD from triplicate experiments. NT cells: non transformed cells. (E) Transwelled cells stained with crystal violet. Invasive cells were observed under a ×40 objective lens and the number of cells in five random fields were counted and an average calculated. Scale bar: 100 μm.
Protrusion-localized Stat3 interacts with platelet-derived growth factor receptor tyrosine kinase
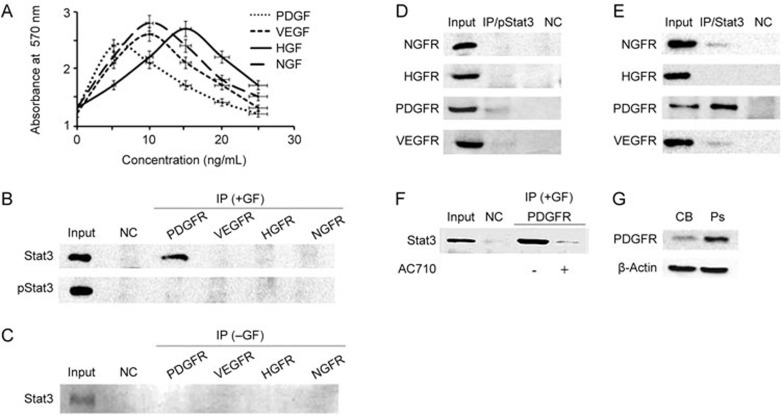
Previous studies have shown that Stat3 can be directly phosphorylated by receptor protein tyrosine kinases (RPTKs) located at the cell membrane after extracellular signal stimulation by cell growth factors25. To better understand the role of protrusion-localized Stat3, we explored a possible interaction between Stat3 and several hepatic-related growth factor receptors: vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR), hepatocyte growth factor receptor (HGFR) and the non-hepatic related nerve growth factor receptor (NGFR). Cells were first treated with growth factors at their optimal concentration: PDGF=5 ng/mL, VEGF=10 ng/mL, NGF=10 ng/mL, and HGF=15 ng/mL (Figure 4A). Following cell extraction, co-immunoprecipitation (co-IP) was performed to detect an interaction between either Stat3 or phospho-Stat3 and the various growth factor receptors. We found no interaction between phosphorylated Stat3 and PDGFR, VEGFR, HGFR or NGFR (Figure 4B). However, non-phosphorylated Stat3 interacted with PDGFR (Figure 4B), and the interaction was growth factor-dependent (Figure 4C). Using a reverse co-IP experiment, we further confirmed that non-phosphorylated Stat3 interacts with PDGFR (Figure 4D and 4E). Interestingly, upon treatment with the PGDFR inhibitor, AC710, this interaction was reduced (Figure 4F). Meanwhile, Western blot analysis showed that PDGFR was present at higher levels in the protrusion fraction than in the cell body (Figure 4G). Altogether, these results suggest that protrusion-localized Stat3 interacts with PDGFR, and this interaction depends on the activation of PDGFR.
Figure 4.
Protrusion-localized Stat3 interacts with platelet-derived growth factor receptor tyrosine kinase, PDGFRTK. (A) HCCLM3 cells were treated with 0, 5, 10, 20 and 25 ng/mL VEGF, PDGF, NGF and HGF for 24 h, respectively, and cell viability was measured with an MTT assay (n=4). (B) Immunoprecipitation of receptor tyrosine kinases (RTK). Vascular endothelial growth factor receptor (VEGFR); Platelet-derived growth factor receptor (PDGFR); Nerve growth factor receptor receptor (NGFR) and Hepatocyte growth factor receptor (HGFR), followed by detection of Stat3 or phospho-Stat3 (pStat3) by Western blot. Input: Stat3 from total HCCLM3 cell extract; NC: IgA beads without antibody; IP: immunoprecipitated growth factor receptors from cells treated with each growth factor, Western blot for Stat3/pStat3; GF: growth factor. (C) Stat3 does not co-immunoprecipitates with PDGFR in the absence of growth factor. Input: Stat3 from total HCCLM3 cell extract; NC: IgA beads without antibody; IP: immunoprecipitated growth factor receptors, Western blot for Stat3; GF: growth factor. (D) Immunoprecipitation of phospho-Stat3 followed by Western blot detection of VEGFR, PDGFR, NGFR or HGFR. IP: phospho-Stat3, Input: VEGFR, PDGFR, NGFR and HGFR from total HCCLM3 cell extract; NC: IgA beads without antibody. (E) Immunoprecipitation of Stat3 followed by Western blot detection of VEGFR, PDGFR, NGFR or HGFR. IP: phospho-Stat3, Input: VEGFR, PDGFR, NGFR and HGFR from total HCCLM3 cell extract; NC: IgA beads without antibody. (F) The interaction between Stat3 and PDGFR is reduces in the presence of the PDGFR-family kinase inhibitor AC710. Input: Stat3 from total HCCLM3 cell extract; NC: IgA beads without antibody. (G) Western blot analysis of PDGFR in cell body (CB) or protrusions (Ps) fractions from HCCLM3 cells. Western blotting was performed on pooled material from three independent Boyden chamber experiments representing cell body fraction (CB) and protrusions (Ps). β-Actin was used as loading control.
Discussion
Only a few studies have addressed the issue of mRNA localization in protein sorting at the cellular protrusions of astrocytes, neurons, fibroblasts, and cancer cells16,17,18. The lack of efficient large scale methods of protein and RNA collection has been the main problem. Here, we used a Boyden chamber assay to isolate and collect total RNA from the cell body and protrusions of HCCLM3 and SMMC-2271 cells and characterized the transcriptome of these fractions with high sensitivity and low bias using direct RNA sequencing. To validate the Boyden chamber assay, total cytoplasmic fluorescent staining was used to confirm the migration of the cell protrusion, and DAPI-stained nuclei were used to confirm that the cell bodies remained on the upper-side of the insert membrane. Our results suggest that protrusions can be purified from the Boyden chamber assay without contamination from trans-migrated cell bodies. Western blot analysis of the Ps and CB fractions for histone H3 confirmed that there was no considerable contamination of cell bodies in the Ps fraction. This was a crucial assessment of the quality of the Boyden chamber isolation assay because even a small migration of cell bodies would introduce a severe contamination of cell body material to the protrusion fraction.
The direct RNA sequencing method was used to detect the RNA in the HCCLM3 Ps and CB fractions. Quantitative RT-qPCR on the mRNA of three tumor-related genes was used to validate the results of DRS. The RT-qPCR results were concordant with those obtained from DRS, which confirms that protrusion-localized RNA on a genome wide scale can be found using the Boyden chamber assay and DRS. Most mRNAs localized in the cell protrusions of HCCLM3 were related to functional pathways such as cancer and cell motility. Interestingly, a similar analysis performed on the non-metastatic cell line SMMC-7721 revealed a lack of overlap in the population of protrusion-localized mRNAs between metastatic HCCLM3 cells and non-metastatic SMMC-7721 cells.
The majority of pseudopod-enriched mRNAs are involved in cell growth, differentiation, adhesion and migration. These biological processes are closely associated with cancer progression, especially cell migration capacity. The mRNA for Stat3 was among the identified mRNAs from DRS analysis. We confirmed the localization of STAT3 mRNA in cell protrusions using RT-qPCR and RNA FISH analysis, and the localization of Stat3 protein in cell protrusions using immunofluorescence and the Western blot assay. To study the functional role of localized mRNA in tumor cellular invasiveness, an RNA interference approach targeting STAT3 was used. The MTT assay showed almost no difference in proliferation ability between target-specific siRNA transfected cells and control group. However, this knockdown clearly affected the invasive and metastatic ability of these cells. The results of these assays indicate that STAT3 mRNA is not a critical factor for the proliferation of HCCLM3 cells but, rather, plays a more significant role in cell motility.
Stat3, which is expressed in most tissues, plays a dual molecular role: signal transducer and transcription activator. Studies have shown that Stat3 is upregulated in many cancers, including glioblastoma, head and neck cancer, prostate cancer, and breast cancer26. Phosphorylation of Stat3 at specific residues (Tyr705 or Ser727) in response to ligand stimulation determines its molecular role. Tyr705 phosphorylation promotes Stat3 homodimerization, resulting in its nuclear translocation and binding to DNA. Ser727 phosphorylation of Stat3 takes place at the C-terminal transactivation domain and allows for maximal activation of transcription of Stat3's target genes27. Our results suggest that protrusion-localized Stat3 interacts with PDGFR in a growth factor-dependent manner. Western blot analysis shows that PDGFR is more abundant in the Ps fraction than in the CB fraction. High expression of platelet-derived growth factor receptor-α (PDGFR α), which plays many critical roles in embryonic and postnatal development, was found in early liver development28. Moreover, overproduction of PDGF may be involved in the growth stimulation of human tumors29. From these data we propose that the localization of STAT3 mRNA at a cellular protrusion promotes the local synthesis and accumulation of Stat3 protein, where it can interact with PDGFR located at the cell membrane. Following the stimulation of PDGFR, the tyrosine kinase activity of this receptor could phosphorylate Stat3 and promote its dimerization, resulting in its nuclear localization and activation of its target genes30 (Figure 5).
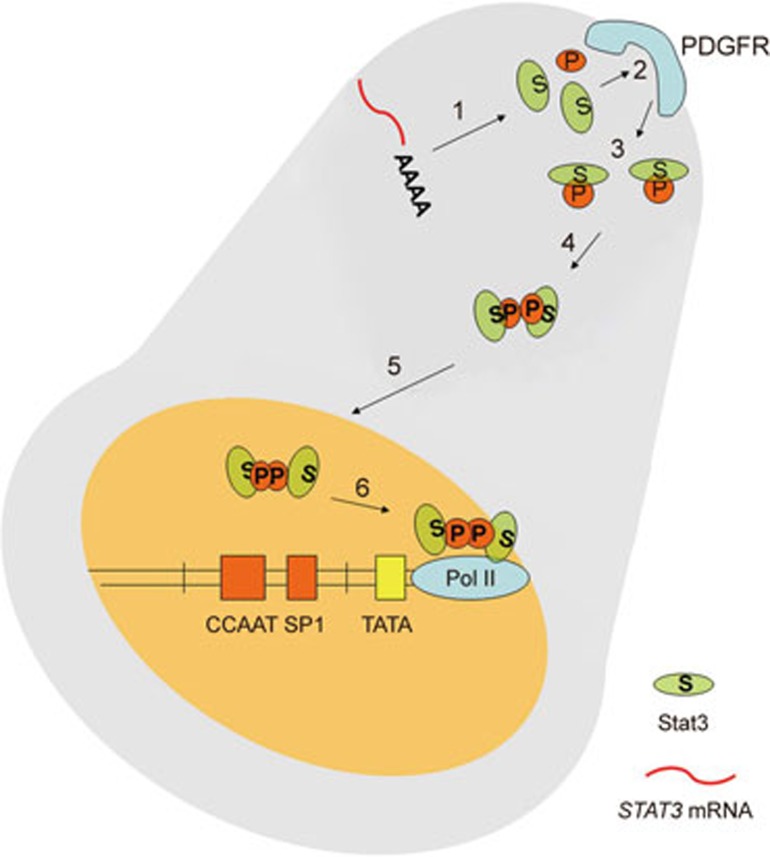
Figure 5.
Model for the local synthesis and local phosphorylation of Stat3 in cellular protrusions. 1) Protrusion-localized STAT3 mRNA is translated into Stat3 protein; 2) After PDGF stimulation, Stat3 interacts with PDGFR located at the membrane of protrusions; 3) This results in the phosphorylation of Stat3; 4) Phosphorylated Stat3 can form dimers; 5) Phospho-Stat3 dimer is imported in the nucleus; 6) Phospho-Stat3 dimers activate the transcription of their target genes.
In summary, our study reports a genome-wide identification of asymmetrically distributed mRNAs in HCCLM3 cell protrusions. The protein product of the protrusion-localized STAT3 mRNA interacts with PDGFR tyrosine kinase, which is highly enriched in the protrusion fraction. We propose that the local accumulation of Stat3 in cellular protrusions plays an important role in the invasive and metastatic ability of HCC cells.
Author contribution
Zhi-fa SHEN, Yan-hong LIU, Yu-zhe WANG, Pascal CHARTRAND and Yu-yang JIANG designed the experiments; Yan-hong LIU, Jia-lu JIN, Yu-zhe WANG, Ying TAN, Ying-ying ZHOU, Ting PENG, Feng LI, and Wan-dong LIANG performed the experiments; Yan-hong LIU, Jia-lu JIN, and Yu-zhe WANG analyzed the data; Zhi-fa SHEN, Pascal CHARTRAND and Yan-hong LIU wrote the manuscript.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (21302108), Shenzhen Municipal Government (JCYJ20140827150509058, 20150113A0410006, JSGG20141016150327538, and SZSTI CXB201104210013A) and Zhejiang Province Natural Science Foundation of China (LY16C070002).
Footnotes
Supplementary information is available at the Acta Pharmacologica Sinica's website.
Supplementary Information
Supplementary methods
The total number of mRNAs with RPKM values larger than 5 in both Ps and CB.
793 mRNAs identified to be localized in the cell protrusions.
Top 50 protrusion-localized mRNAs in SMMC-7721 cells.
References
- Yuen MF, Lai CL. Serological markers of liver cancer. Best Pract Res Clin Gastroenterol 2005; 19: 91–9. [DOI] [PubMed] [Google Scholar]
- Kew MC. Hepatocellular cancer. A century of progress. Clin Liver Dis 2000; 4: 257–68. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Mason AC, Key C. Trends in survival of patients with hepatocellular carcinoma between 1977 and 1996 in the United States. Hepatology 2001; 33: 62–5. [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002; 2: 563–72. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science 2003; 302: 1704–9. [DOI] [PubMed] [Google Scholar]
- Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol 2010; 189: 541–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding SJ, Wang W, Jacobs JM, Qian WJ, Moore RJ, et al. Profiling signaling polarityin chemotactic cells. Proc Natl Acad Sci U S A 2007; 104: 8328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Mingle L, Van De Water L, Liu G. Control of cell migration through mRNA localization and local translation. Wiley Interdiscip Rev RNA 2015; 6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardarheh FK, Paul A, Kumper S, Sadok A, Paterson H, Mccarthy A, et al. Global analysis of mRNA, translation, and protein localization: Local translation is a key regulator of cell protrusions. Dev Cell 2015; 35: 344–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Macara IG. RNA localization and polarity: from A(PC) to Z(BP). Trends Cell Biol 2009; 19: 156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res 2010; 70: 3780–90. [DOI] [PubMed] [Google Scholar]
- Jia Z, Barbier L, Stuart H, Amraei M, Pelech S, Dennis JW, et al. Tumor cell pseudopodial protrusions. Localized signaling domains coordinating cytoskeleton remodeling, cell adhesion, glycolysis, RNA translocation, and protein translation. J Biol Chem 2005; 280: 30564–73. [DOI] [PubMed] [Google Scholar]
- Beckner ME, Chen X, An J, Day BW, Pollack IF. Proteomic characterization of harvested pseudopodia with differential gel electrophoresis and specific antibodies. Lab Invest 2005; 85: 316–27. [DOI] [PubMed] [Google Scholar]
- Stuart HC, Jia Z, Messenberg A, Joshi B, Underhill TM, Moukhles H, et al. Localized Rho GTPase activation regulates RNA dynamics and compartmentalization in tumor cell protrusions. J Biol Chem 2008; 283: 34785–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Goren A, Gymrek M, Guttman M, Regev A, Bernstein BE, et al. Digital transcriptome profiling from attomole-level RNA samples. Genome Res 2010; 20: 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, et al. Direct RNA sequencing. Nature 2009; 461: 814–8. [DOI] [PubMed] [Google Scholar]
- Thomsen R, Lade Nielsen A. A Boyden chamber-based method for characterization of astrocyte protrusion localized RNA and protein. Glia 2011; 59: 1782–92. [DOI] [PubMed] [Google Scholar]
- Smit M, Leng J, Klemke RL. Assay for neurite outgrowth quantification. Biotechniques 2003; 35: 254–6. [DOI] [PubMed] [Google Scholar]
- Chartrand P, Singer RH, Long RM. Sensitive and highresolution detection of RNA in situ. Methods Enzymol 2000; 318: 493–506. [DOI] [PubMed] [Google Scholar]
- Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J, et al. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J Cancer Res Clin Oncol 2003; 129: 43–51. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–8. [DOI] [PubMed] [Google Scholar]
- Jakobsen KR, Sørensen E, Brøndum KK, Daugaard TF, Thomsen R, Nielsen AL. Direct RNA sequencing mediated identification of mRNA localized in protrusions of human MDA-MB-231 metastatic breast cancer cells. J Mol Signal 2013; 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Ross JL, Cowell JK. The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT 2014; 3: e 28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CH, Guo FL, Xu GL, Jia WD, Ge YS. STAT3 activation mediates epithelial-to-mesenchymal transition in human hepatocellular carcinoma cells. Hepatogastroenterology 2014; 61: 1082–9. [PubMed] [Google Scholar]
- Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene 2000; 19: 6613–26. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. STAT3 as an oncogene. Cell 1999; 98: 295–303. [DOI] [PubMed] [Google Scholar]
- Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets 2004; 8: 409–22. [DOI] [PubMed] [Google Scholar]
- Betsholtz C. Insight into the physiological functions of PDGF through genetic studies in mice. Cytokine Growth Factor Rev 2004; 15: 215–28. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev 1999; 79: 1283–316. [DOI] [PubMed] [Google Scholar]
- Vignais ML, Sadowski HB, Watling D, Rogers NC, Gliman M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol Cell Biol 1996; 16: 1759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods
The total number of mRNAs with RPKM values larger than 5 in both Ps and CB.
793 mRNAs identified to be localized in the cell protrusions.
Top 50 protrusion-localized mRNAs in SMMC-7721 cells.