Abstract
Intercommunication between cancer cells and with their surrounding and distant environments is key to the survival, progression and metastasis of the tumour. Exosomes play a role in this communication process. MicroRNA (miRNA) expression is frequently dysregulated in tumour cells and can be reflected by distinct exosomal miRNA (ex-miRNA) profiles isolated from the bodily fluids of cancer patients. Here, the potential of ex-miRNA as a cancer biomarker and therapeutic target is critically analysed. Exosomes are a stable source of miRNA in bodily fluids but, despite a number of methods for exosome extraction and miRNA quantification, their suitability for diagnostics in a clinical setting is questionable. Furthermore, exosomally transferred miRNAs can alter the behaviour of recipient tumour and stromal cells to promote oncogenesis, highlighting a role in cell communication in cancer. However, our incomplete understanding of exosome biogenesis and miRNA loading mechanisms means that strategies to target exosomes or their transferred miRNAs are limited and not specific to tumour cells. Therefore, if ex-miRNA is to be employed in novel non-invasive diagnostic approaches and as a therapeutic target in cancer, two further advances are necessary: in methods to isolate and detect ex-miRNA, and a better understanding of their biogenesis and functions in tumour-cell communication.
Keywords: microRNA, exosomes, exosome isolation, oncogenesis, tumour microenvironment, cell communication
Cell communication is essential for tumorigenesis: individual tumour cells must interact with each other and host cells to survive, progress and metastasise. Through inter-tumour communication, a heterogeneous population of cells can co-operate and advance in a hostile environment. Local tumour-stromal communication facilitates manipulation of the microenvironment, optimising tumour growth, invasion and survival (1,2). Similarly, long-distance communication with stromal cells at distant host sites facilitates pre-metastatic niche formation, to promote colonisation and metastasis (3). However, the signals involved in tumour-stromal cell communication are yet to be fully elucidated. Deciphering these will facilitate the generation of novel, effective biomarkers and possibly provide a therapeutic benefit through targeting of these signals.
It is becoming increasingly clear that tumour-derived exosomes (a form of endosome-derived extracellular vesicle [EV]) play an important role in this communication process through the transport of various proteins, lipids and nucleic acids in their membranous compartments (4). The discovery that exosomes can transfer microRNA (miRNA) between cells (5, Fig. 1) and the accompanying research developments in the miRNA field have sparked great interest in this area. miRNAs are short non-coding RNAs that target and repress complementary mRNAs, with roles in a number of cellular functions, such as differentiation, proliferation and cell cycle regulation (6). Due to the location of miRNA genes in chromosomal regions at cancer-associated genomic regions and fragile sites (unstable genomic regions that are often the sites of chromosomal rearrangements in cancer), expression is frequently dysregulated in tumour cells, leading to the upregulated expression of oncogenic miRNA and downregulation of tumour suppressor miRNA (7).
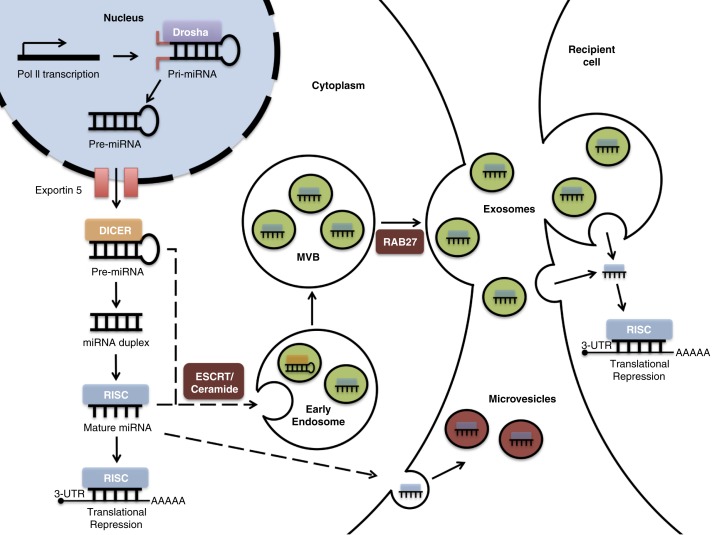
Fig. 1.
miRNA biogenesis and loading into exosomes. miRNA genes are transcribed by RNA polymerase II (Pol II), forming pri-miRNAs in the nucleus. The Drosha complex cleaves pri-RNA to pre-miRNA, which is exported to the cytoplasm via exportin 5. Further cleavage by the Dicer complex generates an intermediary miRNA duplex, of which one strand is incorporated into the RNA-induced silencing complex (RISC) to form mature miRNA, which targets complementary mRNA for translational repression. Inward budding of the early and late endosome forms exosomes. During this process, mature miRNA, some pre-miRNAs and other RNA molecules, proteins, and lipids are loaded into the exosomes. Within exosomes, loaded pre-miRNAs may be processed into mature miRNA. The exosome-loading process involves an endosomal-sorting complex required for transport (ESCRT) or ceramide-dependent mechanisms. The fusion of multivesicular bodies (MVBs) with the plasma membrane releases exosomes. This process is dependent on Rab GTPases (e.g. Rab27). The exosomal fusion with the plasma membrane of the recipient cell, or phagocytosis followed by membrane fusion, leads to the release of miRNA cargo into the cytosol and translational repression.
The aberrant expression of cellular miRNA has been observed in cancer (7), and there is evidence that exosomal miRNA (ex-miRNA) expression is also altered (8). For this reason, could ex-miRNA be a potential cancer biomarker? Furthermore, could this dysregulation play a role in facilitating tumorigenesis? If so, ex-miRNA might provide a novel therapeutic target.
Rapidly emerging methods in the field of exosome isolation and analysis have facilitated the identification of protein markers that distinguish exosomes from other EVs (9). The latter include microvesicles that bud off from the surface of the cell, including very large membrane-bound structures called oncosomes (10). In this review, there will be a specific focus on exosomes, since they are the best-characterised form of EV and most studies of miRNA-containing vesicles have targeted these structures. However, it is important to emphasise that often authors have not clearly distinguished between exosomes and other EVs, because of the current limitations with isolation procedures, although many methodologies employed typically separate away large oncosomes. Here, we will evaluate the technological and clinical feasibility of ex-miRNA as a biomarker. Further, to determine its potential as a therapeutic target, the accuracy and relevance of experiments demonstrating the role of ex-miRNA in tumour progression will be critically discussed.
Ex-miRNAs as diagnostic markers
Detection
Current cancer biomarkers, such as prostate-specific antigen (PSA) and carbohydrate antigens, are plagued by problems such as false-negatives, false-positives and lack of tumour-type specificity (11). Consequently, tumour biopsy, a potentially invasive and damaging method, is the only definitive method of diagnosis. Non-exosomal, extracellular miRNA, bound to protein complexes (12), can be detected in biological fluids including blood, cerebrospinal fluid (CSF), breast milk, saliva and urine (13). Similarly, since their initial detection in secretions from mouse and human mast cell lines (5), ex-miRNAs have also been identified in bodily fluids (14–17). Further, exosomes have been found to provide a stable source of miRNA, preventing RNase degradation (18). In fact, exosomally derived miRNA has been demonstrated to remain stable at −20°C for 5 years, is largely unaffected after 2 weeks at 4°C and is resistant to freeze-thaw cycles (13). Therefore, exosomes are a source of miRNA that enables efficient storage and recovery in conditions that would normally degrade free miRNA.
Due to its ease of access and stability, ex-miRNA has been proposed as a novel, minimally invasive tool for cancer diagnosis, with possible prognostic value. It inevitably suffers from some of the same problems as more conventional tumour markers. For example, it will almost certainly be secreted by other cell types, potentially masking cancer-specific signals. However, it is envisaged that, by profiling multiple ex-miRNA markers and by isolating exosomes using tumour-specific protein markers, it will be possible to improve on sensitivity and specificity, eliminating the issues faced with current cancer biomarkers.
For effective biomarker analysis of exosomes, pure exosome samples are required. Currently, there are difficulties in the isolation of exosomes from other EVs found in bodily fluids. Exosomes are distinguished from other EVs, such as microvesicles, based on membrane composition, size and density. Whereas microvesicles are often 100–1,000 nm in diameter and originate from the plasma membrane, exosomes are around 40–100 nm in diameter (19) and are enriched in proteins that are associated with the endocytic pathway, suggesting endosomal origin (20,21).
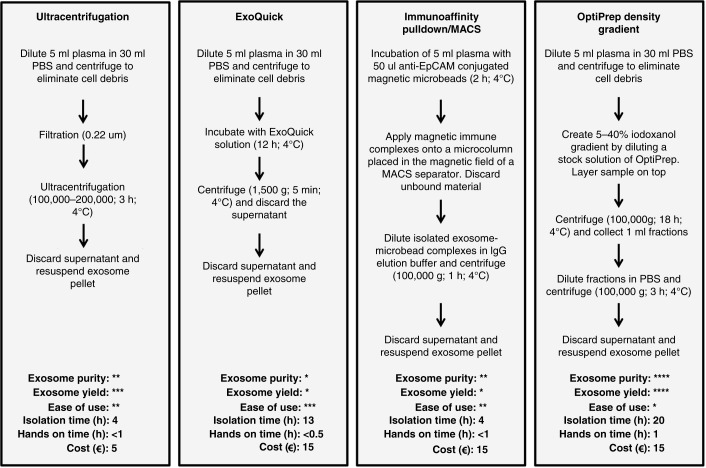
Several methods have been employed to isolate exosomes (Fig. 2), the most common of which involves ultracentrifugation (UC) or ExoQuick precipitation. Both are fast and simple procedures, but relatively crude isolation methods for ex-miRNA measurements due to contaminating soluble proteins (particularly after ExoQuick isolation) and non-exosomal particles (22,23). Another method is immunoaffinity pulldown using anti-EpCAM-coupled microbeads, but not all exosomes express EpCAM (24). With this method, the failure to collect EpCAM-negative subtypes of exosomes and the presence of EpCAM-positive circulating tumour cells are likely to influence subsequent miRNA quantification. Few studies have adopted density gradient centrifugation (DGC), which allows the isolation of vesicles based on buoyant density. This approach has been shown to generate purer exosome samples than other methods (22,25, Fig. 2). However, the suitability of the density gradient method in a clinical setting is questionable, due to difficulties in upscaling and automating such a process.
Fig. 2.
Exosome isolation methods. Example procedures and comparison of 4 commonly used methods, which are ultracentrifugation, ExoQuick precipitation, immunoaffinity pulldown/magnetic-activated cell sorting using anti-EpCAM (MACS) and OptiPrep density gradient to isolate exosomes from a plasma sample. These are compared in terms of purity, exosome yield, ease of use, approximate isolation time, approximate hands-on time and approximate cost per sample. *=low, **=moderate, ***=high, ****=very high. The relative assessment of procedures for exosome purity, yield, ease of use, isolation time, hands-on time and cost are adapted from Van Deun et al. (22).
The search for a more robust, quick and clinically feasible exosome isolation method has led to the development of novel strategies. Size exclusion chromatography (SEC) has been frequently employed for general EV isolation, but has also more recently been used as another method specifically for exosome isolation (26–28). These latter studies have shown SEC to be a simple and low-cost method that can reproducibly isolate exosomes within 20–30 min (26,27). In comparison with UC, these isolated exosomes contain less contaminant plasma proteins. Furthermore, whereas UC and DGC can damage exosomes, exosomes isolated by SEC are morphologically intact and functional (26). SEC is a viable option for isolating exosomes, but it is important to point out that although isolates are within the correct size category for exosomes and western blotting has confirmed the presence of exosomal markers, further investigations are needed to fully characterise the level of purity of these exosomes due to concerns of co-isolation with lipoproteins and other EVs (28).
Another promising method for the isolation of exosomes involves the use of microfluidic technology. Microfluidic immunoisolation and analysis involve the infusion of serum through an inlet into microchambers, which contain antibodies against exosome-specific markers (such as abundantly expressed CD63) and analytical reagents (29). This is a rapid (~100 min), readily scalable technique that employs much smaller samples than other approaches (~1/100 the amounts used in conventional methods) (30). Its one-step isolation system eliminates multiple preparation steps, saving time and reagent costs (31). The potential for high specificity for exosome isolation will also enable the selection of subpopulations of exosomes by targeting both exosomal markers and tumour-associated markers (such as EpCAM, α-IGF-1R and CA125) (30). A key limitation of this feature, however, is that like other immunoisolation methods, it focuses only on the exosomes that express the target surface marker and will therefore fail to identify other subpopulations of exosomes.
An alternative microfluidic technique involves trapping exosomes through microporous silicon nanowires (32). This technique has been shown to selectively trap “exosome-like” vesicles based on size, although no further analysis to confirm exosome identity has been undertaken. This method is prone to saturation at greater sample volumes and is slower than immunological isolation as it is necessary to dissolve the silicon nanowire overnight to collect isolate.
An important benefit of microfluidic techniques is the ability to combine exosome extraction with analysis, further optimising its use in a clinical setting. For instance, Kanwar et al. integrated immunocapture with the fluorescent staining of exosomes in a single device, allowing simultaneous exosome isolation and quantification (29). Similarly, Im et al. developed a nano-plasmonic exosome sensor, enabling label-free detection of immune-captured exosomes in real time (33).
Current miRNA measuring techniques also suffer from problems that limit their clinical effectiveness. Two commonly used methods for measuring ex-miRNA in research are microarray and reverse transcription polymerase chain reaction (RT-PCR). Microarray offers genome-wide expression profiles of miRNA (34), facilitating the detection of a large number of aberrant miRNAs. However, there is difficulty developing probes for some miRNAs, and the same hybridisation conditions do not work for all molecules. Compared to RT-PCR, microarray has higher specificity but a lower sensitivity (35). Despite similar accuracy, microarray has been shown to be unsuitable for quantifying low levels of miRNAs and thus would require larger sample sizes for quantification (35). However, RT-PCR requires a suitable “housekeeping” miRNA control, which is often not reliable. A frequently used control is miR-16 (36), but it is dysregulated in myeloma (37) and rheumatoid arthritis (38). Absolute qPCR overcomes this issue, although a standard calibration curve for specific miRNAs always needs to be constructed and consistently high-quality exosome preparations are needed (39).
The accuracy of these methods could be limited further by the presence of exosomal mRNA (40) and pre-miRNAs (41) that may also be measured within isolated exosomes. Additionally, miRNAs have been identified to exhibit “isomiRs” (sequence heterogeneity at the 3’ and 5’ ends) that may further complicate measurements, particularly for qPCR-based methods (42). A more recently adopted method for measuring expression profiles of miRNA is next-generation sequencing, a powerful approach that may prove useful clinically. This method enables accurate genome-wide quantification of miRNAs and can distinguish between miRNAs that differ by even 1 nucleotide. Since no primers or probes are required, it can detect novel miRNAs. Nevertheless, data analysis and sample preparation are labour intensive and require personnel expertise (43,44).
Ex-miRNA profiles
Tumour-derived cellular miRNAs exhibit different expression patterns in comparison with non-tumour cells of the same cell type (7). Similarly, ex-miRNA in bodily fluids have been shown to exhibit significantly differing expression profiles in comparison with normal control patients (Table I). Through profiling of this abnormal expression in cancers, signature ex-miRNAs in bodily fluids can be used for the detection of early-stage cancers (8). Although it has been shown that some miRNAs are selectively loaded or retained in exosomes, there often is little difference between the cellular miRNA expression profile of tumour cells and the expression profile of their secreted exosomes; therefore, exosomal profiles are potentially suitable as a surrogate for tumour biopsy (8,45). In addition, exosomes contain tissue-specific membrane protein markers that can also aid in detecting tumour origin: for example, prostate cancer-derived exosomes obtained in urine display-specific molecular markers such as PCA-3 or TMPRSS2 (46).
Table I.
Dysregulated exosomal miRNAs in cancer
| Cancer type | Clinical samples | Isolation and quantification method | Dysregulated ex-miRNAs Fold increase (↑)/decrease (↓) |
|---|---|---|---|
| Ovarian cancer (8) | Serum samples from patients with malignant ovarian cancer (n=50) compared to patients with benign ovarian cancer (n=10) | 1. Magnetic-activated cell sorting using anti-EpCAM 2. Microarray |
miR-21 (3↑), miR-141 (3↑), miR-200a (3↑), miR-200c (2.5↑), miR-203 (2↑), miR-205 (3.5↑) and miR-214 (5↑) |
| Lung adenocarcinoma (45) | Plasma from patients with lung adenocarcinoma (n=27) compared to plasma from normal controls (n=9) | 1. Size exclusion chromatography and magnetic-activated cell sorting using anti-EpCAM 2. Microarray |
miR-17-3p (↑), miR-21 (↑), miR-106a (↑), miR-146 (↑), miR-155 (↑), miR-191 (↑), miR-192 (↑), miR-203 (↑), miR-205 (↑), miR-210 (↑), miR-212 (↑) and miR-214 (↑) |
| Nasopharyngeal carcinoma (71) | Serum samples from NPC patients (n=83) compared to healthy controls (n=10) | 1. Ultracentrifugation 2. Microarray |
miR-24-3p (2↑), miR-891a (2↑), miR-106a-5p (3↑), miR-20a-5p (3↑) and miR-1908 (3↑) |
| Breast cancer (49) | Serum samples from breast cancer patients (n=168) and benign breast cancer patients (n=19) compared to healthy controls (n=28) | 1. ExoQuick exosome precipitation 2. RT-PCR |
miR-101 (↑), miR-372 (↑) and miR-373 (↑) |
| Oesophageal squamous cell carcinoma (OSCC) (50,61) | Serum samples from OSCC patients (n=51) compared to patients with benign diseases such as asymptomatic cholecystolithiasis and hernia (n=41) Serum samples from OSCC patients (n=101) compared to normal controls (n=46) Human OSCC cell lines compared to human fibroblast cell lines |
1. ExoQuick exosome precipitation 2. RT-PCR 1. Ultracentrifugation 2. RT-qPCR |
miR-21 (>2↑) miR-1246 (>2↑) |
| Pancreatic cancer (51) | Serum samples from pancreatic cancer patients (n=22), benign pancreatic cancer patients (n=6) and ampullary carcinoma patients (n=7) compared to healthy controls (n=8) | 1. Ultracentrifugation 2. RT-PCR |
miR-21 (6↑) and miR-17-5p (3↑) |
| Brain metastatic melanoma and breast cancer (52) | Brain metastatic melanoma and breast cancer cell lines compared to non-brain metastatic cell lines | 1. ExoQuick exosome precipitation 2. RT-PCR |
miR-210 (3↑), miR-19a (2.5↓) and miR-29c (2↓) |
| Docetaxel-resistant prostate cancer (59) | Docetaxel resistant compared to non-resistant prostate cancer cell lines | 1. Ultracentrifugation 2. RT-PCR |
miR-598 (2↓), miR-148a (2.4↓), miR-34a (11↓) and miR-146a (3↑) |
Fold changes in miRNAs are included in right-hand column where known.
Ex-miRNA may give an insight into the classification or subtype of the tumour. Breast cancer, for instance, is a highly heterogeneous disease with phenotypically different tumour subtypes (47). Expression profiling showed that a number of miRNAs are associated with molecular subtypes of breast cancer (48). Although these are cellular miRNA profiles, since secreted exosomes often reflect the miRNA expression profile of the cell of origin, ex-miRNA may also have similar profiles that reflect breast cancer subtype. Indeed, analysis of exosomal miR-373 showed elevated serum levels in oestrogen receptor-negative and progesterone receptor-negative, compared to receptor-positive breast cancers (49).
Further, ex-miRNAs have been shown to indicate tumour severity, progression and aggressiveness. Serum-derived exosomal miR-21 was significantly increased in oesophageal squamous cell carcinoma (OSCC) patients and correlated with advanced tumour classification, positive lymph node status and the presence of metastasis with inflammation (50). miR-21 is not specific to OSCC and, therefore, could also be a progression biomarker in other cancers including lung, ovarian and pancreatic tumours. However, in spite of an elevated serum exosomal miR-21 level in pancreatic adenocarcinoma, there was no significant correlation with tumour stage. Instead, miR-17-5p was shown to correlate with metastasis and advanced cancer stage (51). Identifying ex-miRNAs that associate with distinct metastasis sites could provide an additional diagnostic tool for clinicians to evaluate disease stage and monitor progression. Ex-miRNA derived from brain metastatic breast cancer and melanoma cell lines shared an upregulation in miRNA-210 and downregulated miR-19a and miR-29c in comparison with non-brain metastatic breast cancer and melanoma cell lines (52). This may represent a profile for metastasis to the brain or could simply represent a common marker for metastasis.
It is important to consider the possible influence cancer treatment could have on ex-miRNA profiles. Although some reports have shown changes in miRNA profiles in response to surgery and chemotherapy (53,54), these studies often involve circulating miRNAs and there are few studies that look specifically at ex-miRNA profiles. In breast cancer patients, neoadjuvant therapy was shown to result in a greater than 2-fold increase in circulating miR-503. The source of this increase was postulated to be endothelial cells, as exosomal miR-503 isolated from human umbilical vein endothelial cells increased 3- and 4-fold in the presence of epirubicin and paclitaxel, respectively, compared to controls (55). These data highlight the important issue that circulating miRNA changes might partly originate from normal tissues in response to cellular stress induced by cancer therapy.
Not all studies have demonstrated significant ex-miRNA profile changes in response to cancer therapy: ex-miRNA levels from acute myeloid leukaemia cell lines have been shown to be resistant to cytarabine and quizartinib, with only minor changes observed (56). Moreover, in glioblastoma cell lines, radiation-induced exosomes showed little change in their miRNA composition (57). These findings suggest that some ex-miRNAs could be used as general biomarkers for cancer, whilst others might be reporters for the effectiveness of therapies.
It has been reported that the number of detected exosomes correlates with the size of the tumour (58). This, however, should be considered in the context of (a) the incomplete and sometimes inconsistent exosome recovery that current isolation techniques provide and (b) the limitations of exosome enumeration techniques (which often do not discriminate between vesicular and non-vesicular events), leading to low accuracy of quantification. Nevertheless, in the future, this may prove useful as one measure for determining the clinical efficacy of a particular treatment. More important, there is a possibility that detection of specific ex-miRNAs could predict drug efficacy and resistance (a major obstacle to successful cancer treatment). For example, reduced expression of exosomal miR-34a was shown to be a predictive marker of docetaxel resistance in castration-resistant prostate cancer (59).
The varied range of information that might be gained from ex-miRNA, such as progression of a tumour, detection of metastasis and possible drug resistance, could be used to influence clinical decisions and treatment strategies, thus facilitating optimised treatments tailored to meet the needs of the patient. These data could also be of prognostic value. In fact, studies have shown a correlation between specific ex-miRNAs and overall survival in castration-resistant prostate cancer (miR-1290 and mir-375) (60), OSCC (miR-1246) (61) and ovarian cancer (miR-21) (8).
However, due to the heterogeneity of methodologies used to isolate and quantify ex-miRNA, current data are not comparable, making it difficult to assess their clinical suitability at present. It has been shown that there can be differences in results obtained from different isolation (22,25) and quantification methods (35), even from different vendors of the same platform. Therefore, although experimental evidence suggests ex-miRNA has potential as a diagnostic marker for clinical use, ex-miRNA profiles must be characterised in patients using the same standard procedure. This procedure should be simple, reproducible and permit relatively pure exosome recovery from large numbers of human specimens.
Roles of ex-miRNAs in cancer
The presence of distinct ex-miRNAs in the circulation may have functional importance. When first discovered, exosomes were believed to be involved in the disposal of cellular “trash” (62). The observation that cell lines release tumour suppressor miRNA via exosomes (63,64) prompted the notion that ex-miRNA could be a mechanism by which tumour cells reduce intracellular concentrations of tumour suppressor miRNA to maintain oncogenesis. Consistent with this idea, Rab27 siRNA knockdown in bladder cancer cells caused a reduced exosomal export of tumour suppressor miR-23b and a rise in intracellular miR-23b, corresponding to attenuated cell invasiveness, an effect that was reversed by miR-23b inhibition. In spite of this, the demonstration that exosomes can activate tumour-related pathways (Fig. 3) and the observation that miRNA can be transferred via exosomes to other cells (5) suggest that ex-miRNAs are also a means of communication. If so, ex-miRNA could provide a novel therapeutic target.
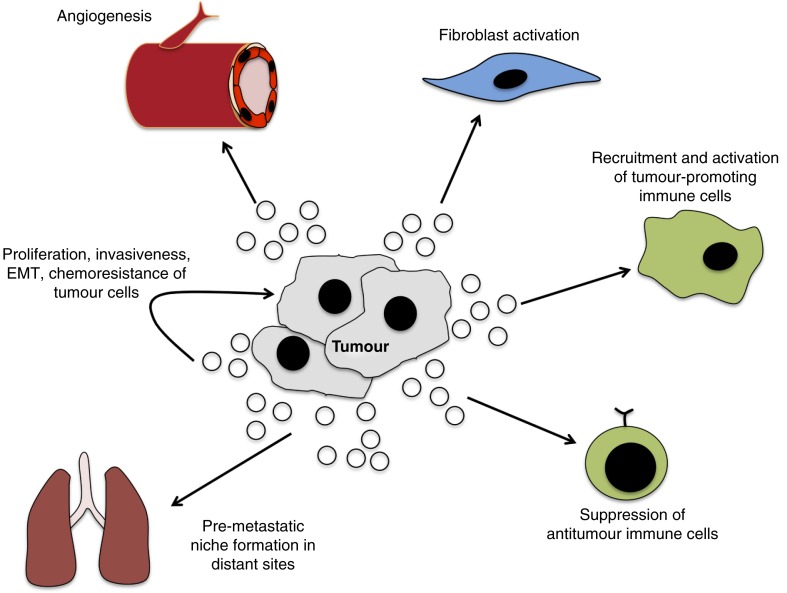
Fig. 3.
Role of exosomes in tumour-related pathways. Through the transfer of oncogenic signals, exosomes promote tumorigenesis through interaction with local and distant cells. The intra-tumour transfer of exosomes can facilitate growth and invasiveness, as well as confer chemoresistance. The exosome-mediated activation of fibroblasts facilitates the remodelling of the microenvironment and angiogenesis to enhance growth and invasion of cancer. The direct transfer of tumour-derived exosomes to vascular endothelial cells can promote angiogenesis. Tumour-derived exosomes can also modulate immune function to increase activity of tumour-promoting immune cells and suppress anti-tumour cells. The activation of these and other pathways in distant sites generates a pre-metastatic niche that facilitates metastasis.
It is important to mention that miRNAs are likely to be at low levels in exosomes. Even the most abundant ex-miRNAs were measured on average at less than one copy per exosome in one study (65). Thus far, there is limited knowledge about the biology of ex-miRNA, and it is important that similar studies are performed to determine whether this is a general phenomenon. However, it has been proposed that there could be miRNA-enriched subtypes of exosomes, or specific targeting of exosomes and fast uptake by target cells that enables low concentrations of miRNA to accumulate at functionally significant amounts locally. Furthermore, in addition to their role in translational repression, miRNAs act via other non-conventional mechanisms, including Toll-like receptor binding (66) and the possible effects on epigenetic regulation (67), which are thought to require lower concentrations of miRNA.
Tumours often consist of a heterogeneous population of cells with varying proliferative, invasive and metastatic potential. Thus, through the intra-tumour transfer of ex-miRNA, tumour cells could co-operate to drive cancer progression. Exosomal miR-10b is upregulated in breast cancer. The transfer of miR-10b, through the co-culture of these isolated exosomes with non-metastatic human breast cancer cells, promotes invasive capacity through targeting mRNAs for tumour suppressors KLF-4 and HOXD10 (68). In another study, metastatic breast cancer and melanoma-derived exosomes were identified to contain increased oncogenic miR-210 and reduced levels of multiple tumour suppressor miRNAs, such as miR-29c, compared to non-metastatic cell lines. The uptake of these exosomes by non-metastatic breast cancer and melanoma cells resulted in an increase in invasive capacity (52). Isolated exosomes from docetaxel-resistant breast cancer cells contain several upregulated miRNAs, and co-culture was shown to reduce chemosensitivity in non-resistant breast cancer cell lines (59). Thus, as well as metastatic and invasive capabilities, tumour cells can also transfer survival adaptation properties through ex-miRNA.
Ex-miRNA can be received by stromal cells in the local microenvironment or at distant sites to promote survival and generate pro-metastatic niches for colonisation. miR-92a and miR-135b target anti-angiogenic mRNAs, encoding the integrin subunit α5 (69) and factor-inhibiting hypoxia-inducible factor 1 (FIH-1) (70), respectively. The exosomal transfer of these miRNAs to human umbilical vein endothelial cells enhances migration and tube formation. In addition to angiogenesis, ex-miRNA has been shown to modulate immune function, promoting immune evasion and cancer progression. Nasopharyngeal carcinoma cells release exosomes that are upregulated in several miRNAs compared to normal cells, including miR-24-3p, miR-891a, miR-106a-5p, miR-20a-5p and miR-1908. The transfer to T-cells leads to a downregulation of the MAPK and JAK/STAT signalling pathways and an impairment of T-cell function by inhibiting T-cell proliferation and Th1 and T17 differentiation, whilst promoting Treg differentiation (71). Although the primary mechanism by which miRNAs exert their effects are through post-transcriptional repression, miR-21 and 29a transferred via exosomes to macrophages at the tumour-stromal interface can bind to TLR8 to trigger inflammatory responses that promote tumour growth and metastasis (66).
As well as receiving ex-miRNA, there is evidence that stromal cells also release exosomes that promote tumour progression and metastasis. Phosphatase and tensin homologue (PTEN) is a tumour suppressor whose miRNA-induced suppression has been demonstrated to prime brain metastasis after tumour cell extravasation (72). The co-culture of tumour cells with astrocytes led to a reduction in PTEN expression, which was rescued by mutating PTEN's miR-19a-binding site (72), suggesting a role for astrocyte-derived miR-19a in reducing tumour suppressor PTEN expression. In agreement, isolated astrocyte exosomes were confirmed to express miR-19a, and treating tumour cells with these exosomes led to a dose-dependent increase in miR-19a and a subsequent decrease of PTEN mRNA. Additionally, the blockade of astrocyte exosome secretion, using siRNA targeting Rab27a, led to a restoration of PTEN mRNA levels in tumour cells (72). Together, these data show that miR-19a-containing exosomes released by astrocytes downregulate the PTEN expression in tumour cells.
Many of the experiments described above used exosome isolation methods that are known to have limitations. UC or ExoQuick isolation methods were predominantly used, and despite verification of the exosome presence using specific markers, it is unlikely that all EVs collected are exosomes. Furthermore, exosomes are known to contain a variety of signalling RNAs, lipids and proteins that could be causing the effects observed. These issues were addressed by Kosaka et al. (73), through the knockdown of nSMase2 (required for the synthesis of ceramide), which impairs exosome release and miRNA transfer, eliminating the metastatic breast cancer-derived exosomes containing proangiogenic miR-210. Although this treatment presumably has several other cell biological effects, it blocked the in vitro and in vivo induction of angiogenesis and metastasis (73). To show more directly that this miRNA contributes to angiogenesis, breast cancer cell lines were generated overexpressing miR-210, which release exosomes containing high levels of miR-210. miR-210-enriched exosomes significantly enhanced angiogenesis and partially overcame angiogenic inhibition in recipient endothelial cells transfected with anti-miR-210. This overexpression, however, could have had effects on exosomes other than simply promoting miR-210 loading.
Linking to the issue of low ex-miRNA concentration, another criticism is that experiments using isolated exosome preparations may not be physiologically relevant. Most reported studies involve the addition of purified exosomes to cultured cells. Thus, ex-miRNAs are unlikely to be at physiological concentration. Nevertheless, co-culture of IL4-activated macrophages with breast cancer cells, at a cell number ratio similar to that observed in samples of breast cancer patients, has been shown to promote an increase in miR-223 uptake and the invasiveness of breast cancer cells through the Mef2c-β-catenin pathway (74). This was also observed when breast cancer cells were cultured with isolated exosomes. To verify that these observations were mediated by exosomal miR-223, in both experiments, the effects were reversed by anti-miR-223 transfection into recipient tumour cells (74). Thus, despite low enrichment, there is evidence that ex-miRNAs can still have an effect, even at physiological concentrations, although one must recognise the in vitro nature of all these experiments.
In an attempt to overcome this issue, studies have utilised a murine Cre-lox-based recombination system to trace tumour–tumour and tumour–stromal EV transfer in vivo. Myeloid-derived suppressor cells were observed to take up EVs containing Cre mRNA, which led to changes in their immunosuppressive phenotype and miRNA profiles (75), whilst tumour cells that took up EVs were shown to have enhanced migratory and metastatic behaviour (76). Although these studies were not focused on miRNA transfer via exosomes, future studies may be able to adopt a similar method to allow in vivo tracking of exosome targeting, so that the effects of ex-miRNA can be monitored specifically in target cells.
The evidence and discussion above shows that oncogenic ex-miRNAs could function in the progression of tumorigenesis and may provide a novel target for therapy. One possible treatment strategy is therapeutic haemofiltration to deplete circulating exosomes (77). Alternatively, drugs could be used to target aspects of exosome biogenesis, such as ESCRT-dependent (78), ceramide-dependent (79) or Rab27-dependent (80) pathways (Fig. 1). Proteins involved in miRNA sorting or loading, such as heterogeneous nuclear ribonucleoprotein A2B1 (81) or proteins in the RISC complex (82), could be targeted. These approaches, however, would have a low selectivity of inhibition, affecting both tumour and non-tumour ex-miRNA, and thus interfering with ex-miRNA's normal physiological functions (6,83). Furthermore, modulating the activities of molecules like Rabs and ESCRTs, that have key roles in multiple cell biological events, is very likely to affect processes other than ex-miRNA secretion. Whilst the pathways for exosome biogenesis and loading are yet to be fully elucidated, further research may reveal altered exosome biosynthetic pathways in tumour cells, providing tumour-specific targets to block altered loading and increased exosome release. Last, cytosolic oncogenic miRNA could be targeted through the delivery of anti-miRNA oligonucleotides to cells, although there remains a risk of other adverse effects since a single miRNA can influence multiple pathways.
Conclusion
In summary, exosomes are efficient and stable transporters of miRNA present in bodily fluids. Furthermore, there are distinct differences in ex-miRNA expression patterns in tumour cells, compared to their normal counterparts. Variations in profiles have also been shown to correlate with tumour characteristics, such as invasiveness, metastasis and chemoresistance. However, data on these cancer ex-miRNA profiles are incomplete. Although advances have been made in ex-miRNA detection methods, for the profiling of miRNA signatures and detection in patients, there remains a need for a rapid and inexpensive standard procedure that generates an effective, pure isolation of exosomes and accurate genome-wide quantification.
In addition, miRNAs are key regulators of gene expression, and consequently, it appears that ex-miRNAs can have a dual role in cancer. On one side, the exosomal transfer of miRNA can alter the behaviour of the recipient tumour or stromal cells, facilitating progression and metastasis. On the other, exosomes provide a pathway to limit the intracellular abundance of tumour suppressor miRNAs, maintaining tumour oncogenesis. Current targets for exosome biogenesis and loading are not specific to tumour cells and would interfere with the normal physiological function of exosomes. An increased understanding of the mechanism of exosome loading and biogenesis may reveal altered pathways in tumour cells, which could be targeted. The use of anti-miRs is another strategy, but pathways by which miRNAs exert their functions must be characterised to minimise the risk of off-target effects.
Thus, with standardised procedures and complete profiling, ex-miRNA could become a novel cancer diagnostic marker. However, its use as a therapeutic target, although a possibility in the future, is a more difficult task hampered by the limited understanding of ex-miRNA biogenesis, loading and mechanism of action.
Acknowledgements
CW gratefully acknowledges the support of the BBSRC (BB/K017462/1, BB/L007096/1), Cancer Research UK (C19591/A19076), the Cancer Research UK Oxford Centre Development Fund (C38302/A12278) and the John Fell Fund, Oxford, as well as studentships and scholarships from the Wellcome Trust, CRUK, Urology Foundation and the MRC in supporting research in this area.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Bruecher BL, Jamall IS. Cell-cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol Biochem. 2014;34:213–43. doi: 10.1159/000362978. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Sceneay J, Smyth MJ, Möller A. The pre-metastatic niche: finding common ground. Cancer Metastasis Rev. 2013;32:449–64. doi: 10.1007/s10555-013-9420-1. [DOI] [PubMed] [Google Scholar]
- 4.Tickner JA, Urquhart AJ, Stephenson SA, Richard DJ, O'Byrne KJ. Functions and therapeutic roles of exosomes in cancer. Front Oncol. 2014;4:3389. doi: 10.3389/fonc.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–2. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 8.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:968–77. doi: 10.1073/pnas.1521230113. doi: http://dx.doi.org/10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins GL, Slater ED, Sanders GK, Prichard JG. Serum tumor markers. Am Fam Physician. 2003;68:1075–88. [PubMed] [Google Scholar]
- 12.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. MiRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1568–75. doi: 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287(6):3842–9. doi: 10.1074/jbc.M111.277061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288:26888–97. doi: 10.1074/jbc.M113.452458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Dov IZ, Brown M, Whalen VM, Tuschl T. 30 Profiling urine cell and exosome microRNA using a barcoded small RNA deep sequencing approach. Am J Kidney Dis. 2011;57:B24. [Google Scholar]
- 18.Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, et al. Exosome can prevent RNase from degrading microRNA in feces. J Gastrointest Oncol. 2011;2:215–22. doi: 10.3978/j.issn.2078-6891.2011.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–9. [PubMed] [Google Scholar]
- 20. Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–18. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 21.Gruenberg J, Maxfield FR. Membrane transport in the endocytic pathway. Curr Opin Cell Biol. 1995;7:552–63. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 22.Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24858. 24858, doi: http://dx.doi.org/10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47:135–8. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122:437–46. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 25.Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–64. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 26.Hong CS, Funk S, Muller L, Boyiadzis M, Whiteside TL. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles. 2016;24:5. doi: 10.3402/jev.v5.29289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranyai T, Herczeg K, Onódi Z, Voszka I, Módos K, Marton N, et al. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One. 2015;10:e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welton JL, Webber JP, Botos LA, Jones M, Clayton A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J Extracell Vesicles. 2015;4 doi: 10.3402/jev.v4.27269. 27269, doi: http://dx.doi.org/10.3402/jev.v4.27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14:1891–900. doi: 10.1039/c4lc00136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773–80. doi: 10.1039/c4lc00662c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10:505–11. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, et al. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip. 2013;13:2879–82. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao JJ, Hua YJ, Sun DG, Meng XX, Xiao HS, Ma X. Genome-wide microRNA profiling in human fetal nervous tissues by oligonucleotide microarray. Childs Nerv Syst. 2006;22:1419–25. doi: 10.1007/s00381-006-0173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–15. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Nat Acad of Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocci A, Hofmeister CC, Geyer S, Talabere T, Gambella M, Cascione L, et al. Circulating Mir-16 and Mir-25 as new prognosticators for multiple myeloma. Blood. 2013;122:1853. doi: 10.1038/leu.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filková M, Aradi B, Šenolt L, Ospelt C, Vettori S, Mann H, et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann Rheum Dis. 2014;73:1898–904. doi: 10.1136/annrheumdis-2012-202815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–8. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rani S, O'Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, et al. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods Mol Biol. 2011;784:181–95. doi: 10.1007/978-1-61779-289-2_13. [DOI] [PubMed] [Google Scholar]
- 41.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, et al. Wang K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–80. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013;98:236–8. doi: 10.1136/archdischild-2013-304340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O'Briant K, Godwin AK, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–6. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–7. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4–13. doi: 10.3121/cmr.2009.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eichelser C, Stückrath I, Müller V, Milde-Langosch K, Wikman H, Pantel K, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–67. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 51.Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:1. doi: 10.1186/1477-7819-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camacho L, Guerrero P, Marchetti D. MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS One. 2013;8:e73790. doi: 10.1371/journal.pone.0073790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frères P, Josse C, Bovy N, Boukerroucha M, Struman I, Bours V, et al. Chemotherapy in breast cancer patients induces miR-34a and miR-122 expression. J Cell Physiol. 2015;230:473–81. doi: 10.1002/jcp.24730. [DOI] [PubMed] [Google Scholar]
- 54.Shapira I, Lee A, Oswald M, Parameswaran J, Sultan K, Budman D. Surgical and chemotherapy influence on circulating developmental microRNA miR17-92 and long term survival in breast and ovarian cancer patients. Cancer Res. 2013;73(Suppl 8):1843. [Google Scholar]
- 55.Bovy N, Blomme B, Frères P, Dederen S, Nivelles O, Lion M, et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget. 2015;6:10253–66. doi: 10.18632/oncotarget.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, et al. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci Rep. 2015;5:11295. doi: 10.1038/srep11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arscott WT, Tandle AT, Zhao S, Shabason JE, Gordon IK, Schlaff CD, et al. Ionizing radiation and glioblastoma exosomes: implications in tumor biology and cell migration. Transl Oncol. 2013;6:638–48. doi: 10.1593/tlo.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corcoran C, Rani S, O'Driscoll L. MiR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014;74:1320–34. doi: 10.1002/pros.22848. [DOI] [PubMed] [Google Scholar]
- 60.Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–52. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnstone RM, Bianchini A, Teng K. Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood. 1989;74:1844–51. [PubMed] [Google Scholar]
- 63.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ostenfeld MS, Jeppesen DK, Laurberg JR, Boysen AT, Bramsen JB, Primdal-Bengtson B, et al. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014;74:5758–71. doi: 10.1158/0008-5472.CAN-13-3512. [DOI] [PubMed] [Google Scholar]
- 65.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Nat Acad Sci USA. 2014;111:14888–93. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Nat Acad Sci. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–9R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 68.Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol cancer. 2014;13:256. doi: 10.1186/1476-4598-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32:2747–55. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 70.Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood. 2014;124:3748–57. doi: 10.1182/blood-2014-05-576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–52. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, Li P, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–4. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–59. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ridder K, Sevko A, Heide J, Dams M, Rupp AK, Macas J, et al. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology. 2015;4:e1008371. doi: 10.1080/2162402X.2015.1008371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–57. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marleau AM, Chen CS, Joyce JA, Tullis RH. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med. 2012;10:134. doi: 10.1186/1479-5876-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 79.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–7. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 80.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 81.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–21. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bobrie A, Colombo M, Raposo G, Théry C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–68. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]