Sir,
The proliferation of carbapenem-resistant Enterobacteriaceae (CRE) over the past two decades signifies an escalating threat to modern healthcare. CRE are typically resistant to most antibiotic classes and produce infections associated with high mortality.1 Carbapenem resistance mediated by blaKPC has been described in all major Enterobacteriaceae, but can also result from alterations in porins combined with overexpression of endogenous AmpC enzymes.2 However, clinical infections have been largely restricted to Klebsiella pneumoniae, especially epidemic blaKPC-harbouring ST258.3
Recent studies report the local emergence of carbapenem-resistant Enterobacter cloacae (CREC) internationally4 and in the USA,5–9 suggesting accelerating carbapenem resistance acquisition in this organism. The prevalence and population structure of CREC remain incompletely understood. Here we report an increase in CREC incidence at a tertiary care hospital in New York City over a 7 year period and characterize its molecular epidemiology.
We retrospectively reviewed the proportion of E. cloacae non-susceptible to carbapenems from 2007 to 2014 at our hospital using antibiogram data. We then investigated the clinical and molecular epidemiology of CREC isolates since 2010 using MLST.10 Isolate susceptibilities were determined using Vitek 2 and interpreted using recent CLSI guidelines,11 accounting for 2010 revisions to carbapenem MIC breakpoints. We PCR-amplified carbapenemase genes blaKPC, blaVIM, blaIMP, blaNDM and blaOXA-48, ampC β-lactamase12 and ompC and ompF porin genes.2 PCR products were sequenced to differentiate subtypes or identify non-functional mutations. Our institutional review board approved this study.
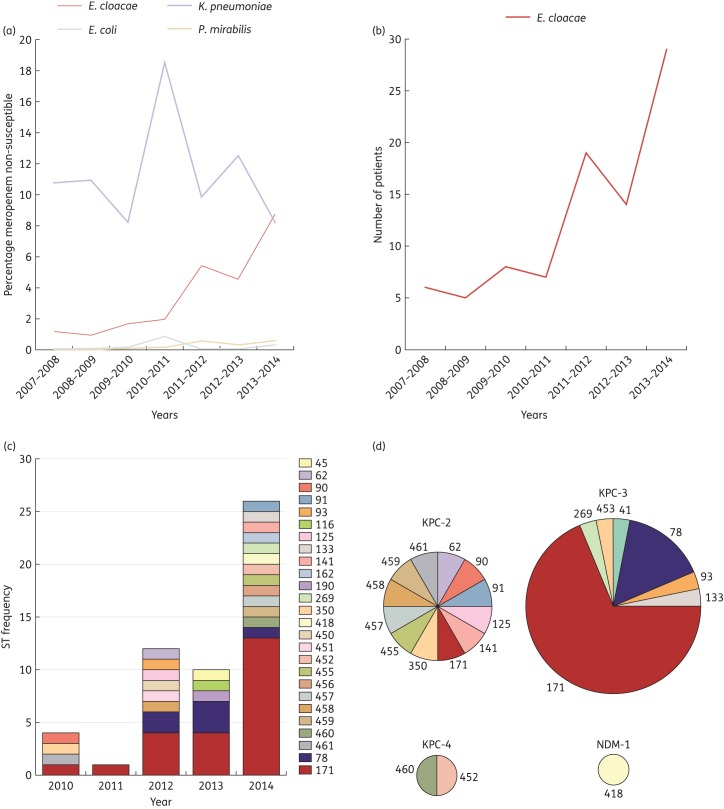
Between 2007 and 2014, we observed a hospital-wide increase in the proportion of carbapenem-non-susceptible E. cloacae from 1.6% to 8.7% of isolates (Figure 1a). This corresponded to a 5-fold absolute increase in incidence (Figure 1b), ranging from 4.6% to 19% in ICU inpatients, 1.3% to 10% in non-ICU inpatients and 0% to 2.6% in outpatients. During the same time period the incidence of carbapenem-resistant K. pneumoniae, Escherichia coli and Proteus mirabilis remained stable or decreased (Figure 1a).
Figure 1.
Emergence of CREC and corresponding multilocus STs. (a) Percentage of single-patient isolates of various Enterobacteriaceae non-susceptible to meropenem between 2007 and 2014 based on antibiogram data. (b) Absolute number of patients infected with meropenem-non-susceptible E. cloacae isolates per year. (c) STs of CREC isolates between 2007 and 2014. (d) Carbapenemase gene carriage amongst CREC isolates according to ST. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
We obtained clinical data for 56 consecutive patients with CREC, of whom 53 had initial isolates available for molecular typing. CREC largely affected patients requiring prolonged or recurrent hospitalizations and most had multiple comorbidities. The most common culture source was the respiratory tract (n = 24), followed by blood (n = 11) and urine (n = 10). Most isolates (79%) had meropenem MICs of ≥16 mg/L. While susceptibility to amikacin (95%) and polymyxin B (100%) was preserved, most isolates were resistant to gentamicin (73%), tobramycin (82%), levofloxacin (77%) and trimethoprim/sulfamethoxazole (89%). Both 30 day mortality (29%) and hospital mortality (46%) were high.
MLST revealed substantial clonal diversity, as CREC isolates belonged to 26 different STs. ST171 accounted for nearly half of all isolates (n = 23, 43%), followed in frequency by ST78 (n = 6) (Figure 1c). All other STs were singletons (n = 24), including 11 novel STs. blaKPC-3 (n = 30), blaKPC-2 (n = 12) and blaKPC-4 (n = 2) were the predominant putative mechanisms of carbapenem resistance, whereas blaNDM occurred in one isolate (Figure 1d). ST171 and ST78 were found to harbour blaKPC-3, while blaKPC-2, blaKPC-4 and blaNDM occurred in unique STs. All carbapenemase-negative isolates harboured ampC genes and four out of eight contained premature stop codons in the ompC porin (Table S1, available as Supplementary data at JAC Online). The mechanism of resistance remains undetermined in four isolates with lower meropenem MICs.
Our data support a highly concerning diversification of CRE beyond K. pneumoniae. While many CREC isolates belonged to diverse lineages, suggesting sporadic acquisition of blaKPC-encoding plasmids, we also observed the emergence and rapid spread of two CREC clones, ST78 and ST171. ST171 was first detected at a Western Pennsylvania hospital7 and has spread in the Midwestern USA.9 ST78 and ST171 also accounted for the majority of 20 CREC isolates at a Boston hospital.8 Based on these findings and recent reports in the literature, we propose that ST171 is emerging as a dominant CREC clone in the USA.
While international CREC isolates primarily harboured metallo-β-lactamases and OXA-48,4 our data support a growing role for blaKPC-mediated CREC in the USA. At least three different blaKPC-encoded carbapenemases have been described in ST171 alone.8 This points to the efficiency of E. cloacae in acquiring a variety of resistance plasmids, potentiating the emergence of new resistant strains. Moreover, E. cloacae ST171 and to a lesser extent ST78 appear to be able to acquire and maintain blaKPC-harbouring plasmids and persist as hospital-associated CRE strains.
This was a single-centre, retrospective study, and thus our findings may have limited generalizability. Notably, the increased incidence was unlikely to result from changes in CLSI breakpoints as only revised guidelines were applied, or from a system-wide breakdown of infection control as it was restricted to E. cloacae.
The alarming increase in CREC infections was driven by diverse clones, suggesting frequent acquisition of blaKPC-encoded resistance as well as an increase in ST78 and ST171. The emergence of blaKPC-harbouring ST171 across the USA suggests its potential as an epidemic CREC clone. Future molecular surveillance studies are urgently needed to further assess the spread of CREC including ST171.
Funding
This work was supported in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (1R01AI116939 and 1R01AI116939-S01 to A.-C. U. and 5T32AI100852 to A. G.-S.) and the Columbia University Irving scholarship (A.-C. U.).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
References
- 1.CDC. Antibiotic Resistance Threats in the United States, 2013. http://www.cdc.gov/drugresistance/threat-report-2013/.
- 2.Doumith M, Ellington MJ, Livermore DM et al. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK . J Antimicrob Chemother 2009; 63: 659–67. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011; 17: 1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girlich D, Poirel L, Nordmann P. Clonal distribution of multidrug-resistant Enterobacter cloacae. Diagn Microbiol Infect Dis 2015; 81: 264–8. [DOI] [PubMed] [Google Scholar]
- 5.Marchaim D, Chopra T, Perez F et al. Outcomes and genetic relatedness of carbapenem-resistant Enterobacteriaceae at Detroit Medical Center. Infect Control Hosp Epidemiol 2011; 32: 861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiedrowski LM, Guerrero DM, Perez F et al. Carbapenem-resistant Enterobacter cloacae isolates producing KPC-3, North Dakota, USA. Emerg Infect Dis 2014; 20: 1583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn C, Syed A, Hu F et al. Microbiological features of KPC-producing Enterobacter isolates identified in a U.S. hospital system . Diagn Microbiol Infect Dis 2014; 80: 154–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pecora ND, Li N, Allard M et al. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. MBio 2015; 6: e01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hargreaves ML, Shaw KM, Dobbins G et al. Clonal dissemination of Enterobacter cloacae harboring blaKPC-3 in the upper midwestern United States. Antimicrob Agents Chemother 2015; 59: 7723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N et al. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 2013; 8: e66358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fifth Informational Supplement M100-S25. CLSI, Wayne, PA, 2015. [Google Scholar]
- 12.Voets GM, Fluit AC, Scharringa J et al. A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int J Antimicrob Agents 2011; 37: 356–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.