Abstract
Background
In small series or individual reports, SNPs within the mprF ORF and dysregulation of its expression in Staphylococcus aureus have been linked to daptomycin resistance (DAP-R) via a proposed gain-in-function mechanism. Similarly, dysregulation of dltABCD has also been associated with DAP-R.
Methods
Using 22 well-characterized, isogenic daptomycin-susceptible (DAP-S)/DAP-R clinical MRSA strain pairs, we assessed potential relationships of the DAP-R phenotype with: (i) regulation of mprF transcription; (ii) regulation of dltABCD transcription; (iii) expression of the two-component regulatory system, graRS (upstream regulator for both mprF and dltABCD transcription); (iv) SNPs within the graRS promoter or its ORF; and (v) altered mprF transcription and lysyl-phosphatidylglycerol (L-PG) synthesis.
Results
Enhanced expression of mprF occurred with SNPs in highly distinct and well-chronicled MprF domain ‘hot spots’ and rarely occurred without such mutations. Increased expression and/or dysregulation of mprF and dltABCD were not uncommon in DAP-R strains, occurring in 27% of strains for each gene. In these latter strains, neither graRS expression profiles nor polymorphic sequences within the graRS promoter or ORF could be significantly linked to altered transcription of mprF or dlt.
Conclusions
Although graRS can co-regulate mprF and dltABCD expression, loci outside of this regulon appear to be involved in dysregulation of these latter two genes and the DAP-R phenotype. Finally, DAP-R strains exhibiting significantly altered mprF transcription profiles produced significantly increased levels of L-PG.
Introduction
Prior investigations in our laboratory and others have linked SNPs within the mprF (multiple peptide resistance factor) locus of S. aureus with DAP-R [note—although the official term is ‘daptomycin non-susceptibility’, ‘daptomycin resistance’ (DAP-R) is used in this paper for ease of presentation].1–3
In a recent investigation,1 using 22 well-characterized isogenic daptomycin-susceptible (DAP-S) and DAP-R MRSA clinical isolate pairs, we showed that all the mprF SNPs observed in the DAP-R strains were clustered within one of the two ‘hot spot’ MprF domains (central bifunctional domain or C-terminal synthase domain). Moreover, these mprF SNPs were correlated with excess cell membrane (CM) synthesis of lysyl-phosphatidylglycerol (L-PG) and enhanced surface positive charge.
In the current study, we utilized the same large well-characterized DAP-S and DAP-R isogenic clinical MRSA strain-pair collection (n = 22 pairs) as that mentioned above to assess: (i) expression profiles of mprF and dltABCD transcription during both exponential-phase and stationary-phase growth; (ii) the relationships of such transcriptional patterns with expression and sequence profiling of the upstream two-component regulator of both mprF and dltA, i.e. graRS;4–6 and (iii) correlation between dysregulation of mprF expression and L-PG production.
Materials and methods
Bacterial strains
We employed the previously described 22 DAP-S/DAP-R MRSA isogenic strain pairs of clinical bloodstream isolates randomly selected from the Cubist Pharmaceuticals Isolate Collection (Table 1).1
Table 1.
Daptomycin MICs, mprF SNPs and phospholipid (PL) profiles among the study strainsa
| Strain | Daptomycin MIC (mg/L)b | mprF SNPc | PL profiles (%, mean ± SD) among total PLs |
||
|---|---|---|---|---|---|
| L-PG | phosphatidylglycerol | cardiolipin | |||
| C1 | 0.19 | ||||
| C2 | 2 | L826F | |||
| C3 | 0.5 | 23.5 ± 3.2 | 68.5 ± 4.2 | 8.0 ± 3.8 | |
| C4 | 4 | P314L | 25.6 ± 5.0 | 65.7 ± 10.5 | 8.7 ± 7.5 |
| C5 | 0.25 | 14.2 ± 1.5 | 81.8 ± 1.5 | 4.0 ± 0.9 | |
| C6 | 3 | T345A | 19.2 ± 3.2* | 77.7 ± 3.7 | 3.0 ± 1.1 |
| C7 | 0.5 | ||||
| C8 | 3 | none | |||
| C9 | 0.5 | 19.3 ± 7.1 | 76.0 ± 5.5 | 4.7 ± 1.7 | |
| C10 | 3 | L826F | 23.9 ± 5.6* | 71.5 ± 6.5 | 4.7 ± 3.4 |
| C13 | 0.75 | ||||
| C14 | 4 | T472K | |||
| C15 | 0.75 | ||||
| C16 | 4 | M347R | |||
| C17 | 0.5 | ||||
| C18 | 4 | L341S | |||
| C19 | 0.38 | 14.9 ± 1.7 | 75.1 ± 3.2 | 10.0 ± 2.0 | |
| C21 | 4 | L826F | 30.9 ± 3.2* | 50.5 ± 3.1* | 18.6 ± 2.0* |
| C22 | 0.5 | ||||
| C23 | 4 | none | |||
| C24 | 0.5 | ||||
| C25 | 3 | S295L | |||
| C26 | 0.38 | 31.8 ± 13.9 | 62.9 ± 14.9 | 5.3 ± 2.5 | |
| C27 | 2 | T345K | 28.7 ± 0.8 | 68.9 ± 2.6 | 2.4 ± 1.8 |
| C30 | 0.25 | ||||
| C31 | 2 | L826F | |||
| C32 | 0.5 | 21.9 ± 3.6 | 71.2 ± 2.5 | 6.9 ± 3.8 | |
| C33 | 2 | S337L | 24.1 ± 3.1 | 73.1 ± 4.0 | 2.7 ± 2.0 |
| C34 | 0.38 | ||||
| C35 | 4 | none | |||
| C36 | 0.5 | 15.1 ± 4.7 | 81.5 ± 5.2 | 3.4 ± 2.6 | |
| C37 | 3 | V351E | 26.1 ± 4.6* | 69.0 ± 4.3* | 5.0 ± 2.3 |
| C38 | 0.75 | ||||
| C39 | 3 | L826F | |||
| C40 | 0.25 | 26.0 ± 5.2 | 68.4 ± 4.4 | 5.6 ± 1.9 | |
| C41 | 3 | M347R | 30.0 ± 3.1 | 59.7 ± 3.3* | 10.3 ± 3.7* |
| C42 | 0.75 | ||||
| C43 | 3 | S337L | |||
| C46 | 0.38 | ||||
| C47 | 3 | L826F | |||
| C48 | 0.5 | ||||
| C49 | 2 | T345I | |||
| C50 | 0.5 | ||||
| C51 | 2 | T345I | |||
aPairs of isolates are represented by alternate no shading and shading, with the first strain in each pair being the DAP-S parental strain and the second being the DAP-R strain.
bThese data have been published before.1
cPositions of nucleotide change within mprF ORFs.
*P < 0.05 versus respective DAP-S parental strain.
All strains were grown in either Tryptic Soy Broth (TSB; Difco Laboratories, Detroit, MI, USA) or Mueller–Hinton broth (MH; Difco Laboratories) depending on the individual experiments. Liquid cultures were grown in Erlenmeyer flasks at 37°C with shaking (250 rpm) in a volume that was ≤10% of the flask volume. DAP-R was defined as an Etest MIC of ≥2 mg/L.7
DNA isolation and sequencing
Genomic DNA was isolated from the S. aureus study strains using the method of Dyer and Iandolo.8 PCR amplification of the graRS ORFs was performed as previously described4,6 and DNA sequencing of the graRS ORFs was kindly performed at City of Hope, Duarte, CA, USA. Sequencing of the mprF promoter (∼500 bp upstream of the ATG start codon) was performed in the selected strain pairs using the primer pair mprF-pro-F (5′-CCCGAATTCTATGGTAATGATGTAGGTGAATATG-3′) and mprF-pro-R (5′-CCCTCTAGAGCTGTAGCAAACGTAATT-3′).
RNA isolation and quantitative real-time PCR
Fresh overnight cultures of S. aureus strains were used to inoculate TSB to an OD600 of 0.1. Total RNA was then obtained from either exponential (2.5 h) or late stationary (12 h) growth-phase cultures using previously described methods.6
For quantitative real-time PCR analyses, 2 μg of DNase-treated RNA was reverse transcribed using the SuperScript III First-Strand Synthesis kit (Invitrogen) according to the manufacturer's protocols. Quantification of cDNA levels was performed following the instructions of the Power SYBR® Green Master Mix kit (Applied Biosystems) on an ABI PRISM 7000 Sequence Detection System in triplicate samples. The mprF, dltA and gyrB genes were detected using respective specific primers as described before.6,9–11 Expression of the graS gene was detected using the primer pair qRT-F-graS (5′-GATGGGTTTTATTGGTGCAGA-3′) and qRT-R-graS (5′-CATTGGTATAGAACGG TTTTTGC-3′). Fold changes in expression levels of each target gene were quantified in relation to gyrB. A minimum of two independent runs were performed for each RNA sample.
CM phospholipid composition
The three major S. aureus CM phospholipids are phosphatidylglycerol, L-PG and cardiolipin.12 To quantify the relative proportions of these three phospholipids in our strain sets, CM phospholipids were extracted from the selected S. aureus strain pairs as described previously.13,14 The target phospholipids were separated and identified via two-dimensional thin-layer chromatography, then quantified by spectrophotometric assay, as described before.13,14 At least three independent experiments were performed.
Statistical analysis
The Kruskal–Wallis ANOVA test with the Tukey post hoc correction for multiple comparisons was utilized where indicated. Significance was accepted at P < 0.05.
Results and discussion
Expression of mprF and L-PG synthesis among DAP-R strains
As shown in Figure 1(a), quantitative real-time PCR analyses of mprF transcripts revealed that 10/22 DAP-R strains had enhanced mprF expression versus their respective isogenic DAP-S parental strains during either the exponential growth phase or the stationary growth phase. Among these 10 strains, this increase in mprF expression was statistically significant in 5 (50%). Of note, only the C37 strain exhibited significantly enhanced expression of mprF during both growth phases. These results suggest that, although altered expression of mprF among DAP-R MRSA isolates is relatively frequent, significantly altered mprF expression is less common. Although the C6, C8, C10, C25 and C37 strains had significantly increased mprF transcription, the mprF promoter sequences of these strains were identical to those of their respective DAP-S parental strains (data not shown). These data suggested that the increased mprF expression in these five DAP-R strains was not related to mutations within the mprF promoter.
Figure 1.
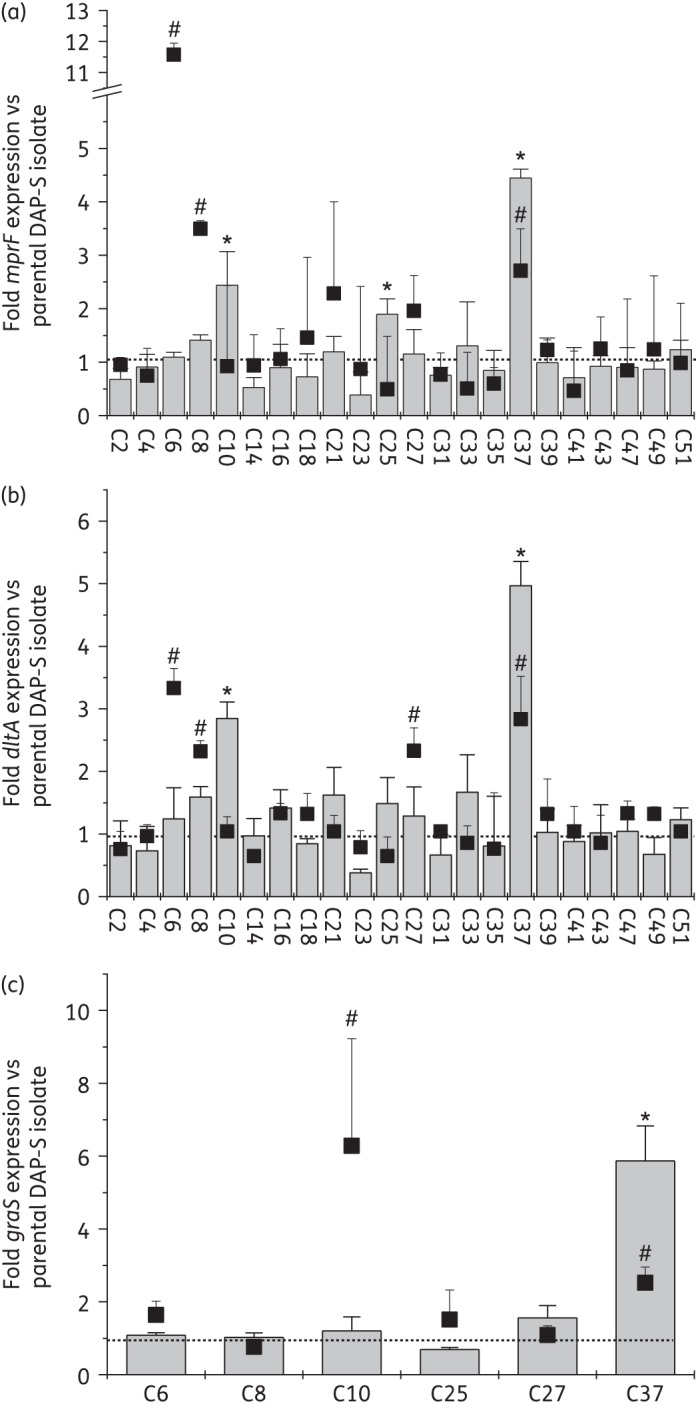
Transcription of mprF (a), dltA (b) and graS (c) genes among the study strains during exponential growth phase (bar graphs) and stationary growth phase (overlaid scatter plots with filled squares). Total cellular RNA samples from the strains grown in TSB medium were isolated at 2.5 h (exponential growth phase) or 12 h (stationary phase) post-inoculation and subjected to quantitative real-time PCR analyses. Each DAP-S parental S. aureus strain was normalized to 1. *P < 0.01 versus respective DAP-S parental strains during exponential growth phase. #P < 0.01 versus respective DAP-S parental strains during stationary phase.
Next, to assess the correlation between altered expression of mprF and L-PG synthesis, eight DAP-S and DAP-R pairs were selected for phospholipid compositional analyses depending on their mprF expression profiles (Figure 1a): (i) DAP-R strains expressing significantly increased mprF transcripts in the exponential and/or stationary phase; (ii) DAP-R strains displaying no changes in mprF expression; (iii) DAP-R strains expressing marginally increased mprF transcripts during both growth phases; and (iv) DAP-R strains expressing marginally increased mprF transcript only during the exponential growth phase.
The proportion of total L-PG within the overall phospholipid content was significantly increased in the DAP-R strains (C6, C10 and C37) exhibiting significantly increased mprF transcription in exponential, stationary or both growth phases versus the DAP-S parental strains (Figure 1a and Table 1). In contrast, no significant changes in L-PG synthesis were observed in the two DAP-R strains (C4 and C41) that did not show any changes in mprF transcription in either growth phase versus the respective parental DAP-S strains. Of the two DAP-R strains (C21 and C27) that exhibited a marginal increase in mprF transcription during both growth phases, only C21 displayed significantly increased L-PG synthesis. The C33 strain, which displayed a marginal increase in mprF transcription only during the exponential growth phase, showed no increase in L-PG production versus the DAP-S parental strain (C32) (Table 1).
Our current data were generally in line with prior observations,2,11,15 showing that only when mprF expression is significantly enhanced in either growth phase is L-PG production significantly increased. Interestingly, when mprF transcription was increased by an insignificant amount, increased L-PG production was occasionally observed among DAP-R isolates.
Expression of dltABCD among DAP-R strains
The dltABCD operon also contributes to the staphylococcal net positive surface charge by d-alanylating cell-wall teichoic acids through distinct effector mechanisms.16 As shown in Figure 1(b), 14 of the 22 DAP-R strains exhibited enhanced dltA expression profiles during exponential or stationary growth phases as compared with their respective DAP-S parental strains. This increased dltA expression in DAP-R versus DAP-S strains was significant in five isolates (23%) (C6, C8, C10, C27 and C37). Importantly, in four of these five isolates (C6, C8, C10 and C37) enhanced mprF expression was also seen, suggesting a co-regulation scenario by the upstream transcriptional regulator(s), graRS.4–6
Overall analyses of mprF and dltA expression in the 22 clinical DAP-R MRSA strains revealed that, among the 22 strain pairs, six DAP-R isolates (∼27%) exhibited significantly altered expression of mprF and/or dltABCD during exponential- and/or stationary-phase growth. Of interest, five of these six strains contained mprF SNPs within the central bifunctional domains (n = 4) or within the synthase domain (n = 1). One isolate had no mprF SNP (Table 1).
The six DAP-R strains with significantly enhanced mprF and/or dltA expression were grouped and compared for surface positive charge profiles using the previously published cytochrome c binding data1 versus the remaining 16 DAP-R strains; there was no significant difference in surface charge profiles between the two groups (P = 0.6; data not shown). These data suggest that dysregulation of mprF and/or dltABCD expression alone is not enough to explain the enhanced surface positive charge observed in some clinical DAP-R strains.
Expression of graRS among DAP-R strains
Since 6/22 DAP-R strains exhibited enhanced expression of mprF and/or dltA genes, we assessed the potential role of graRS induction in the increased mprF and dltABCD transcription in these 6 DAP-R isolates. As shown in Figure 1(c), only strains C10 and C37 showed enhanced expression of graS during either exponential- or stationary-phase growth. To assess whether the enhanced expression of graRS in these two DAP-R strains was linked to altered promoter structure, graRS promoter regions were sequenced; this revealed that the graRS promoter sequences of these two DAP-R strains were identical to those of their respective DAP-S parental strains (data not shown).
We next sequenced the graRS ORFs of these six DAP-R strains to assess the potential correlation of DAP-R, enhanced mprF/dltABCD expression and the presence of SNPs within the graRS ORF. ORF sequencing analyses revealed that these six DAP-R strains displaying increased mprF/dltABCD expression had graRS ORF sequences identical to those of their respective DAP-S parental strains (data not shown). Thus, dysregulation and enhanced expression of mprF and dlt could not be linked to mutations within either the promoter or the ORF regions of graRS. Therefore, future studies should endeavour to extend knowledge of regulatory mechanisms/factors affecting mprF and dltABCD expression beyond the graRS system.
Funding
This research was supported by grants from the National Institutes of Health (5RO1-AI-39108-17 to A. S. B. and 1RO1-AI-091801-05 to A. L. C.). This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015R1D1A1A01057442 to S.-J. Y.).
Transparency declarations
None to declare.
Acknowledgements
We thank Kuan-Tsen, Steven N. Ellison, Rachelle Gonzales and Danya N. Alvarez for excellent technical assistance with quantitative real-time PCR analyses, graRS sequencing and chemical analyses of phospholipids.
References
- 1.Bayer AS, Mishra NN, Chen L et al. Frequency and distribution of single-nucleotide polymorphisms within mprF in methicillin-resistant Staphylococcus aureus clinical isolates and their role in cross-resistance to daptomycin and host defense antimicrobial peptides. Antimicrob Agents Chemother 2015; 59: 4930–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer AS, Mishra NN, Sakoulas G et al. Heterogeneity of mprF sequences in methicillin-resistant Staphylococcus aureus clinical isolates: role in cross-resistance between daptomycin and host defense antimicrobial peptides. Antimicrob Agents Chemother 2014; 58: 7462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murthy MH, Olson ME, Wickert RE et al. Daptomycin non-susceptible methicillin-resistant Staphylococcus aureus USA 300 isolate. J Med Microbiol 2008; 57: 1036–8. [DOI] [PubMed] [Google Scholar]
- 4.Cheung AL, Bayer AS, Yeaman MR et al. Site-specific mutation of the sensor kinase GraS in Staphylococcus aureus alters the adaptive response to distinct cationic antimicrobial peptides. Infect Immun 2014; 82: 5336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meehl M, Herbert S, Gotz F et al. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother 2007; 51: 2679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S-J, Bayer AS, Mishra NN et al. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun 2012; 80: 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher HW, Sakoulas G. Antimicrobial resistance: perspectives on daptomycin resistance, with emphasis on resistance in Staphylococcus aureus. Clin Infect Dis 2007; 45: 601–8. [DOI] [PubMed] [Google Scholar]
- 8.Dyer DW, Iandolo JJ. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol 1983; 46: 283–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertsche U, Weidenmaier C, Kuehner D et al. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and d-alanylation. Antimicrob Agents Chemother 2011; 55: 3922–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang SJ, Kreiswirth BN, Sakoulas G et al. Enhanced expression of dltABCD is associated with development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J Infect Dis 2009; 200: 1916–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang SJ, Xiong YQ, Dunman PM et al. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus. Antimicrob Agents Chemother 2009; 53: 2636–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst C, Staubitz P, Mishra NN et al. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 2009; 5: e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra NN, Yang SJ, Sawa A et al. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus (MRSA). Antimicrob Agents Chemother 2009; 53: 2312–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay K, Whitmire W, Xiong YQ et al. In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 2007; 153: 1187–97. [DOI] [PubMed] [Google Scholar]
- 15.Yang S-J, Nast CC, Mishra NN et al. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob Agents Chemother 2010; 54: 3079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weidenmaier C, Peschel A, Kempf VAJ et al. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect Immun 2005; 73: 8033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


