Abstract
Objectives
Escherichia coli is the most common agent of bacteraemia, bacterial gastroenteritis and urinary tract infections (UTIs). Lineages causing UTIs and gastrointestinal disease are well defined, but less is known about those causing bacteraemia. We therefore investigated the population structure of E. coli from bacteraemia in the UK and Ireland between 2001 and 2010.
Methods
E. coli isolates (n = 2166) were submitted to the BSAC Bacteraemia Surveillance Programme from 18 UK and Irish centres from 2001 to 2010. Genotypes were analysed by MLST using the Achtman scheme; MICs, blaCTX-M group and patient demographics were previously determined in the BSAC surveillance.
Results
Four hundred and forty-eight STs were identified, but five of these, and their associated clonal complexes (CCs), accounted for 58.4% (1264 of 2166) of isolates: CC73 was the most common (20.7%), followed by CC131 (13.9%), CC95 (11.3%), CC69 (6.9%) and CC12 (5.5%). All these, except CC69 (group D), belong to phylogenetic group B2. CC131 isolates were much more often MDR than other STs were: they rose from 2.9% of isolates in 2001 to 20.5%–20.7% in 2007–08 and then declined to 14.3% in 2010. Resistance rates to cephalosporins, aminoglycosides and fluoroquinolones remained below 10% in other major CCs throughout.
Conclusions
The five most prevalent bacteraemia STs have all been associated previously with UTIs. They dominated in all years, but their proportions fluctuated, most notably for ST131, a globally disseminated high-risk clone that is often MDR.
Introduction
The incidence of Escherichia coli bacteraemia has recently increased progressively and, by 2013, the species accounted for 32% of all bacteraemias reported to Public Health England in the UK except Scotland.1 Reporting became mandatory in England in June 2011, with 34 275 cases in fiscal 2013–14.2 Mortality is 10%–30%,3 rising to 60% for MDR infections.4
Understanding the distribution of E. coli strains in bacteraemia could elucidate routes to improve control, for example by tailoring antibiotic use to the strain, or developing vaccines against major lineages. Here, we studied the population structure of bloodstream E. coli from 18 hospitals in England, Ireland, Northern Ireland and Wales over a decade.
Materials and methods
Bacterial strains
E. coli isolates (n = 2166) from patients with bacteraemia were collected under the aegis of the BSAC Bacteraemia Surveillance Programme.5,6 They comprised all the E. coli submitted from 18 centres that participated in the surveillance throughout, with ∼10 isolates per centre in 2001–07, 20 per centre in 2008 and 2009 and 14 per centre in 2010. MICs and blaCTX-M groups were determined by published methods,7,8 and supplied by the BSAC along with basic demographic information. Susceptibility data were reviewed against 2014 EUCAST/BSAC breakpoints, which may differ from those when the isolates were collected. Amoxicillin/clavulanate had been tested as a 2:1 ratio and was reviewed against the previous 8+4 mg/L breakpoint, not the current 8 + 2 (fixed) mg/L criterion.
DNA extraction
DNA extraction was performed using a QIAxtractor robot, with DX reagents and plasticware, according to the manufacturer's instructions (QIAxtractor; Qiagen, Crawley, UK).
Phylotyping and MLST
Isolates were assigned to the major phylogenetic groups of E. coli (A, B1, B2 and D) by multiplex PCR.9 MLST was performed as described.10 Data were assembled and analysed in BioNumerics (v6.1; Applied Maths, Keistraat, Belgium); isolates were assigned allele numbers and STs via the Warwick database.11
Statistical analysis
Clonal complexes (CCs) were grouped as CC73, CC131, CC95, CC69, CC12 and ‘others’ for analysis, with CC73 (the largest group) as ‘baseline’. Factors predicting CC were explored in multinomial logistic regression models. Intermediate and resistant isolates were combined as ‘non-susceptible’ for analysis. Candidate models were compared by Akaike information criterion. The final models used cluster-robust standard errors to allow for clustering by collection centre.
Results and discussion
Molecular typing
The 2166 isolates belonged to 448 different STs (Table S1, available as Supplementary data at JAC Online), 230 of which had new alleles or new allelic combinations not described in the Warwick database. Nevertheless 47% (1020 of 2166) belonged to five major STs, namely ST73 (16.8%; n = 363), ST131 (12.0%; n = 260), ST95 (8.5%; n = 184), ST69 (5.4%; n = 117) and ST12 (4.4%; n= 96). If single locus variants and double locus variants were included, the proportion belonging to the five major CCs rose to 58.4% (1264 of 2166) (Table 1).
Table 1.
Non-susceptibility to antimicrobials in relation to CC
| CC | Total no. | Patient sex (%) |
Age (years) (%) |
Onset (%) |
Non-susceptibility (%) |
β-Lactamase (%) |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N/R | female | male | N/R | 0–19 | 20–49 | 50–69 | 70+ | N/R | ≤48 h | >48 h | AMX | AMC | TZP | CTXa | CAZ | CIP | GEN | TGCa | IPM | CTX-M otherb | CTX-M-1 | CTX-M-9 | ESBL, non-CTX-M | non-ESBL | ||
| 73 | 449 | 0.2 | 59.5 | 40.3 | 0.0 | 3.6 | 12.5 | 20.7 | 63.3 | 1.3 | 64.4 | 34.3 | 55.9 | 27.8 | 9.1 | 1.5 | 1.8 | 1.3 | 1.6 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 | 98.9 |
| 131 | 302 | 0.3 | 42.7 | 57.0 | 0.7 | 3.0 | 6.6 | 25.8 | 63.9 | 0.0 | 50.0 | 50.0 | 83.4 | 59.3 | 22.2 | 35.0 | 29.5 | 64.2 | 20.2 | 0.0 | 0.0 | 0.3 | 32.5 | 0.3 | 1.0 | 65.9 |
| 95 | 245 | 0.0 | 63.7 | 36.3 | 0.4 | 9.4 | 13.5 | 23.7 | 53.1 | 0.8 | 64.5 | 34.7 | 45.3 | 13.9 | 2.9 | 0.0 | 0.0 | 0.4 | 2.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100 |
| 69 | 149 | 0.0 | 72.5 | 27.5 | 0.0 | 2.7 | 18.1 | 23.5 | 55.7 | 0.7 | 65.8 | 33.6 | 81.9 | 32.2 | 10.1 | 1.4 | 2.0 | 6.7 | 4.0 | 0.7 | 0.0 | 0.0 | 0.7 | 0.0 | 0.0 | 99.3 |
| 12 | 119 | 0.0 | 49.6 | 50.4 | 0.8 | 3.4 | 11.8 | 22.7 | 61.3 | 0.0 | 68.9 | 31.1 | 71.4 | 28.6 | 7.6 | 1.8 | 5.0 | 0.8 | 3.4 | 0.0 | 0.0 | 0.0 | 0.8 | 0.0 | 1.7 | 97.5 |
| Otherc | 902 | 0.2 | 50.4 | 49.3 | 0.4 | 4.5 | 11.2 | 27.3 | 56.5 | 1.1 | 57.0 | 41.9 | 60.9 | 27.7 | 9.3 | 5.3 | 5.3 | 15.2 | 6.4 | 0.1 | 0.2 | 0.2 | 2.2 | 0.1 | 1.0 | 96.5 |
| Total | 2166 | 0.2 | 54.2 | 45.6 | 0.4 | 4.5 | 11.6 | 24.8 | 58.8 | 0.9 | 59.6 | 39.5 | 63.3 | 30.9 | 10.3 | 7.9 | 7.1 | 16.1 | 6.6 | 0.1 | 0.1 | 0.1 | 5.5 | 0.1 | 0.9 | 93.4 |
Onset ≤48 h considered as community onset and onset >48 h considered as hospital onset.
Values shaded grey are significantly different (P ≤ 0.0001) compared with the baseline CC73 group.
Values in bold are significant (P ≤ 0.05) compared with the baseline CC73 group.
N/R, not reported; AMX, amoxicillin; AMC, amoxicillin/clavulanate; TZP, piperacillin/tazobactam; CTX, cefotaxime; CAZ, ceftazidime; CIP, ciprofloxacin; GEN, gentamicin; TGC, tigecycline; IPM, imipenem.
aCefotaxime and tigecycline were tested in 2002–10 only.
b‘CTX-M other’ indicates an unidentified CTX-M group.
c‘Other’ CCs includes 902 isolates representing 443 STs, with no more than 65 isolates belonging to any CC.
CC12, CC73, CC95 and CC131 isolates all belonged to phylogenetic group B2, and CC69 isolates belonged to group D. More generally, phylogroups B2 (68.4%; 1481 of 2166) and D (19.0%; 412 of 2166) dominated the entire collection and are the E. coli groups typically associated with virulent extra-intestinal infections.
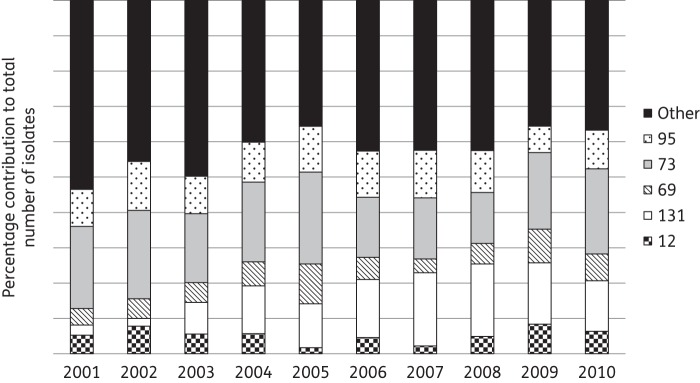
Overall, the proportion of isolates belonging to the five major CCs rose from 46.5% (80 of 172) in 2001 to 63.3% (150 of 237) in 2010 (P = 0.001 for trend). The prevalence of the five individual major CCs fluctuated, most noticeably for CC131, which rose from 2.9% (5 of 172 isolates) in 2001 to a peak of 20.5%–20.7% (37 of 179 and 71 of 346) in 2007 and 2008 (Figure 1). CC73 was most prevalent between 2001 and 2006, but was surpassed by CC131 in 2007–08, only to become the most prevalent CC again in 2009 and 2010. The decline of ST131 may reflect the change in prescribing practice in the UK away from cephalosporins and quinolones.12
Figure 1.
Proportions of different CCs among bloodstream E. coli over time. ‘Other’ CCs includes 902 isolates representing 443 STs, with no more than 65 isolates belonging to any CC and with 96 isolates being sole representatives of unique STs.
Antibiotic resistance
Except for amoxicillin and amoxicillin/clavulanate there was little resistance (≤10%) to tested antimicrobials in CC12, CC69, CC73 or CC95, but resistance to cephalosporins, fluoroquinolones and aminoglycosides was prevalent in CC131 (Table 1). Other CCs showed smaller, but still significant (P ≤ 0.02), differences from CC73, which served as the reference; thus CC69 and ‘other’ CC isolates were more often resistant to ciprofloxacin and gentamicin, and CC69 and CC12 isolates were more often resistant to amoxicillin, whereas CC95 isolates were less often resistant to β-lactams and their inhibitor combinations (Table 1).
Non-susceptibility generally rose from 2001 to 2006–08, including for amoxicillin/clavulanate, cefotaxime, ceftazidime, ciprofloxacin and gentamicin, though with some subsequent decline in 2009–10 for cephalosporins and ciprofloxacin, as outlined elsewhere.12,13 These trends were not detected within any of the five major CC groups or ‘others’, but, rather, reflected the fluctuating proportion of the frequently MDR CC131 isolates.
ESBL phenotypes were seen for 144 (6.6%) isolates. Among these, 86.8% (125 of 144) had blaCTX-M genes, 96.0% (120 of 125) of them had blaCTX-M-group-1 types. Predictably, non-susceptibility to other agents was more prevalent in ESBL producers than those without ESBLs. blaCTX-M-group-1 genes were present in 98 of 302 CC131 isolates (32.5%) (Table 1), with CC131 accounting for 81.7% (98 of 120) of all blaCTX-M-group-1 genes. The remaining 22 isolates with blaCTX-M-group-1 genes were spread across 11 CCs, with CC23 (5 of 120; 4.2%) and CC10 (4 of 120; 3.3%) accounting for the largest (but still small) shares. Much less common were blaCTX-M-group-9 genes, found in single ST131 and ST405 isolates, and non-CTX-M ESBLs, with 19 producers spread across 11 CCs, but most often in CC73 (5 isolates) (Table 1). Only two isolates were non-susceptible to imipenem, with MICs of 8 and 16 mg/L; both belonged to CC405 and were from different hospitals in southern England, they were isolated in 2006 and had a CTX-M ESBL with impermeability.13 No carbapenemase producers were collected.
CC distribution in relation to patient demographics
There was an association between patient sex and CC (P < 0.0001); specifically, and compared with CC73, CC131 and ‘other’ CCs were significantly more often associated with men, whereas CC69 was more associated with women, as noted previously by Gibreel et al.14 Age, by contrast, had little association with CC after accounting for sex. Patients with CC131 isolates divided equally between those with onset ≤48 or >48 h after hospitalization, whereas onset at ≤48 h was twice as frequent for patients with CC12, 69, 73 or 95 isolates. Hospital onset thus was a significant predictor of CC (P < 0.0001), with CC131 and ‘others’ being more associated with hospital-onset bacteraemias. Previous studies have shown greater 30 day mortality for hospital than community-acquired E. coli bacteraemias (28% versus 14%),15 though it is unclear if this reflects the strains, their resistances or the underlying health status of hospital versus community cases.
Presumed-source-of-bacteraemia data were available for 1446 isolates (66.8%). Overall, and for each of the top five CCs individually, urinary tract infections were the most common source (66.4%; 960 of 1446) followed by intra-abdominal infection (17.6%; 254 of 1446). Presumed source (genitourinary versus other) was a strong predictor of CC (P < 0.0001), with CC69 significantly more associated with a genitourinary source than CC73, whereas ‘others’ were significantly less associated with a genitourinary source.
Conclusions
Overall, these results show the dominance, for a decade, of the same five E. coli CCs as agents of bacteraemia in the UK and Ireland, namely CC73, CC131, CC95, CC69 and CC12. This dominance increased over time or, looked at another way, these five types accounted for much of the overall increase in E. coli bacteraemias during the decade. In addition, these five CCs have been associated with urinary tract infections in (i) north-west England (2007–09) and their proportions were 16.6% (ST73), 13.3% (ST131), 9% (ST69), 6% (ST95) and 4.3% (ST10),14 and (ii) Yorkshire and Humber (2010–12) where ST131, ST73 and ST95 were predominant.16 ST131 (59%)—which is an internationally distributed ‘high-risk clone’ notorious for fluoroquinolone resistance and producing CTX-M ESBLs—dominated among cephalosporin-resistant urinary isolates in north-west England in 2004/2005,17 whereas, as here, resistance rates in ST73 and ST95 were low, both at 6.8%.
The association of bacteraemia with just a few E. coli lineages, and of MDR with just one of these (CC131), along with the increased mortality associated with resistance and treatment inadequacy, suggest that it may be both practicable and beneficial to identify strain lineages rapidly, predicting the likelihood of resistance to standard therapies. This could be done, e.g. by PCR from early growth in blood culture bottles,18 allowing swifter adaptation of therapy than is possible based on conventional testing. The low rates of resistance in the other top CCs besides CC131 imply that their success reflects some other selective advantage and STs 131, 69, 12 and 127 have all been reported to have unusually high virulence scores.14,19,20
Funding
This work was supported by the National Institute of Health Strategic Research and Development Grant.
Transparency declarations
D. M. L. is partly self-employed and consults for numerous pharmaceutical and diagnostic companies, including Achaogen, Adenium, Allecra, Astellas, AstraZeneca, Bayer, Basilea, bioMérieux, bioVersys, Cubist, Curetis, GSK, Longitude, Merck, Meiji Seika, Pfizer, Roche, Tetraphase, VenatoRx and Wockhardt, he holds grants from AstraZeneca, Basilea, Cubist, Meiji Seika, Merck and VenatoRx, he has received lecture honoraria or travel reimbursement from AstraZeneca, Curetis, GSK, J&J, Leo, Meiji, Merck, Novartis, Pfizer and Tetraphase, and he holds shares in Dechra, GSK, Merck and Pfizer, collectively amounting to <10% of portfolio value.
N. W. has no personal interests to declare. However, PHE's Antimicrobial Resistance and Healthcare Associated Infections Reference Unit has received financial support from numerous sources, including: Achaogen Inc., Allecra Antiinfectives GmbH, Amplex, AstraZeneca UK Ltd, Becton Dickinson Diagnostics, bioMérieux, Bio-Rad Laboratories Ltd, BSAC, Cepheid, Check-Points B.V., Cubist Pharmaceuticals, Department of Health, Enigma Diagnostics Ltd, Food Standards Agency, Glaxo SmithKline Services Ltd, Henry Stewart Talks, IHMA Ltd, Merck Sharpe & Dohme Corp., Meiji Seika Kiasya Ltd, Melinta Therapeutics Inc., Mobidiag, Momentum Bioscience Ltd, Nordic Pharma Ltd, Norgine Pharmaceuticals, Rempex Pharmaceuticals Ltd, Rokitan Ltd, Smith & Nephew UK Ltd, Tetraphase Pharmaceuticals, Trius Therapeutics, VenatoRx and Wockhardt Ltd.
All other authors: none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
We would like to thank BSAC for the provision of isolates and related data for this study and the University of Oxford Zoology sequencing service for sequence determination.
References
- 1.PHE. Escherichia coli Bacteraemia, Voluntary Surveillance. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/339992/Ecoli_bacteraemia_in_Eng_Wales_and_NI_2009_to_2013.pdf.
- 2.PHE. Quarterly Counts of E. coli Bacteraemias by Clinical Commissioning Group. https://www.gov.uk/government/statistics/escherichia-coli-e-coli-bacteraemia-annual-data.
- 3.Gransden WR, Eykyn SJ, Phillips I et al. Bacteremia due to Escherichia coli: a study of 861 episodes. Clin Infect Dis 1990; 12: 1008–18. [DOI] [PubMed] [Google Scholar]
- 4.Melzer M, Petersen I. Mortality following bacteraemic infection caused by extended spectrum β-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J Infect 2007; 55: 254–9. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds R, Hope R, Williams L. Survey, laboratory and statistical methods for the BSAC Resistance Surveillance Programmes. J Antimicrob Chemother 2008; 62(Suppl 2): ii15–28. [DOI] [PubMed] [Google Scholar]
- 6.BSAC Resistance Surveillance Project. http://www.bsacsurv.org/.
- 7.Andrews JM. Determination of Minimum Inhibitory Concentration. http://bsac.org.uk/wp-content/uploads/2012/02/Chapter-2-Determination-of-MICs-2006updated.pdf. [DOI] [PubMed] [Google Scholar]
- 8.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother 2006; 57: 154–5. [DOI] [PubMed] [Google Scholar]
- 9.Doumith M, Day MJ, Hope R et al. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J Clin Microbiol 2012; 50: 3108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirth T, Falush D, Lan R et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006; 60: 1136–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warwick MLST Database. http://mlst.warwick.ac.uk/mlst/dbs/Ecoli.
- 12.Livermore DM, Hope R, Reynolds R et al. Declining cephalosporin and fluoroquinolone non-susceptibility among bloodstream Enterobacteriaceae from the UK: links to prescribing change? J Antimicrob Chemother 2013; 68: 2667–74. [DOI] [PubMed] [Google Scholar]
- 13.Livermore DM, Hope R, Brick G. Non-susceptibility trends among Enterobacteriaceae from bacteraemias in the UK and Ireland, 2001-06. J Antimicrob Chemother 2008; 62(Suppl 2): ii41–54. [DOI] [PubMed] [Google Scholar]
- 14.Gibreel TM, Dodgson AR, Cheesbrough J et al. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 2012; 67: 346–56. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JR, Nicolas-Chanoine MH, DebRoy C et al. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967-2009. Emerg Infect Dis 2012; 18: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horner C, Fawley W, Morris K et al. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010–12). J Antimicrob Chemother 2014; 69: 91–100. [DOI] [PubMed] [Google Scholar]
- 17.Lau SH, Reddy S, Cheesbrough J et al. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 2008; 46: 1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doumith M, Day M, Ciesielczuk H et al. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J Clin Microbiol 2015; 53: 160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco J, Mora A, Mamani R et al. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother 2011; 66: 2011–21. [DOI] [PubMed] [Google Scholar]
- 20.Alghoribi MF, Gibreel TM, Dodgson AR et al. Galleria mellonella infection model demonstrates high lethality of ST69 and ST127 uropathogenic E. coli. PLoS One 2014; 9: e101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.