Abstract
Objectives
The objective of this study was to assess the presence of mcr-1 in Shigella sonnei isolated in Vietnam.
Methods
WGS data were analysed for the presence of the mcr-1 gene sequence. The association of mcr-1 with a plasmid was assessed by PCR and by conjugation.
Results
Through genome sequencing we identified a plasmid-associated inactive form of mcr-1 in a 2008 Vietnamese isolate of Shigella sonnei. The plasmid was conjugated into Escherichia coli and mcr-1 was activated upon exposure to colistin, resulting in highly colistin-resistant transconjugants.
Conclusions
This is the first description of the mcr-1 gene in Shigella, which is atypical given that colistin is not ordinarily used to treat diarrhoea. Our data suggest the mcr-1 gene has been circulating in human-restricted pathogens for some time but likely carries a selective fitness cost.
Introduction
The mcr-1 gene, which confers resistance to colistin, was recently described in Enterobacteriaceae in China.1 The index isolate, Escherichia coli SHP45, was isolated at a pig farm in Shanghai in July 2013 and displayed a colistin MIC of 8 mg/L. mcr-1 was associated with a transposable element located on an IncI2 plasmid, pHNSHP45. Subsequent PCR screening using primers targeting the 5′ end of mcr-1 detected the gene in ∼20% of E. coli isolated from pigs at slaughter and 15% of E. coli isolated from retail meat in China. The gene was also detected amongst clinical isolates of various Gram-negative bacteria cultured from inpatients in Chinese hospitals (1.4% of E. coli and 0.7% of Klebsiella pneumoniae).
By screening available isolate collections via PCR, or mining WGS data, many research groups have now reported the presence of mcr-1 from widespread geographical locations and sources (food, animal and humans in South-East Asia, Europe and Africa, and in travellers returning to Europe from South-East Asia, South America and Africa).2–4 The gene has been detected in multiple Salmonella serotypes as well as numerous E. coli sequence types2,5–7 and can be carried on various plasmid backbones, including IncI2, IncX4, IncHI2 and IncP.2,7,8 We previously published a WGS study of >200 Vietnamese Shigella sonnei isolated from children with dysenteric diarrhoea between 1995 and 2010.9 Here we report the detection of an inactivated form of the mcr-1 gene in one of these S. sonnei isolates and its selective re-activation, resulting in high-level transferrable colistin resistance.
Methods
Screening for mcr-1 and IncI2 plasmids
Raw WGS data (56 bp paired-end Illumina HiSeq reads) generated previously from genomic DNA extracted from Vietnamese S. sonnei9 were screened for mcr-1 using SRST2,10 which allows the detection of genes of interest direct from short reads and with higher sensitivity than assembly-based approaches. IncI2 plasmids were detected by mapping reads to the pEG430-1 plasmid sequence using Bowtie2.11 Nucleotide divergence was assessed by SNP calling using SAMtools as previously described.9
Plasmid DNA extraction and sequencing
Plasmid DNA was extracted from S. sonnei EG430 using the Qiagen Plasmid Midi Kit (Qiagen, Germany) and sequenced via Illumina MiSeq (Illumina, USA) to generate 250 bp paired-end reads, following the manufacturer's recommendations. Assemblies were generated using SPAdes v3.6.212 and the resulting plasmid sequences were annotated using Prokka v1.11.13 ACT (Artemis Comparison Tool)14 was used to compare the pEG430-1 sequence with that of the reference plasmid pHNSHP45 (accession number KP347127)1 and to perform manual curation of the plasmid annotation. The annotated plasmid sequence was deposited in the ENA database under accession number LT174530.
Bacterial conjugation was performed by combining equal volumes (5 mL) of overnight LB cultures (∼5 × 108 cfu/mL) of S. sonnei EG430 and E. coli J53 (sodium azide resistant). Bacteria were conjugated for 12 h in LB broth at 37°C and E. coli transconjugants were selected on medium containing sodium azide (100 mg/L) and azithromycin (24 mg/L). Azithromycin- and sodium azide-resistant E. coli were subjected to mcr-1 PCR amplification to identify mcr-1-positive transconjugants. Etests (AB Biodisk, Sweden) were used to determine the MIC of colistin for S. sonnei EG430, S. sonnei EG430 derivatives, E. coli J53 and E. coli J53 transconjugants. Susceptibility to colistin was determined using CLSI breakpoints for Pseudomonas spp. (susceptible, ≤2 mg/L; intermediate, 4 mg/L; resistant ≥8 mg/L).15
PCR and sequencing
PCRs were performed using HotStar Taq DNA polymerase (Qiagen, Germany) with the recommended concentrations of reagents under the following conditions: 95°C for 15 min, 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 1 min and 72°C for 5 min. PCR amplification of mcr-1 to identify transconjugants was performed using previously published primers.1 PCR amplification of mcr-1 to confirm copy number of the 22 bp tandem repeat in colistin-resistant strains was performed using the published forward primer CLR5-F1 (5′-CGGTCAGTCCGTTTGTTC-3′) and a custom reverse primer, MCR-indel-R1 (5′-TGGCTTACGCATATCAGG-3′); amplicons were sequenced using the amplification primers, using BigDye Terminators in both directions on an ABI 3130 sequencing machine (ABI, USA).
Results
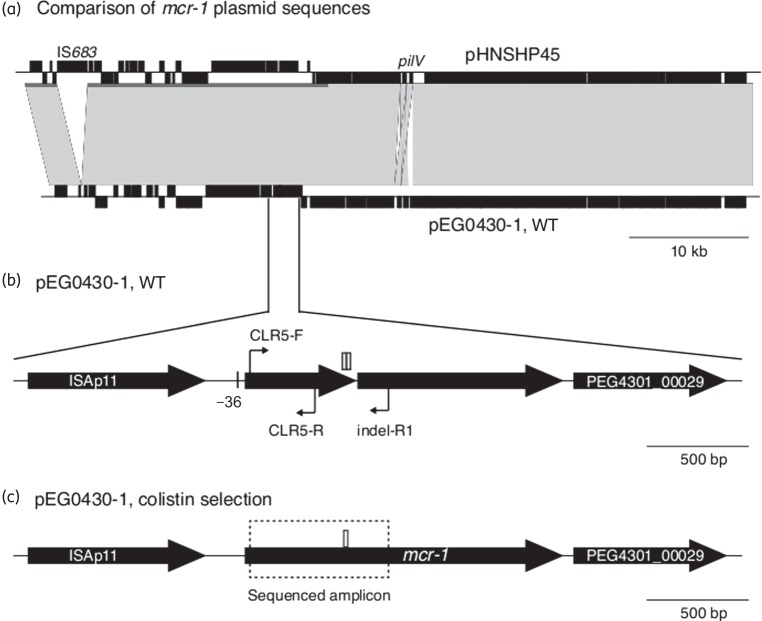
The mcr-1 gene was detected in the genome sequence of a single S. sonnei strain (EG430) that was isolated in 2008 from a hospitalized child with diarrhoea in Ho Chi Minh City, Vietnam. All other sequenced S. sonnei isolates from the same study were negative for mcr-1. Assembly analysis showed that the mcr-1 gene in EG430 was associated with an IncI2 plasmid backbone; however, the entire plasmid sequence could not be fully resolved using the 56 bp paired end reads.9 Upon antimicrobial susceptibility testing S. sonnei EG430 was found to be susceptible to colistin (MIC 0.094 mg/L) but resistant to azithromycin (MIC 24 mg/L) via an erm(B) gene. We attributed colistin susceptibility to a 22 bp duplication of bases 503–525 of the mcr-1 ORF (GAACGCCACCACAGGCAGTAAA), which induces a frameshift resulting in a truncated product (193 amino acids in length, compared with the 541 amino acid product encoded in pHNSHP45) (Figure 1). It is also possible that a single SNP upstream of mcr-1 (−36) in pE0G430-1 may affect expression of the gene.
Figure 1.
Schematic of mcr-1 plasmid sequences. (a) Comparison of novel mcr-1 plasmid pEG430-1 from S. sonnei EG430 (Vietnam, 2008) with mcr-1 plasmid pHNSHP45 from E. coli (China, 2013). Black blocks indicate protein-coding genes; genes on the forward and reverse strands are indicated above and below the line, respectively. Grey blocks indicate regions of sequence homology between the two plasmids. (b) Zoomed-in view of the mcr-1 mobile element in pEG430-1. Large arrows indicate ORFs (all are encoded on the forward strand). Small arrows indicate binding sites for PCR primers. Open blocks indicate the position of 22 bp tandem repeats, present in two copies in pEG430-1; the line marked ‘−36’ indicates the position of a point mutation relative to pHNSHP45. (c) Mobile element with restored mcr-1 sequence, identified in EG430 and E. coli transconjugants carrying pEG430-1 following selection on colistin. The dashed box shows the region that was amplified and sequenced using PCR (with primers CLR5-F and indel-R1), which confirmed restoration of the ORF via deletion of one copy of the 22 bp repeat.
Assuming mcr-1 and erm(B) were located on the same plasmid we performed conjugation of S. sonnei EG430, using E. coli J53 as a recipient, and selected for transconjugants using azithromycin as a marker. PCR screening for mcr-1 on azithromycin/sodium azide-resistant E. coli identified multiple mcr-1 PCR amplification-positive organisms. We subcultured the E. coli transconjugant, S. sonnei EG430 and E. coli J53 on LB medium containing a range of colistin concentrations (0.05–8 mg/L). Several colonies were identified and the organisms on the highest concentration of colistin were again subcultured on medium containing increasing colistin concentrations, from 8 to 32 mg/L. We were able to isolate both E. coli transconjugants and S. sonnei EG430 variants that were resistant to colistin (MIC 16 and 32 mg/L, respectively); no colonies were recovered from E. coli J53 at colistin concentrations above 4 mg/L. PCR amplification and sequencing of mcr-1 using a custom primer (previously published mcr-1 primers amplify upstream of the tandem repeat region) showed that, in the colistin-resistant E. coli and S. sonnei strains, one copy of the 22 bp tandem repeat had been deleted, restoring the ORF of mcr-1. We repeated this experiment multiple times and found that the re-activation of mcr-1 was consistently reproducible.
To investigate the genetic context of mcr-1 in detail, we isolated plasmid DNA from an E. coli transconjugant and sequenced it on an Illumina MiSeq to generate 250 bp paired-end reads. Combined assembly of the two read sets yielded two circular plasmid sequences—pEG430-1 carrying mcr-1 and pEG430-2 carrying erm(B)—and not just one plasmid with both determinants, as previously assumed. The pEG430-2 erm(B)-encoding plasmid sequence was 68 999 bp in size, carried an IncFII repA gene and shared close homology with E. coli plasmid pHK17a (accession number JF779678.1). The pEG430-1 mcr-1-encoding plasmid sequence was 61 826 bp in size and nearly identical to the previously described mcr-1-encoding plasmid pHNSHP45, differing by: (i) the lack of a 2704 bp insertion of IS683 downstream of the repA gene; (ii) a reorganization of the pilV shufflon; (iii) four single-base substitutions, including one 36 bp upstream of mcr-1; and (iv) the 22 bp tandem repeat in mcr-1 (Figure 1). IncI2 plasmid sequences were detected in 19 of the other Vietnamese S. sonnei isolates; however, these all lacked mcr-1 and displayed 0.3%–1.3% nucleotide divergence from pEG430-1.
Discussion
This is the first report describing mcr-1 in Shigella and the earliest example yet of mcr-1 in a human clinical isolate. The only earlier example of mcr-1 reported to date was in E. coli isolated from calves in France in 2005;6 however, the genetic context is not known. E. coli carrying mcr-1 were recently reported in Vietnam, isolated from a pig farm in Hanoi in 2014, 6 years after the isolation of EG430.16 These isolates carried mcr-1 in distinct, non-IncI2, plasmid backbones. However, our data show the acquisition of mcr-1 into an IncI2 plasmid backbone and its presence in the human population in Vietnam occurred at least as early as 2008.
Notably, the interruption we detected occurs downstream of the PCR primers used by Liu et al.1 and others to detect mcr-1,1 so it is possible that some isolates that test positive by PCR but not confirmed to be phenotypically resistant to colistin may carry this inactivated form of the gene. However, as we have shown, the gene can be restored again upon colistin exposure, so its presence even in inactive form could be problematic in settings where colistin is heavily used.
The fact that the mcr-1 gene was disrupted in S. sonnei EG430 is concerning. S. sonnei is a human-restricted pathogen with no known animal reservoir and is not likely to be regularly exposed to colistin. While the activity of Mcr-1 is not yet well understood, it appears to be a membrane-anchored enzyme with phosphoethanolamine transferase activity that likely confers resistance to colistin by modifying lipid A. We hypothesize that in the absence of colistin exposure, these modifications carry a fitness cost and impair interactions with the human host, such that the gene may be under negative selection. The MIC we observed in S. sonnei carrying mcr-1 (32 mg/L) is substantially higher than that reported previously for mcr-1-positive WT and transconjugant strains, which display a wide range (0.5–8 mg/L).1 We speculate that this variability in protection against colistin might be associated with the diversity of lipid A structures found in Enterobacteriaceae.
In conclusion, we have identified a deactivated version of the colistin resistance gene mcr-1 in the human-restricted pathogen S. sonnei, which was isolated from a Vietnamese child in 2008. Our data suggest this gene has likely been circulating in the human population in Asia in an inducible form, suggesting a fitness cost for the active mcr-1 gene.
Funding
This work was funded by the Wellcome Trust. D. P. T. is funded as a leadership fellow through the Oak Foundation. S. B. is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (100087/Z/12/Z). K. E. H. is supported by fellowship #1061409 from the NHMRC of Australia.
Transparency declarations
None to declare.
References
- 1.Liu YY, Wang Y, Walsh TR et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 2.Hasman H, Hammerum AM, Hansen F et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 2015; 20: pii = 30085. [DOI] [PubMed] [Google Scholar]
- 3.Arcilla MS, van Hattem JM, Matamoros S et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 147–9. [DOI] [PubMed] [Google Scholar]
- 4.Olaitan AO, Chabou S, Okdah L et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 147. [DOI] [PubMed] [Google Scholar]
- 5.Stoesser N, Mathers AJ, Moore CE et al. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis 2016; 16: 285–6. [DOI] [PubMed] [Google Scholar]
- 6.Haenni M, Poirel L, Kieffer N et al. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis 2016; 16: 281–2. [DOI] [PubMed] [Google Scholar]
- 7.Webb HE, Granier SA, Marault M et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 144–5. [DOI] [PubMed] [Google Scholar]
- 8.Tse H, Yuen KY. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 145–6. [DOI] [PubMed] [Google Scholar]
- 9.Holt KE, Thieu Nga TV, Thanh DP et al. Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc Natl Acad Sci USA 2013; 110: 17522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inouye M, Dashnow H, Raven LA et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 2014; 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bankevich A, Nurk S, Antipov D et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. [DOI] [PubMed] [Google Scholar]
- 14.Carver TJ, Rutherford KM, Berriman M et al. ACT: the Artemis Comparison Tool. Bioinformatics 2005; 21: 3422–3. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. CLSI, Wayne, PA, USA, 2014. [Google Scholar]
- 16.Malhotra-Kumar S, Xavier BB, Das AJ et al. Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect Dis 2016; 16: 286–7. [DOI] [PubMed] [Google Scholar]