Abstract
Pancreatic ductal adenocarcinoma is characterized by a prominent fibroinflammatory stroma with both tumor-promoting and tumor-suppressive functions. The pancreatic stellate cell (PSC) is the major cellular stromal component and the main producer of extracellular matrix proteins, including collagens, which are degraded by metalloproteinases (MMPs). PSCs interact with cancer cells through various factors, including transforming growth factor (TGF)β and interleukin (IL)-1α. The role of TGFβ in the dual nature of tumor stroma, i.e., protumorigenic or tumor suppressive, is not clear. We aimed to investigate the roles of TGFβ and IL-1α in the regulation of MMP profiles in PSCs and the subsequent effects on cancer cell migration. Human PSCs isolated from surgically resected specimens were cultured in the presence of pancreatic cancer cell lines, as well as IL-1α or TGFβ. MMP production and activities in PSCs were quantified by gene array transcripts, mRNA measurements, fluorescence resonance energy transfer–based activity assay, and zymography. PSC-conditioned media and pancreatic cancer cells were included in a collagen matrix cell migration model. We found that production of IL-1α by pancreatic cancer cells induced alterations in MMP and tissue inhibitors of matrix metalloproteinase (TIMP) profiles and activities in PSCs, upregulated expression and activation of MMP1 and MMP3, and enhanced migration of pancreatic cancer cells in the collagen matrix model. TGFβ counteracted the effects of IL-1α on PSCs, reestablished PSC MMP and TIMP profiles and activities, and inhibited migration of cancer cells. This suggests that tumor TGFβ has a role as a suppressor of stromal promotion of tumor progression through alterations in PSC MMP profiles with subsequent inhibition of pancreatic cancer cell migration.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal forms of cancer, with a mortality rate approaching the rate of incidence and with a 5-year survival rate below 5% [1]. Despite advances in the understanding of the genomics of the disease and identification of novel potential therapeutic targets [2], a real improvement in clinical outcome has not been achieved [3]. PDAC is largely resistant to all forms of conventional chemotherapy and radiation therapy. Surgical resection remains the only treatment with a potential for cure, although only 15% to 20% of the patients are eligible for resection [1]. Even following a successful resection, most patients develop disease recurrence within a year, indicating that additional treatment strategies are needed to improve survival [4].
A characteristic feature of PDAC is a prominent desmoplastic reaction, characterized by a dense fibroinflammatory stroma that surrounds the malignant cells [5]. The tumor stroma is composed of cellular elements and extracellular matrix (ECM) proteins. The pancreatic stellate cell (PSC) is the principal cell type of the desmoplastic stroma in PDAC and the major producer and regulator of the ECM [6], [7]. Upon activation, PSCs secrete excessive amounts of collagen, fibronectin, and matrix-degrading enzymes such as matrix metalloproteinases (MMPs) and their inhibitors, tissue inhibitors of matrix metalloproteinases (TIMPs) [8], [9], [10]. It has recently become clear that the tumor stroma has both tumor-promotive and tumor-suppressive effects in PDAC [11]. Although the precise mechanisms of the mutual interactions between cancer cells and stromal elements are only partially known, proteolytic and structural modifications of the ECM are believed to be involved [12], [13]. Matrix-degrading factors such as MMPs are the most important enzymes that cause ECM remodeling [14] and may provide favorable conditions for migration of cancer cells and as such facilitate invasive cancer growth [15].
Interleukin (IL)-1α is abundantly present in the tumor microenvironment and exerts multiple effects in the tumor stroma, including tumor-promoting effects [16], [17]. In PDAC, IL-1α is expressed exclusively by the malignant cells of the tumor and is immunohistochemically detected in the majority of tumors [18], [19]. IL-1α–positive PDAC cancer cell lines were previously shown to induce a specific inflammatory profile of the PSCs, and under IL-1α stimulation, PSCs induce migration of PDAC cells in vitro [18], [20]. Moreover, induction of IL-1α expression in PDAC cell lines has been shown to promote metastatic and invasive behavior in an orthotopic mouse model [21].
Transforming growth factor (TGF)β controls numerous biological functions and has both pro- and antitumor activities depending on the context. In early stages of carcinogenesis, the effect is usually tumor suppressive, whereas in later stages of cancer progression, TGFβ has tumor-promotional effects [22]. In PDAC, TGFβ is a key signaling mediator involved in stroma-tumor cross talk, epithelial-to-mesenchymal transition, and tumor invasion [23], [24], [25]. The overexpression of TGFβ and its receptors on cancer cells and in the tumor microenvironment has made this cytokine an attractive target for therapeutic intervention [26]. However, few data exist regarding distinct effects of TGFβ targeted therapy on cancer cells and stroma cells, respectively [27].
In the present study, we examined MMP and TIMP production by human PSCs and its regulation by IL-1α and TGFβ. We found that IL-1α induced a specific MMP/TIMP profile in the PSCs, which facilitated migration of pancreatic cancer cells in a collagen matrix model, and that these effects were suppressed by TGFβ. The data suggest an important role for TGFβ as a suppressor of PSC-induced migration of pancreatic cancer cells and add to the complexity of stroma-tumor interactions with potential implications for stroma-targeted therapies in pancreatic cancer.
Material and Methods
Patients
The study protocol and patient consent documents were approved by the Regional Committee for Medical and Health Research Ethics (REC South East, project number 2010/694a) and were in compliance with the Helsinki Declaration. Written informed consent was obtained from all study participants. The study included only adults.
Cells, Isolation, and Culture
Human PSCs were isolated from pancreatic tumor tissue obtained during pancreatic surgery (pancreatoduodenectomies) from patients with resectable pancreatic ductal adenocarcinoma. PSCs were cultured by the outgrowth method as developed by Bachem et al. [28] and described elsewhere [29]. The purity of the PSCs was assessed by morphology and cytofilament staining of α-SMA and vimentin. None of the cells were positive for CK7 or CK20. All experiments were performed using cell populations between passage 4 and 8. The primary PDAC cell line PC013 was propagated from PDAC tumor tissue samples as described elsewhere [18]. BxPC-3 and CAPAN2 were purchased from ATCC (Manassas, VA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium containing 4.5 g/l of glucose (DMEM). The media were supplemented with 100 μg/ml of Pen-Strep, Glutamax, and 10% fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA). For the co-culture assays, 6 × 105 CAPAN2, PC013, or BxPC-3 cells were seeded per Transwell insert (Corning Incorporated, Corning, NY) and cultured for 24 hours before moving the inserts into 6-well plates containing a confluent layer of PSCs. Cancer cells and PSCs were co-cultured for 72 hours before analyzing gene expression. For IL-1α (Biolegend, San Diego, CA), recombinant interleukin-1 receptor antagonist IL-1RA (Anakinra; gift from Swedish Orphan Biovitrum AS, Norway) and TGFβ stimulation, the PSCs were cultured to confluence, washed with NaCl, and cultured in serum-free DMEM supplemented with 1 ng/ml of IL-1α, and/or 2 ng/ml of TGFβ and 10 μg/ml of IL-1Ra. Supernatants were harvested after 4 days of culture, centrifuged, and stored at − 30°C until use.
Chemicals
Dulbecco’s modified Eagle's medium, glutamine, and Pen-Strep (10,000 U/ml) were obtained from Lonza (Verviers, Belgium). Amphotericin was purchased from Life Technologies (Carlsbad, CA). Recombinant human transforming growth factor-β1 was obtained from R&D Systems Europe, Ltd (Abingdon, England). [6-3H] thymidine (20-30 Ci/mmol) was purchased from PerkinElmer (Boston, MA). Antibodies against IL-1R1 were obtained from Millipore (Billerica, MA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was obtained from Cell Signaling Technology (Boston, MA). Secondary antibodies were purchased from Bio-Rad Laboratories (Hercules, CA), LI-COR Biosciences (Lincoln, NE), and Jackson Immunoresearch (Baltimore, PA).
Gene Expression Analysis
Gene transcription-profiling data set was obtained from previous studies (Tjomsland, E-MEXP-2826, https://www.ebi.ac.uk/arrayexpress). Subsequent data analysis involved the normalization of array values using RMA software. The gene array data were obtained from single-cultured and fluorescence-activated cell sorting–separated PSCs cultured in direct contact with the primary PDAC cell line PC013 [18].
RNA Extraction and Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction (qPCR)
Total RNA was prepared from the samples using RNA Easy Mini kit (Qiagen Inc., Valencia, CA), and cDNA was synthesized with SuperScript III Reverse Transcriptase First-Strand cDNA Synthesis kit according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA). Quantitative PCR was performed with Platinum SYBR Green Master Mix (Life Technologies, Carlsbad, CA) on 7900 Real-Time PCR system with 7900 System SDS 2.3 Software (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. Specific primers for MMP1, MMP2, MMP3, TIMP1, TIMP2, TIMP3, and IL-1α (Life Technologies, Carlsbad, CA) were used. GAPDH was used as a housekeeping gene. The primers were designed using Primer-BLAST [30]. All reactions were performed in triplicates including nontemplate controls. The results were analyzed using the ΔΔCt method [31]. The relative gene expression raw data were normalized to GAPDH and presented as relative gene expression for each gene.
ELISA
The levels of IL-1α in supernatants from PC013, BxPC-3, and CAPAN2 cells were assessed after culturing the cells for 3 days in DMEM supplemented with 1% FBS. The supernatants were harvested, and the concentration of IL-1α was measured by ELISA according to the manufacturer’s protocol (Biolegend, San Diego, CA).
Casein Zymography
MMP3 proteolytic activities were detected by casein substrate zymography. Conditioned media from control, IL-1α (1 ng/ml), and a combination of IL-1α and TGFβ (2 ng/ml) were concentrated by Amicon Ultracel 3K centrifugal filter device (Millipore, Billerica, MA). The concentrated conditioned media were mixed in nonreducing sample buffer containing SDS, glycerol, and bromophenol blue and subsequently subjected to electrophoresis on 12% polyacrylamide SDS gels containing 0.1% casein (Sigma Aldrich, Oslo, Norway). After electrophoresis, the polyacrylamide gels were washed three times in 2.5% Triton X-100 for 30 minutes to remove all traces of SDS. The gels were rinsed in distilled water for 5 minutes and finally incubated at 37°C for 20 hours in developing buffer containing 50 mM Tris HC (pH 7.5), 5 mM CaCl2, 0.02% Brij-35, and 200 mM NaCl3. The gels were stained with 0.125% Coomassie brilliant blue G-250 (Sigma Aldrich, Oslo, Norway) in 40% methanol and 10% acetic acid solution for 2 hours and destained with 40% methanol and 10% acetic acid solution. The MMP3 level was identified as clear zones of lysis against a blue background. For the quantification of MMP3 levels, images were acquired using ChemiDoc XRS (Bio-Rad Laboratories, Hercules, CA) and processed using FIJI software as described by Schindelin et al. [32].
MMP1 Activity Assay
The MMP1 activity in conditioned media of control, IL-1α (1 ng/ml), and a combination of IL-1α and TGFβ (2 ng/ml) was determined using the SensoLyte Plus 520 MMP-1 Assay Kit (AnaSpec, San Jose, CA). Briefly, the conditioned media were collected from PC013, BxPC-3, and CAPAN2 cells after 3 days of culture in DMEM supplemented with 1% FBS. One hundred microliters of conditioned media was added to MMP1 antibody precoated microplate wells. The MMP1 substrate 5-FAM/QXL520 fluorescence resonance energy transfer (FRET) peptide was added to the wells, and after 16 hours of incubation, the fluorescence intensity representing the MMP1 activity was measured at 490/520-nm wavelength using EnVision Multilabel Reader (Perkin Elmer, Waltham, MA).
Immunohistochemistry and Immunocytochemistry
Formalin-fixed, paraffin-embedded PDAC tissue and cultured cancer cell lines were immunostained with anti-human IL-1α antibodies (ab7632, ABcam, Cambridge, UK), visualized by Alkaline Phosphatase Red (Biocare Medical, Concord, CA), and counterstained with methyl green (Sigma Aldrich, Oslo, Norway) as described elsewhere [18].
Collagen I Migration Assay
Collagen I infiltration and migration assay was assessed by seeding 2 × 106 PC013, BxPC-3, or CAPAN2 cells in 10% FBS DMEM overnight. The medium was replaced the next day with fresh serum-free DMEM supplemented with 2 μCi/ml [6-3H] thymidine. Collagen I matrix gels were prepared by diluting bovine collagen I (Life Technologies, Carlsbad, CA) in serum-free control medium and supernatants from PSCs cultured in the presence of control (no supplements), IL-1α (1 ng/ml), and TGFβ (2 ng/ml) as single agents or in combination, and 10 μM of the broad-spectrum MMP inhibitor batimastat (R&D Systems Europe, Ltd, Abingdon, England). Cancer cells were added to a final concentration of 1 × 106 cells/ml and a collagen I concentration of 0.3%. Fifty microliters of collagen I gel mixture containing 5 × 104 cells was distributed to cell culture inserts (pore size 8.0 μM) placed in 24-well plates. The inserts were incubated at 37°C for 30 minutes before adding DMEM supplemented with 10% FBS to the bottom compartment. After 24 hours of incubation at 37°C, the cells were harvested by applying a cotton stick on each side of the insert surface, and [6-3H] thymidine incorporation was determined by liquid scintillation (Packard Tri-Carb 1900 TR). Infiltration and migration of cells were calculated as [6-3H] thymidine incorporation detected on the bottom side divided by total incorporation.
Statistical Analysis
The statistical analysis was performed with GraphPad Prism 5 (GraphPad Software). P < .05 was considered statistically significant, and error bars indicate standard error of the mean. The data were analyzed by paired t test, and multiple comparisons were analyzed using analysis of variance including Bonferroni correction.
Results
MMP and TIMP Profiles in PSCs
To establish MMP and TIMP expression profiles in PSCs, we analyzed gene array data from PSCs cultured alone and PSCs co-cultured with the primary pancreatic cancer cell line PC013. The gene array data indicate expression primarily of MMP1, MMP2, and MMP3 in PSCs (Table 1). Direct co-culture with PC013 cells increased the expression of MMP1 and MMP3 and decreased expression of MMP2 in PSCs. Noteworthy, according to the gene array data, expression of MMP1, MMP2, or MMP3 was not detected in the pancreatic cancer cells (data not shown). Expression of TIMP1, TIMP2, and TIMP3 was detected in PSCs, and only minor changes in expression were observed after co-culture with PC013 cancer cells. The gene array data did not indicate any expression of TIMP4 (Table 1).
Table 1.
MMP and TIMP Gene Profiles in PSCs and in PSCs Co-Cultured with Cancer Cells
| MMPs |
TIMPs |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene Expression Values |
Gene Expression Values |
Gene Expression Values |
||||||
| Gene Name | Single PSCs | Co-Cultured PSCs | Gene Name | Single PSCs | Co-Cultured PSCs | Gene Name | Single PSCs | Co-Cultured PSCs |
| MMP1 | 1267 | 7238 | MMP16 | ND | ND | TIMP1 | 14,279 | 15,101 |
| MMP2 | 8068 | 5018 | MMP17 | 105 | 110 | TIMP2 | 4206 | 4797 |
| MMP3 | 6500 | 10,180 | MMP19 | 114 | ND | TIMP3 | 1517 | 1313 |
| MMP7 | ND | ND | MMP20 | ND | ND | TIMP4 | ND | ND |
| MMP8 | ND | ND | MMP21 | ND | ND | |||
| MMP9 | ND | ND | MMP23 | ND | ND | |||
| MMP10 | 235 | 390 | MMP24 | ND | ND | |||
| MMP11 | 177 | 377 | MMP25 | ND | ND | |||
| MMP12 | ND | ND | MMP26 | ND | ND | |||
| MMP14 | 576 | 490 | MMP27 | ND | ND | |||
| MMP15 | ND | ND | MMP28 | 130 | 235 | |||
Gene transcription-profiling data set was obtained from previous studies. The data show mean expression values for each MMP and TIMP in single-cultured PSCs and fluorescence-activated cell sorting–separated PSCs cultured in direct contact with the primary PDAC cell line PC013. The cutoff gene expression value was set to 100, and values below cutoff were regarded as negative expression. ND, not detectable.
PDAC Cell Lines Express and Secrete IL-1α
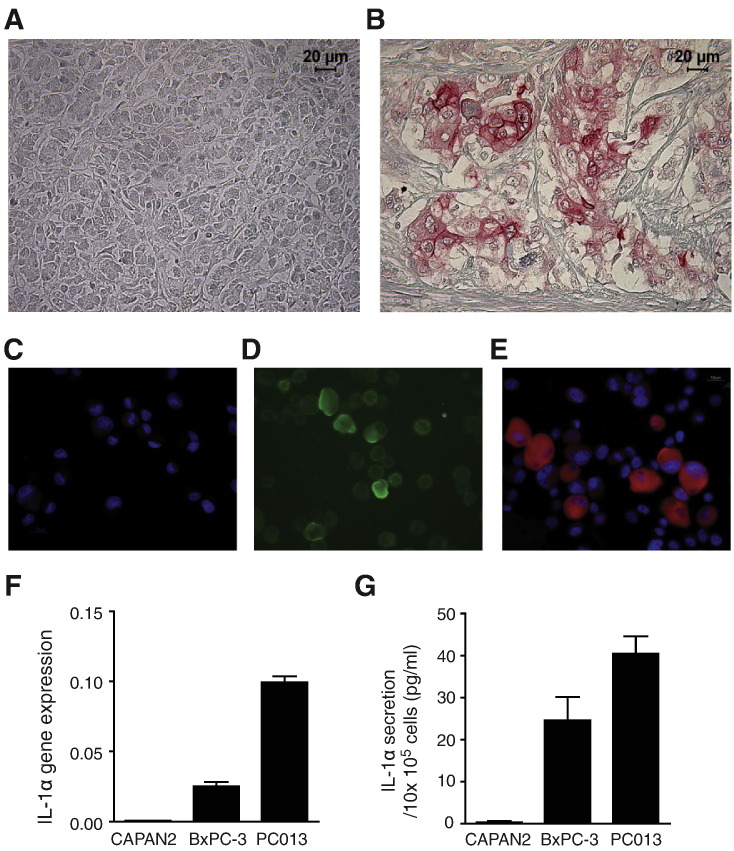
IL-1α was immunohistochemically confirmed to be absent in healthy pancreatic tissue (Figure 1A) and found to be exclusively expressed by malignant cells in PDAC tissue (Figure 1B). PC013, a primary PDAC cell line established from the same tumor as shown in Figure 1B, was investigated by immunocytochemistry for further characterization. The results demonstrated negative binding of secondary fluorescent antibodies (control) (Figure 1C), positive expression of the epithelial cell marker Epcam (Figure 1D), and intracellular expression of IL-1α, primarily in the cell cytoplasm (Figure 1E). Moreover, the PDAC cell lines were assessed for gene expression and secretion of IL-1α. The primary PDAC cell line PC013 showed strong gene expression, BxPC-3 expressed moderate levels, whereas CAPAN2 was found to be IL-1α negative (Figure 1F). The secreted levels of IL-1α confirmed the gene expression data after 3 days of culture, showing the highest levels in supernatants from PC013, lower levels in supernatants from BxPC-3 cells, and absence of IL-1α in supernatants from CAPAN2 cells (Figure 1G).
Figure 1.
IL-1α expression in pancreatic cancer tissue and cell lines.
Expression and localization of IL-1α (red) was analyzed by immunochemistry in healthy pancreatic tissue (A) and PDAC tissue (B). A primary PDAC cell line (established from the same patient as shown in figure B), PC013, was stained by immunocytochemistry to confirm tissue originality; negative control (C), Epcam (green) (D), IL-1α (red) (E), and cell nucleus visualized by DAPI (blue). The pancreatic cancer cell lines CAPAN2, BxPC-3, and PC013 were cultured for 3 days, and IL-1α gene expression (F) and IL-1α protein levels in supernatants (G) were measured.
IL-1α Regulates Expression of MMPs and TIMPs in PSCs
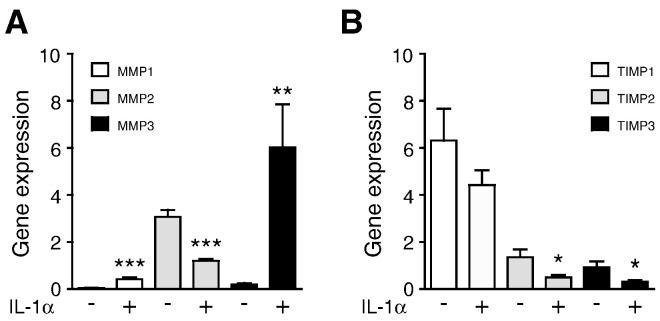
To evaluate the role of IL-1α in the expression of MMPs and TIMPs, PSCs were cultured in the presence of 1 ng/ml of IL-1α. Gene expression levels of MMP1 (P < .005) and MMP3 (P = .006) increased significantly by 9- and 33-fold, respectively, whereas the expression of MMP2 decreased significantly (P < .005) by 2.5-fold compared with untreated PSCs (Figure 2A). Gene expression of MMP9 was not detected in PSCs (data not shown). IL-1α reduced the expression levels of TIMP2 (P = .02) and TIMP3 (P = .01) significantly by three-fold each, whereas expression of TIMP1 was not significantly reduced (Figure 2B).
Figure 2.
IL-1α regulates MMP and TIMP expression in PSCs.
PSCs were cultured in the presence or absence of IL-1α. The relative gene expression of (A) MMP1, MMP2, and MMP3 and (B) TIMP1, TIMP2, and TIMP3 were analyzed and compared with nonstimulated PSCs. *P < .05, **P < .005, and ***P < .001.
IL-1α from PDAC Cell Lines Regulates MMP and TIMP Expression in PSCs
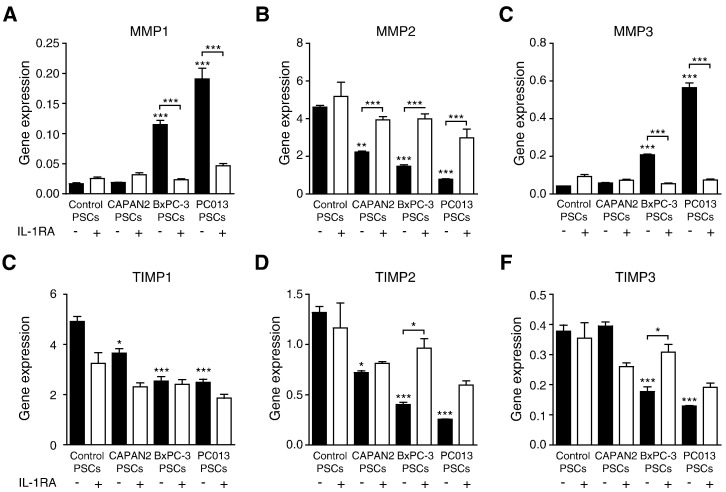
To investigate the involvement of IL-1α in PDAC cell induction of MMPs and TIMPs, IL-1α–positive (PC013 and BxPC-3) and IL-1α–negative (CAPAN2) cancer cell lines were cultured alone or in co-culture with PSCs. PC013 and BxPC-3 cells induced the expression of MMP1 and MMP3 in PSCs, whereas CAPAN2 did not affect the expression. All tested PDAC cell lines reduced the gene expression levels of MMP2 in the PSCs (Figure 3, A–C). Co-culture with PC013 and BxPC-3 reduced PSC gene expression of TIMP1 to 3, whereas co-culture with CAPAN2 caused a decrease only in the expression of TIMP1 and 2 (Figure 3, D–F). IL-1α–expressing PDAC cell lines exerted the most pronounced effects on the PSCs gene expression profiles of MMPs and TIMPs. To investigate the involvement of IL-1α in the regulation of MMP1 and TIMP expression, we neutralized the effect of IL-1α in the medium by adding rhIL-1RA to the co-cultures. By blocking the effects of IL-1α, the cancer cells lost their ability to induce PSC expression of MMP1 and MMP3. Moreover, IL-1α inhibition also increased the expression of MMP2 in the PSCs. IL-1RA did not affect the reduced expression of TIMP1 but enhanced the expression of TIMP2.
Figure 3.
IL-1α–positive PDAC cell lines affect PSC expression of MMP1, MMP2, MMP3, TIMP2, and TIMP3.
The PDAC cell lines PC013, BxPC-3, and CAPAN2 were co-cultured with PSCs in the presence or absence of IL-1RA. The relative gene expression of MMP1 (A), MMP2 (B), MMP3 (C), TIMP1 (D), TIMP2 (E), and TIMP3 (F) was analyzed in PSCs after 3 days of co-culture. *P < .05, **P < .005, and ****P < .001.
TGFβ Inhibits IL-1α Regulated PSC Expression of MMP1, MMP2, MMP3, TIMP2, and TIMP3
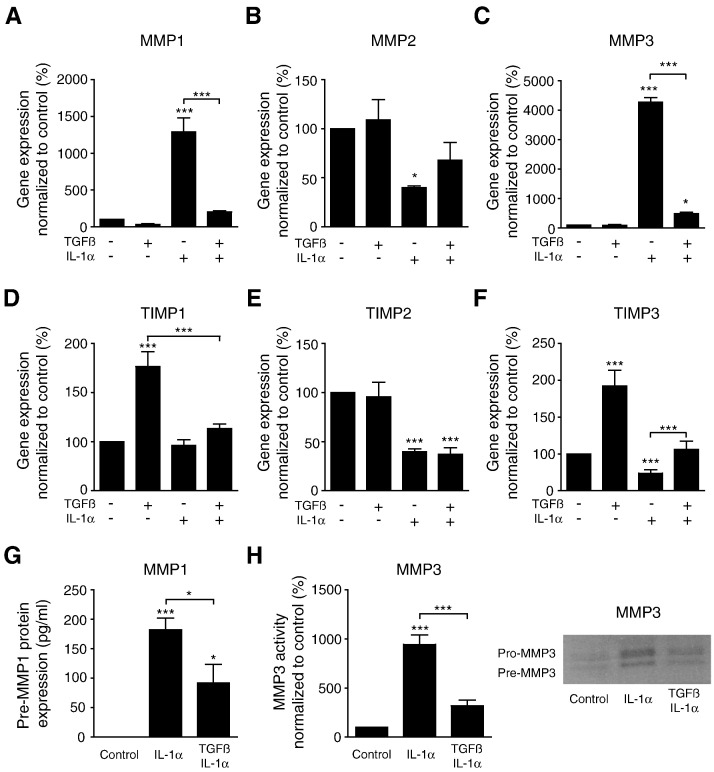
To investigate the influence of TGFβ on IL-1α–induced MMP regulation, we incubated PSCs in the presence of IL-1α (1 ng/ml) and TGFβ (2 ng/ml), alone or in combination. TGFβ significantly inhibited the effects of IL-1α on PSC expression of MMP1 (P < .001) and MMP3 (P < .001), whereas it reduced the inhibiting effects of IL-1α on PSCs expression of MMP2 (Figure 4, A–C). As a single agent, TGFβ significantly induced the expression of TIMP1 (P < .001) and TIMP3 (P < .001), whereas IL-1α as a single agent reduced the levels of TIMP2 and TIMP3 (P < .001). When combining IL-1α and TGFβ, no effects on TIMP1 and TIMP3 expression were observed compared with unstimulated PSCs. In contrast, the combination of the two factors did not affect the inhibiting effects of IL-1α on the expression of TIMP2 (Figure 4, D–F). To further confirm the regulatory effects of TGFβ on IL-1α signaling in PSCs, we analyzed the MMP1 and MMP3 activity in supernatants from PSCs cultured in the presence of 1 ng/ml of IL-1α or a combination of 1 ng/ml of IL-1α and 2 ng/ml of TGFβ. Collagen type I zymography revealed expression of one strong band, which could indicate predominant expression of pre-MMP1 (data not shown). MMP-1 substrate 5-FAM/QXL520 FRET peptide assay was employed to further evaluate the expression of active MMP in the samples. The results showed no expression of active MMP1 in supernatants from untreated PSCs. Activated MMP1 was detected in supernatant from PSCs exposed to 1 ng/ml of IL-1α, whereas the presence of 2 ng/ml of TGFβ significantly (P < .05) reduced this expression (Figure 4G). Active MMP3 was detected by quantification of casein zymograms, which revealed significantly increased MMP3 activity in supernatants from IL-1α–stimulated PSCs compared with unstimulated PSCs. Supernatants from PSCs exposed to a combination of 1 ng/ml of IL-1α and 2 ng/ml of TGFβ showed significantly decreased MMP3 activity compared with PSCs stimulated with IL-1α only (Figure 4H).
Figure 4.
TGFβ inhibits the effects of IL-1α on PSC expression of MMPs and TIMPs.
PSCs were cultured in the presence of IL-1α (1 ng/ml), TGFβ (2 ng/ml), or a combination of both cytokines. Gene expression of MMP1 (A), MMP2 (B), MMP3 (C), TIMP1 (D), TIMP2 (E), and TIMP3 (F) was analyzed and normalized to untreated controls. Supernatants were harvested from PSCs incubated with IL-1α (1 ng/ml), a combination of IL-1α (1 ng/ml) and TGFβ (2 ng/ml), and untreated control PSCs. MMP-1 activity was detected by substrate 5-FAM/QXL520 FRET peptide assay (G), and data are shown in pg/ml of supernatant. Active MMP3 was detected by casein zymography and quantified by FIJI software (H). A representative zymogram from one experiment is shown. *P < .05, **P < .005, and ***P < .001.
TGFβ Inhibits IL-1α–Induced Cancer Cell Migration in Collagen Type I Matrix
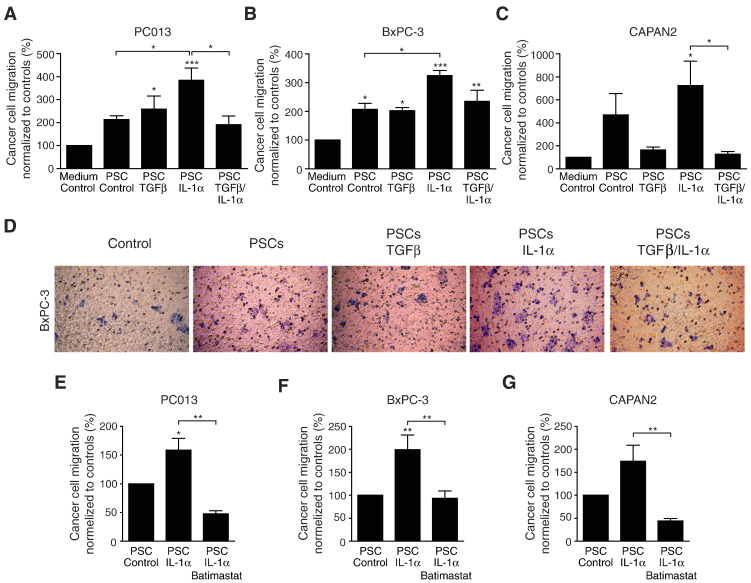
MMP1 has previously been shown to play a major role in collagen type I degradation [33]. Our findings show increased expression of MMP1 after stimulating PSCs with IL-1α or IL-1α–positive PDAC cell lines. We investigated whether IL-1α–stimulated PSCs influence the migration of cancer cells in a collagen type I matrix and whether TGFβ can counteract the effect of IL-1α (Figure 5, A–D). The collagen matrix gels containing PSC-conditioned medium increased the migration of PC013, BxPC-3, and CAPAN2 cells compared with control matrix. Collagen type I matrix containing conditioned medium from IL-1α–stimulated PSCs enhanced the migration of the PDAC cell lines. In contrast, collagen type I matrix containing conditioned medium from TGFβ-stimulated PSCs had no additional effects on PDAC cell migration compared with unstimulated PSCs. However, conditioned medium from PSCs stimulated with a combination of both TGFβ and IL-1α significantly reduced cancer cell migration. To confirm the role of MMPs in the migration of the PDAC cell lines, the broad-spectrum MMP inhibitor batimastat was added to the collagen type I matrix (Figure 5, E–G). Batimastat virtually abolished the stimulatory effects exerted by IL-1α–stimulated PSCs on migration of PC013, BxPC-3, and CAPAN2 in a collagen type I matrix.
Figure 5.
TGFβ inhibits IL-1α–induced cancer cell migration in collagen type I matrix.
[6-3H] thymidine-incorporated pancreatic cancer cell lines (PC013, BxPC-3, or CAPAN2) were added to the collagen matrix migration system. Collagen type I matrix gels were prepared with either serum-free medium or PSC-conditioned medium. The conditioned medium was prepared by culturing the PSCs in the presence or absence of 1 ng/ml of IL-1α and 2 ng/ml of TGFβ as single agents or in combination (A–D). Representative crystal violet staining of BxPC-3 cells from one experiment is shown in figure D. To confirm the role of MMPs in the migration of the PDAC cell lines, the broad-spectrum MMP inhibitor batimastat was added to the migration system (E–F). Pancreatic cancer cell migration is presented as percent [6-3H] thymidine incorporation from the bottom side of the membrane compared with total [6-3H] thymidine incorporation (top side and bottom side) and normalized to respective controls. *P < .05, **P < .005, and ***P < .001.
Discussion
In the present study, we demonstrate that pancreatic cancer cells through an IL-1α signaling mechanism are able to induce a tumor-promotive MMP expression profile in human PSCs. The IL-1α–induced MMP profile, i.e., increased MMP1 and MMP3, is optimal for degradation of the dominating type I collagen present in the ECM of the desmoplastic tumor stroma. Contrary to the prevailing paradigm that TGFβ supports tumor progression, we found that TGFβ suppressed the IL-1α–induced MMP profile in PSCs, which reduced cancer cell migration in a tumor stroma collagen type I model.
We hypothesized that IL-1α has a role in the remodeling of extracellular matrix in pancreatic cancer, such that malignant infiltration is facilitated. Collagen type I is the major component of ECM and provides structural integrity and mechanical flexibility to the tumor stroma [34], [35]. MMP1 is considered to be the rate-limiting enzyme in the degradation of collagen type I [33], and MMP1 derived from cancer-associated fibroblasts appears to facilitate cancer cell migration and invasive behavior in breast cancer [36]. Furthermore, recent observations in cervical cancer implicate stromal fibroblasts in cancer cell migration through MMP-2 production and ECM remodeling [37]. Gene expression of MMP1 and MMP3 was detected in PSCs, and the expression was further induced by IL-1α or IL-1α–positive PDAC cell lines. Noteworthy, the cleaved active form of MMP1 was only detected when PSCs were incubated in the presence of IL-1α. This is in accordance with previous findings in cancer tissue, showing MMP1 expression by tumor cells or adjacent fibroblasts in response to stimulating factors produced by the tumor cells [38]. Notably, MMP1 is detected at the invasive tumor front, and overexpression has been associated with poor prognosis in a variety of advanced cancers, including pancreatic adenocarcinoma [38], [39]. The IL-1α–dependent expression of active MMP1 could be explained by increased expression of MMP3, which is able to cleave pro-MMP1 [40]. In the presence of IL-1α, a specific expression profile was induced in PSCs, which was characterized by increased expression of MMP1 and MMP3 as well as reduced levels of MMP2, TIMP2, and TIMP3. TIMP3 has previously been found to preferentially inhibit the activity of MMP1 and MMP3 [41], and reduced expression of TIMP3 could enhance their proteolytic activity, resulting in remodeling of the tumor stroma ECM.
During the first step of cancer cell invasion, the basement membrane primarily consisting of collagen IV is degraded by extracellular proteases such as MMP2 and MMP9 [42]. However, collagen type I is the major ECM component in the tumor stroma, and ECM remodeling is mandatory for further neoplastic invasion [34]. Moreover, IL-1α is a pivotal inducer of both MMP1 and MMP3 expression [43], and induction of IL-1α expression in PDAC cell lines has been shown to favor their metastatic and invasive behavior in vitro and in preclinical models [44]. IL-1α has been detected in the majority of PDAC tumors, and high expression is associated with poor clinical outcome [18]. This suggests a role for MMP-dependent degradation in carcinoma invasion. However, all clinical trials targeting MMPs in pancreatic cancer, such as tanomastat (inhibiting MMP2, MMP3, MMP9, and MMP13) and marimastat (inhibiting MMP1, MMP2, MMP7, MMP9, and MMP14), have remained without clinical benefit when compared with treatment with gemcitabine alone [45], [46]. The lack of specific MMP inhibitors and a growing list of non-ECM protein substrates for the MMPs may contribute to their ineffectiveness. However, MMP inhibitors are suggested to be more efficacious if used in the early stage of tumor development as MMPs contribute to driving the disease progression [47]. The failure of MMP inhibitors in the clinic prompts alternative strategies to target MMP-induced tumor progression. Because we found that IL-1α induced a specific MMP and TIMP activity profile in PSCs, inhibitors of this process might possibly represent a novel strategy for suppression of invasive tumor behavior.
In the present study, we verified a critical role for the IL-1α–induced MMP and TIMP profiles in a collagen type I migration model. When adding a broad-spectrum inhibitor of MMP activity (batimastat), the stimulatory effect on migration exerted by IL-1α/PSC signaling was abolished, suggesting that the effects of PSCs on migration are mediated through an altered activity of MMPs. Furthermore, the major finding in the present study is that TGFβ inhibited IL-1α/PSC-induced cancer cell migration. This is in accordance with the notion that TGFβ exerts tumor-suppressive effects through the regulation of stromal functions and, furthermore, that the ability of TGFβ to inhibit ECM remodeling through IL-1α could be an essential tumor-protective mechanism.
The role of TGFβ in the tumor stroma is highly complex [48]. TGFβ activates PSCs and is a key player in pancreatic fibrosis in both pancreatitis and pancreatic cancer [49]. TGFβ is also essential in the upregulation of the characteristic marker αSMA in PSCs [50]. Recently, high expression of αSMA in the tumor stroma was found to correlate with better overall survival in PDAC patients, which further supports a cancer protective role of the tumor stroma in PDAC [51]. TGFβ regulates the stroma by production, degradation, and accumulation of ECM proteins; inhibits the synthesis of matrix-depleting proteins such as MMPs in desmoplastic pancreatic cancer tissue; and upregulates collagens I to V, suggesting a protective physiological response in the stroma [49], [52], [53]. In contrast, collagen I secreted from PSCs has also been shown to promote pancreatic cancer cell migration through upregulation of cell adhesion molecules, such as N-cadherin and α2b1 integrins [54], [55]. This emphasizes the complexity of pleiotropic factors and their diverse roles in cancer progression related to stroma composition.
Several lines of evidence indicate that tumor stroma has a dual nature with respect to cancer progression. In contrast to the previously dominating view suggesting that PDAC stroma, including PSCs, has largely tumor-promoting effects, recent evidence also supports a tumor-suppressor function of PDAC stroma in pancreatic cancer. Ozdemir et al. showed that the tumor stroma might act to restrain, rather than support, pancreatic cancer progression [51]. Furthermore, two studies employing different strategies for depletion of stromal cells in pancreatic tumors through targeting of the sonic hedgehog pathway resulted in poorly differentiated tumor histology, accelerated rather than reduced tumor progression, and reduced survival [56], [57]. In contrast to extensive ligand-induced stromal disruption, reprogramming of PSCs resulting in stroma remodeling represents an alternative therapeutic approach [58]. These observations support the notion that the impact of stroma in pancreatic cancer might be highly circumstantial, depending on temporal and spatial events and the functional status of stromal components and malignant cells. Furthermore, the paradigm that tumor stroma is cancer promoting with stroma depletion as a possible therapeutic approach now appears more suitably replaced by the concept that tumor stroma has a dual nature and that stroma remodeling rather than stromal ablation might represent a preferred therapeutic approach.
In the present study, we have explored one mechanism involved in stromal suppression of tumor cell migration. MMP and TIMP profiles and activities are crucial factors in tumor stroma remodeling. This is mediated by IL-1α from the tumor cells. Our data show that TGFβ counteracts the tumor-supportive stromal MMP and TIMP profiles and thereby inhibits cancer cell migration by inhibiting IL-1α–induced effects in PSCs. This suggests that tumor TGFβ has a role as a suppressor of stromal promotion of tumor progression. These findings underscore the notion that tumor stroma in pancreatic cancer is a multicomponent entity where results of intervention strategies are highly dependent on context. Furthermore, this notion could at least partly explain the lack of clinical benefit of therapies targeting tumor stroma.
Authors’ Contributions
V. T. and I. P. G. conceived and designed the project. E. P. and C. S. V. were responsible for sampling and logistics of human pancreatic cancer tissue. V. T. and M. A. performed the experiments. V. T., D. S., and I. P. G. analyzed the data. V. T. and I. P. G. wrote the manuscript. I. P. G. and D. S. supervised the project. All authors commented on and approved of the final manuscript.
Conflict of Interest
The authors declare they do not have any conflict of interest.
Acknowledgements
This study has been supported by the University of Oslo and by grants from the Norwegian Cancer Society, AstraZeneca Norway, Merchant Einar Unsgaard and Mrs. Kitty Unsgaard Award, Gunnar Kristian Olsen and Randi Andresen Scientific Award to Medical Research, and County Governor H.B. Guldahl and Mrs. Lucy Guldahl Award.
Footnotes
This study has been supported by the University of Oslo and by grants from the Norwegian Cancer Society, AstraZeneca Norway, Merchant Einar Unsgaard and Mrs. Kitty Unsgaard Award, Gunnar Kristian Olsen and Randi Andresen Scientific Award to Medical Research, and County Governor H.B. Guldahl and Mrs. Lucy Guldahl Award.
References
- 1.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 2.Reznik R, Hendifar AE, Tuli R. Genetic determinants and potential therapeutic targets for pancreatic adenocarcinoma. Front Physiol. 2014;5:87. doi: 10.3389/fphys.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zijlstra M, Bernards N, de Hingh IH, van de Wouw AJ, Goey SH, Jacobs EM, Lemmens VE, Creemers GJ. Does long-term survival exist in pancreatic adenocarcinoma? Acta Oncol. 2016;55:259–264. doi: 10.3109/0284186X.2015.1096020. [DOI] [PubMed] [Google Scholar]
- 4.Labori KJ, Katz MH, Tzeng CW, Bjornbeth BA, Cvancarova M, Edwin B, Kure EH, Eide TJ, Dueland S, Buanes T. Impact of early disease progression and surgical complications on adjuvant chemotherapy completion rates and survival in patients undergoing the surgery first approach for resectable pancreatic ductal adenocarcinoma—a population-based cohort study. Acta Oncol. 2016;55:265–277. doi: 10.3109/0284186X.2015.1068445. [DOI] [PubMed] [Google Scholar]
- 5.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, Lolkema MP, Buchholz M, Olive KP, Gress TM. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 6.Apte MV, Wilson JS, Lugea A, Pandol SJ. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology. 2013;144:1210–1219. doi: 10.1053/j.gastro.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yen TNF, Aardal NP, Bronner MP, Thorning DR, Savard CE, Lee SP, Bell RH. Myofibroblasts are responsible for the desmoplastic reaction surrounding human pancreatic carcinomas. Surgery. 2002;131:129–134. doi: 10.1067/msy.2002.119192. [DOI] [PubMed] [Google Scholar]
- 8.Phillips PA, McCarroll JA, Park S, Wu MJ, Pirola R, Korsten M, Wilson JS, Apte MV. Rat pancreatic stellate cells secrete matrix metal loproteinases: implications for extracellular matrix turnover. Gut. 2003;52:275–282. doi: 10.1136/gut.52.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apte MV, Park S, Phillips PA, Santucci N, Goldstein D, Kumar RK, Ramm GA, Buchler M, Friess H, McCarroll JA. Desmoplastic reaction in pancreatic cancer—role of pancreatic stellate cells. Pancreas. 2004;29:179–187. doi: 10.1097/00006676-200410000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida S, Yokota T, Ujiki M, Ding XZ, Pelham C, Adrian TE, Talamonti MS, Bell RH, Denham W. Pancreatic cancer stimulates pancreatic stellate cell proliferation and TIMP-1 production through the MAP kinase pathway. Biochem Biophys Res Commun. 2004;323:1241–1245. doi: 10.1016/j.bbrc.2004.08.229. [DOI] [PubMed] [Google Scholar]
- 11.Falasca M, Kim M, Casari I. Pancreatic cancer: current research and future directions. Biochim Biophys Acta. 2016;1865:123–132. doi: 10.1016/j.bbcan.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol Cancer Res. 2012;10:1403–1418. doi: 10.1158/1541-7786.MCR-12-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu PF, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Wever O, Demetter P, Mareel M, Bracke M. Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer. 2008;123:2229–2238. doi: 10.1002/ijc.23925. [DOI] [PubMed] [Google Scholar]
- 16.Apte RN, Dotan S, Elkabets M, White MR, Reich E, Carmi Y, Song XP, Dvozkin T, Krelin Y, Voronov E. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 2006;25:387–408. doi: 10.1007/s10555-006-9004-4. [DOI] [PubMed] [Google Scholar]
- 17.Xu D, Matsuo Y, Ma J, Koide S, Ochi N, Yasuda A, Funahashi H, Okada Y, Takeyama H. Cancer cell–derived IL-1alpha promotes HGF secretion by stromal cells and enhances metastatic potential in pancreatic cancer cells. J Surg Oncol. 2010;102:469–477. doi: 10.1002/jso.21530. [DOI] [PubMed] [Google Scholar]
- 18.Tjomsland V, Spangeus A, Valila J, Sandstrom P, Borch K, Druid H, Falkmer S, Falkmer U, Messmer D, Larsson M. Interleukin 1alpha sustains the expression of inflammatory factors in human pancreatic cancer microenvironment by targeting cancer-associated fibroblasts. Neoplasia. 2011;13:664–675. doi: 10.1593/neo.11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tjomsland V, Bojmar L, Sandstrom P, Bratthall C, Messmer D, Spangeus A, Larsson M. IL-1alpha expression in pancreatic ductal adenocarcinoma affects the tumor cell migration and is regulated by the p38MAPK signaling pathway. PLoS One. 2013;8:e70874. doi: 10.1371/journal.pone.0070874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melisi D, Niu J, Chang Z, Xia Q, Peng B, Ishiyama S, Evans DB, Chiao PJ. Secreted interleukin-1alpha induces a metastatic phenotype in pancreatic cancer by sustaining a constitutive activation of nuclear factor-kappaB. Mol Cancer Res. 2009;7:624–633. doi: 10.1158/1541-7786.MCR-08-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massague J. “TGF beta in”. Cancer Cell. 2008;134:215–230. [Google Scholar]
- 23.Birnbaum DJ, Mamessier E, Birnbaum D. The emerging role of the TGF beta tumor suppressor pathway in pancreatic cancer. Cell Cycle. 2012;11:683–686. doi: 10.4161/cc.11.4.19130. [DOI] [PubMed] [Google Scholar]
- 24.Ellenrieder V, Hendler SF, Boeck W, Seufferlein T, Menke A, Ruhland C, Adler G, Gress TM. Transforming growth factor beta 1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–4228. [PubMed] [Google Scholar]
- 25.Whatcott C, Han H, Posner RG, Von Hoff DD. Tumor-stromal interactions in pancreatic cancer. Crit Rev Oncog. 2013;18:135–151. doi: 10.1615/critrevoncog.v18.i1-2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AL, Robin TP, Ford HL. Molecular pathways: targeting the TGF-beta pathway for cancer therapy. Clin Cancer Res. 2012;18:4514–4521. doi: 10.1158/1078-0432.CCR-11-3224. [DOI] [PubMed] [Google Scholar]
- 27.Neuzillet C, de Gramont A, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. Perspectives of TGF-beta inhibition in pancreatic and hepatocellular carcinomas. Oncotarget. 2014;5:78–94. doi: 10.18632/oncotarget.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachem MG, Schneider E, Gross H, Weidenbach H, Schmid RM, Menke A, Siech M, Beger H, Grunert A, Adler G. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115:421–432. doi: 10.1016/s0016-5085(98)70209-4. [DOI] [PubMed] [Google Scholar]
- 29.Pomianowska E, Sandnes D, Grzyb K, Schjolberg AR, Aasrum M, Tveteraas IH, Tjomsland V, Christoffersen T, Gladhaug IP. Inhibitory effects of prostaglandin E2 on collagen synthesis and cell proliferation in human stellate cells from pancreatic head adenocarcinoma. BMC Cancer. 2014;14:413. doi: 10.1186/1471-2407-14-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(− Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNulty M, Mahmud A, Spiers P, Feely J. Collagen type-I degradation is related to arterial stiffness in hypertensive and normotensive subjects. J Hum Hypertens. 2006;20:867–873. doi: 10.1038/sj.jhh.1002015. [DOI] [PubMed] [Google Scholar]
- 34.Bachem MG, Schunemann M, Ramadani M, Siech M, Beger H, Buck A, Zhou SX, Schmid-Kotsas A, Adler G. Pancreatic carcinoma cells induce fibrosis by stimulating proliferation and matrix synthesis of stellate cells. Gastroenterology. 2005;128:907–921. doi: 10.1053/j.gastro.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 35.Kakkad SM, Solaiyappan M, O'Rourke B, Stasinopoulos I, Ackerstaff E, Raman V, Bhujwalla ZM, Glunde K. Hypoxic tumor microenvironments reduce collagen I fiber density. Neoplasia. 2010;12:608–617. doi: 10.1593/neo.10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boire A, Covic L, Agarwal A, Jacques S, Sherifl S, Kuliopulos A. PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell. 2005;120:303–313. doi: 10.1016/j.cell.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Fullar A, Dudas J, Olah L, Hollosi P, Papp Z, Sobel G, Karaszi K, Paku S, Baghy K, Kovalszky I. Remodeling of extracellular matrix by normal and tumor-associated fibroblasts promotes cervical cancer progression. BMC Cancer. 2015;15:256. doi: 10.1186/s12885-015-1272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ala-Aho R, Kahari VM. Collagenases in cancer. Biochimie. 2005;87:273–286. doi: 10.1016/j.biochi.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Ito T, Ito M, Shiozawa J, Naito S, Kanematsu T, Sekine I. Expression of the MMP-1 in human pancreatic carcinoma: relationship with prognostic factor. Mod Pathol. 1999;12:669–674. [PubMed] [Google Scholar]
- 40.Vincenti MP, White LA, Schroen DJ, Benbow U, Brinckerhoff CE. Regulating expression of the gene for matrix metalloproteinase-1 (collagenase): mechanisms that control enzyme activity, transcription, and mRNA stability. Crit Rev Eukaryot Gene Expr. 1996;6:391–411. doi: 10.1615/critreveukargeneexpr.v6.i4.40. [DOI] [PubMed] [Google Scholar]
- 41.Amour A, Slocombe PM, Webster A, Butler M, Knight CG, Smith BJ, Stephens PE, Shelley C, Hutton M, Knauper V. TNF-alpha converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998;435:39–44. doi: 10.1016/s0014-5793(98)01031-x. [DOI] [PubMed] [Google Scholar]
- 42.Clark AG, Vignjevic DM. Modes of cancer cell invasion and the role of the microenvironment. Curr Opin Cell Biol. 2015;36:13–22. doi: 10.1016/j.ceb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Turner NA, Warburton P, O'Regan DJ, Ball SG, Porter KE. Modulatory effect of interleukin-1 alpha on expression of structural matrix proteins, MMPs and TIMPs in human cardiac myofibroblasts: role of p38 MAP kinase. Matrix Biol. 2010;29:613–620. doi: 10.1016/j.matbio.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melisi D, Niu JG, Chang Z, Xia QH, Peng BL, Lshiyama S, Evans DB, Chiao PJ. Secreted interleukin-1 alpha induces a metastatic phenotype in pancreatic cancer by sustaining a constitutive activation of nuclear factor-kappa B. Mol Cancer Res. 2009;7:624–633. doi: 10.1158/1541-7786.MCR-08-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bramhall SR, Schulz J, Nemunaitis J, Brown PD, Baillet M, Buckels JA. A double-blind placebo-controlled, randomised study comparing gemcitabine and marimastat with gemcitabine and placebo as first line therapy in patients with advanced pancreatic cancer. Br J Cancer. 2002;87:161–167. doi: 10.1038/sj.bjc.6600446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore MJ, Hamm J, Dancey J, Eisenberg PD, Dagenais M, Fields A, Hagan K, Greenberg B, Colwell B, Zee B. Comparison of gemcitabine versus the matrix metalloproteinase inhibitor BAY 12-9566 in patients with advanced or metastatic adenocarcinoma of the pancreas: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2003;21:3296–3302. doi: 10.1200/JCO.2003.02.098. [DOI] [PubMed] [Google Scholar]
- 47.Cathcart J, Pulkoski-Gross A, Cao J. Targeting matrix metalloproteinases in cancer: bringing new life to old ideas. Genes Dis. 2015;2:26–34. doi: 10.1016/j.gendis.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickup M, Novitskiy S, Moses HL. The roles of TGF beta in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rane SG, Lee JH, Lin HM. Transforming growth factor-beta pathway: role in pancreas development and pancreatic disease. Cytokine Growth Factor Rev. 2006;17:107–119. doi: 10.1016/j.cytogfr.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Apte MV, Haber PS, Darby SJ, Rodgers SC, McCaughan GW, Korsten MA, Pirola RC, Wilson JS. Pancreatic stellate cells are activated by proinflammatory cytokines: implications for pancreatic fibrogenesis. Gut. 1999;44:534–541. doi: 10.1136/gut.44.4.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papageorgis P, Stylianopoulos T. Role of TGF beta in regulation of the tumor microenvironment and drug delivery. Int J Oncol. 2015;46:933–943. doi: 10.3892/ijo.2015.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawai H, Okada Y, Funahashi H, Matsuo Y, Takahashi H, Takeyama H, Manabe T. Interleukin-1alpha enhances the aggressive behavior of pancreatic cancer cells by regulating the alpha6beta1-integrin and urokinase plasminogen activator receptor expression. BMC Cell Biol. 2006;7:8. doi: 10.1186/1471-2121-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shintani Y, Hollingsworth MA, Wheelock MJ, Johnson KR. Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH2-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res. 2006;66:11745–11753. doi: 10.1158/0008-5472.CAN-06-2322. [DOI] [PubMed] [Google Scholar]
- 55.Lu J, Zhou S, Siech M, Habisch H, Seufferlein T, Bachem MG. Pancreatic stellate cells promote hapto-migration of cancer cells through collagen I-mediated signalling pathway. Br J Cancer. 2014;110:409–420. doi: 10.1038/bjc.2013.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, Fitamant J, Jones PD, Ghanta KS, Kawano S. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091–E3100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, Collisson EA, Connor F, Van Dyke T, Kozlov S. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]