Abstract
An important fact in studying link prediction is that the structural properties of networks have significant impacts on the performance of algorithms. Therefore, how to improve the performance of link prediction with the aid of structural properties of networks is an essential problem. By analyzing many real networks, we find a typical structural property: nodes are preferentially linked to the nodes with the weak clique structure (abbreviated as PWCS to simplify descriptions). Based on this PWCS phenomenon, we propose a local friend recommendation (FR) index to facilitate link prediction. Our experiments show that the performance of FR index is better than some famous local similarity indices, such as Common Neighbor (CN) index, Adamic-Adar (AA) index and Resource Allocation (RA) index. We then explain why PWCS can give rise to the better performance of FR index in link prediction. Finally, a mixed friend recommendation index (labelled MFR) is proposed by utilizing the PWCS phenomenon, which further improves the accuracy of link prediction.
The research of link prediction mainly focuses on forecasting potential relations between nonadjacent nodes, including the prediction of the unknown links or the further nodes1. Owing to the wide range of applications of link prediction, such as recommending friends in online social networks2, exploring protein-to-protein interactions3, reconstructing airline network4, and boosting e-commerce scales, study on link prediction has attracted much attention recently5,6,7,8. The probabilistic model and machine learning were mainly introduced in link prediction. The notion of probabilistic link prediction and path analysis using Markov chains method were first proposed and evaluated in ref. 9, and then Markov chains method was further studied in adaptive web sites10; in ref. 11, Popescul et al. studied the application of statistical relational learning to link prediction in the domain of scientific literature citations.
However, the mentioned methods for link prediction were mainly based on attributes of nodes. It is known that the structure of the network is easier to be obtained than the attributes of nodes, as a result, the network-structure-based link prediction have attracted increasing attention. Along this line, Liben-Nowell et al. developed approaches to link prediction based on measures for analyzing the “proximity” of nodes in a network12. Since hierarchical structure commonly exists in the food webs, biochemical networks, social networks and so forth, a link prediction method based on the knowledge of hierarchical structure was investigated in ref. 13, and they found that such a method can provide an accurate performance. Zhou et al. proposed a local similarity index—Resource Allocation (RA) index to predict the missing links, and their findings indicate that RA index has the best performance of link prediction14. Given that many networks are sparse and very huge, Liu et al. presented a local random walk method to solve the problem of missing link prediction, and which can give competitively good prediction or even better prediction than other random-walk-based methods while has a lower computational complexity15. In view of the local community features in many networks, Cannistraci et al. proposed an efficient computational framework called local community paradigm to calculate the link similarity between pairs of nodes3. Liu et al. designed a parameter-free local blocking predictor to detect missing links in given networks via local link density calculations, which performs better than the traditional local indices with the same time complexity16.
Since the structural properties of networks have significant effects on the performance of algorithms in link predictions, some methods have been proposed by making use of the structural properties of networks. Such as the algorithms by playing the roles of hierarchical structure13, clustering17, weak ties5, local community paradigm3 or multiple structural features18. However, current advances in incorporating structural properties into link prediction are still not enough. In this paper, by investigating the local structural properties in many real networks, we find a typical phenomenon: nodes are preferentially linked to the nodes with weak clique structure (PWCS). Then based on the observed phenomenon, a friend recommendation (FR) index is proposed. In this method, when a node j introduces one of his friends to a node i, he does not introduce their common neighbors to node i. Our results show that the performance of FR index is significantly better than CN, AA and RA indices since FR index can make good use of the PWCS in networks. At last, to further play the role of PWCS, we define a mixed friend recommendation (MFR) method, leading to the better performance of link prediction.
Results
Typical PWCS phenomenon
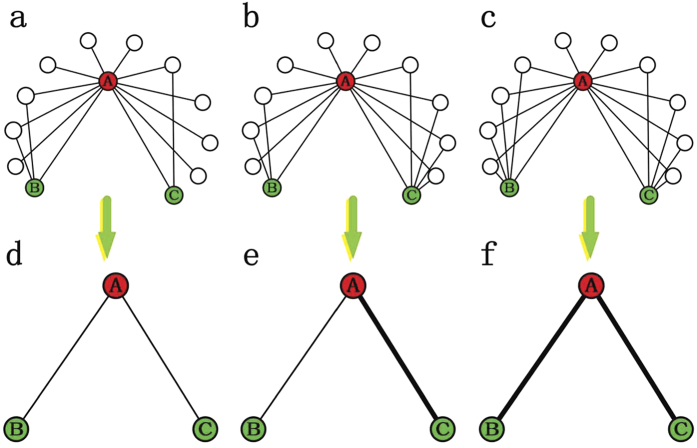
To check whether the PWCS phenomenon commonly exists in real networks, we divide all links into common links or strong-tie links by judging whether the number of common neighbors between the two endpoints is larger than a threshold β. Take Fig. 1 as an example, when we choose β = 3, the links {A, B} and {A, C} in Fig. 1(a–c) can be correspondingly degenerated to the sketches in Fig. 1(d–f), where common links and strong-tie links are marked by thin links and thick links, respectively.
Figure 1. Degenerating the upper sketches into the lower cases by judging whether two links {A, B} and {A, C} are strong-tie link or common link.
Here we assume that if the number of common neighbors between A and B (or A and C) is larger than β = 3, then the link is strong-tie link; otherwise, the link is common link in the opposite case. Thin lines and thick lines in (d–f) are the common links and the strong-tie links, respectively.
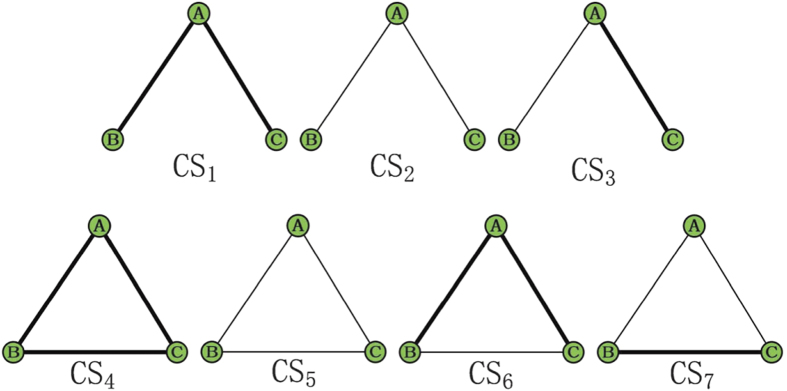
In this paper, the threshold β is chosen such that the number of common links and the number of strong-tie links are approximately equal in each network. Once the value of β is fixed, there are seven possible configurations for the connected subgraphs with 3 nodes (i.e., triples19), all the seven configurations are plotted in Fig. 2, where the thick links and thin lines denote strong-tie links and common links, respectively. Let Ni, i = 1, ···, 7 be the number of CSi, i = 1, ···, 7 (each CS represents a configuration in Fig. 2) in networks. If {A, B} and {A, C} are strong-tie links, then the probability of node B connecting node C is defined as20:
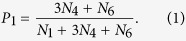 |
Eq. (1) can be understood in the following ways: from Fig. 2, one can find that only CS1, CS4 and CS6 have at least two strong-tie links, but CS1 does not form a triangle. There are three possible combinations of two strong-tie links for CS4, that is, {A, B} -{A, C}, {B, A}- {B, C} and {C, A}- {C, B}. However, there only exists one possible case ({A, B} -{A, C}) for CS6. As a result, N4 and N6 in Eq. (1) are multiplied by 3 and 1, respectively. The following Eq. (2) and Eq. (3) can be explained in a similar way.
Figure 2. Seven possible configurations of connected subgraphs with three nodes.
Thin lines and thick lines are the common links and the strong-tie links, respectively.
If only one of links {A, B} or {A, C} is strong-tie link, then the probability of node B connecting node C is defined as:
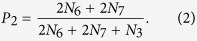 |
If neither of them is strong-tie link, then the probability of node B connecting node C is:
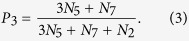 |
We define a subgraph with n nodes be a weak clique if the number of links among the n nodes is rather dense, which is an extended definition of n-clique where all pairs of nodes are connected. Next, by calculating the probability of node B connecting C, we can judge whether the phenomenon that nodes are preferentially linked to the nodes with weak clique structure (i.e., PWCS phenomenon) commonly exists in a network. We say that the PWCS phenomenon exists in the network if P1 > P2 and P1 > P3. Moreover, we say that the PWCS phenomenon is significant if P1 > P2 > P3, otherwise, the PWCS phenomenon is weak when P1 > P3 ≥ P2.
Table 1 reports the values of P1, P2 and P3 in the twelve real networks (labelled as RN) and the values on the corresponding null networks (labelled NN) are also comparatively shown. One can find that P1 > P2 and P1 > P3 in eleven networks except for Metabolic network (P1 < P3, emphasized by underlines). However, in the corresponding null networks, P1 ≈ P2 ≈ P3. Also, for C. celegans, FWEW, FWFW, Power, Router and PB networks, where P1 > P2 > P3. As a result, we can state that PWCS phenomenon is more significant in these six networks. Meanwhile, the values of P1, P2 and P3 for other 15 real networks are summarized in table. S1 in Supplementary Information, and where P1 > P2 and P1 > P3 for all of these real networks, which again validates that PWCS is a typical phenomenon.
Table 1. The values of P 1, P 2 and P 3 in 12 real networks (RN) and the corresponding null networks (NN) are reported.
| Network | Network | P1 | P2 | P3 |
|---|---|---|---|---|
| C. elegans | RN | 0.2351 | 0.1654 | 0.1519 |
| NN | 0.0483 | 0.0485 | 0.0487 | |
| NS | RN | 0.9292 | 0.2392 | 0.5970 |
| NN | 0 | 0.002 | 0.0022 | |
| FWEW | RN | 0.5998 | 0.4832 | 0.2504 |
| NN | 0.3691 | 0.3737 | 0.3761 | |
| FWFW | RN | 0.4191 | 0.3532 | 0.1230 |
| NN | 0.2545 | 0.2555 | 0.2554 | |
| USAir | RN | 0.7008 | 0.1519 | 0.2355 |
| NN | 0.0385 | 0.0387 | 0.0390 | |
| Jazz | RN | 0.6902 | 0.3968 | 0.4503 |
| NN | 0.14 | 0.1406 | 0.141 | |
| Tap | RN | 0.7862 | 0.2969 | 0.3673 |
| NN | 0.0069 | 0.0073 | 0.0073 | |
| Power | RN | 0.2781 | 0.0854 | 0.0686 |
| NN | 0 | 0 | 0 | |
| Metabolic | RN | 0.1630 | 0.0760 | 0.1643 |
| NN | 0.02 | 0.0198 | 0.0198 | |
| Yeast | RN | 0.5945 | 0.1498 | 0.1530 |
| NN | 0.0043 | 0.0042 | 0.0042 | |
| Router | RN | 0.1992 | 0.0254 | 0.0022 |
| NN | 0 | 0 | 0 | |
| PB | RN | 0.3998 | 0.1247 | 0.0855 |
| NN | 0.0224 | 0.0224 | 0.0224 |
Results in NN are marked in Italic. Results in networks with significant PWCS, i.e., P1 > P2 > P3 are shown in blue color, and results in Metabolic are marked by red color due to its specificity.
Friend recommendation model
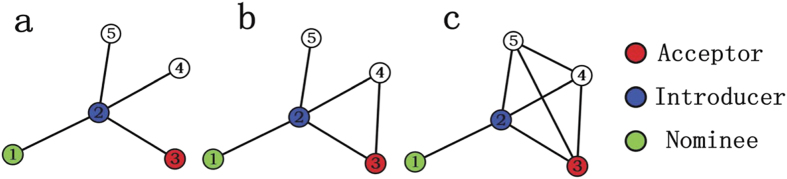
Given that PWCS phenomenon commonly exists in real networks, whether can we design an effective link prediction method based on this phenomenon. Considering the cases in Fig. 3, where node 3 asks its neighbor node 2 to introduce a friend to it. Since the number of common neighbors between node 2 and node 3 in Fig. 3(c) is larger than that of in Fig. 3(b) and is further larger than that of in Fig. 3(a), in other words, the strength of link {2, 3} in Fig. 3(c) is the strongest. According to PWCS phenomenon, the probability (labelled by f123) of node 1 (call nominee, green color) being introduced to node 3 (call acceptor, red color) by node 2 (call introducer, blue color) in Fig. 3(c) should be larger than that of in Fig. 3(b), and then further larger than that of in Fig. 3(a). To reflect the mentioned fact, we define filj be the probability of i being introduced to j by their common neighbor l, which is given as:
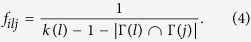 |
Based on the definition in Eq. (4), the values of f123 in Fig. 3(a–c) are 1/3, 1/2 and 1, respectively. That is to say, the probability filj can reflect the PWCS phenomenon in real networks.
Figure 3. The role of PWCS on the probability of f123.
Node 2 (blue color, call introducer) wants to introduce node 1 (green color, call nominee) to node 3 (red color, call acceptor). The number common neighbor between node 2 and node 3 in (a–c) is 0, 1 and 2, respectively. According to Eq. (4), one has (a) f123 = 1/3; (b) f123 = 1/2; (c) f123 = 1. Namely, the probability of node 1 being introduced to node 3 in (c) is larger than (b) and is further larger than in (a).
More importantly, Eq. (4) addresses two important facts: first, since node l will not introduce node j to j, as a result, 1 is subtracted in denominator of Eq. (4); second, in social communication, when a friend introduces one of his friends to me, he should introduce his friends but excluding the common friends. Therefore, the common neighbors set between j and l (i.e., Γ(l) ∩ Γ(j)) should be subtracted in denominator of Eq. (4). For instance, in Fig. 3(c), node 2 will not introduce node 3 to node 3, and nodes 4 and 5 should not be introduced to node 3.
Let fij be the weight of node i being introduced to node j (we use weight rather than probability since fij may larger than 1), which is written as:
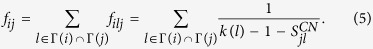 |
Here the value of fij increases with the number of common neighbors, and 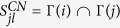 is CN index between node j and node l [see the definition of CN index in Methods section].
is CN index between node j and node l [see the definition of CN index in Methods section].
With the above preparations, the similarity index 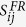 for a pair of nodes i and j is defined as
for a pair of nodes i and j is defined as
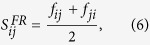 |
which guarantees 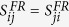 .
.
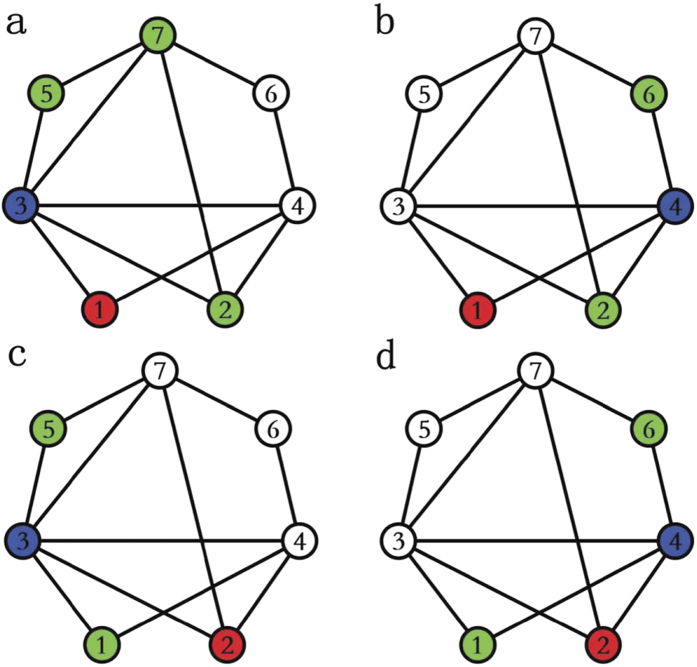
The sketches in Fig. 4 are given to show how to calculate the similarity between node 1 and node 2 based on the FR index. Also, the red, blue and green nodes denote the acceptors, introducers and nominees, respectively. Node 2 can be introduced to node 1 by node 3 (see Fig. 4(a)) or node 4 (see Fig. 4(b)). When node 3 is an introducer (see Fig. 4(a)), who will introduce nodes 2, 5 and 7 (green color) to node 1 with equal probability, but excludes node 4, i.e., f231 = 1/3. Similarly, when node 4 is an introducer (see Fig. 1(b)), who just introduces nodes 2 and 6 (green color) to node 1 with equal probability, but excludes node 3, i.e., f241 = 1/2. Therefore, the weight f21 = 1/3 + 1/2 = 5/6. Likely, from Fig. 5(c,d), the value of f12 = 1/2 + 1/2 = 1. Therefore, the FR similarity index is 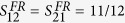 .
.
Figure 4. Calculation of the similarity.
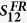 between node 1 and node 2. Nodes 1 and 2 can be introduced by their common neighbors 3 and 4. (a) Node 3 introduces his friends to node 1. Only neighbor nodes 2, 5, 7 can be introduced to node 1 but excludes node 4, since node 4 has been a friend of node 1. Thus, the probability of node 3 introducing node 2 to node 1 is: f231 = 1/3; (b) node 2 is introduced to node 1 by node 4, here only nodes 2 and 6 can be introduced to node 1. As a result, the probability f241 = 1/2; (c) node 1 is introduced to node 2 by node 3, here only nodes 1 and 5 can be introduced to node 1. As a result, the probability f132 = 1/2; (d) node 1 is introduced to node 2 by node 4, here only nodes 1 and 6 can be introduced to node 1. As a result, the probability f142 = 1/2. We have f21 = 1/3 + 1/2 by combing (a,b), and f12 = 1/2 + 1/2 by combing (c,d). So the FR similarity index is
between node 1 and node 2. Nodes 1 and 2 can be introduced by their common neighbors 3 and 4. (a) Node 3 introduces his friends to node 1. Only neighbor nodes 2, 5, 7 can be introduced to node 1 but excludes node 4, since node 4 has been a friend of node 1. Thus, the probability of node 3 introducing node 2 to node 1 is: f231 = 1/3; (b) node 2 is introduced to node 1 by node 4, here only nodes 2 and 6 can be introduced to node 1. As a result, the probability f241 = 1/2; (c) node 1 is introduced to node 2 by node 3, here only nodes 1 and 5 can be introduced to node 1. As a result, the probability f132 = 1/2; (d) node 1 is introduced to node 2 by node 4, here only nodes 1 and 6 can be introduced to node 1. As a result, the probability f142 = 1/2. We have f21 = 1/3 + 1/2 by combing (a,b), and f12 = 1/2 + 1/2 by combing (c,d). So the FR similarity index is 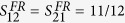 .
.
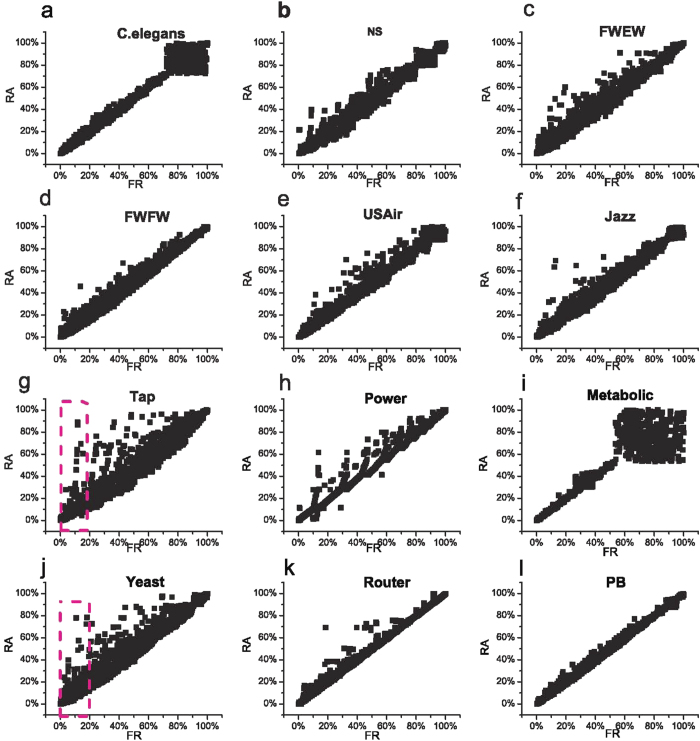
Figure 5. The correlation of ranking values between FR index and RA index based on Precision.
The percentage values in x-axis and y-axis are the top percentage ranking values of FR index and RA index, respectively. The regions marked by pink dash boundary in subfigures (g,j) correspond to the cases in which some links have higher FR ranking values but have lower RA ranking values.
Combing Eqs (4), (5) and (6), the advantages of FR index can be summarized as: (1) similar to many local similarity indices, the similarity between a pair of nodes increases with the number of common neighbors; (2) like AA index and RA index, FR index depresses the contribution of the high-degree common neighbors; (3) most importantly, FR index can make use of the PWCS phenomenon in many real networks; (4) FR index has higher resolution than other local similarity indices. For instance, the similarities 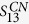 ,
, 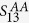 [see the definition of Eq. (14) in Methods section] and
[see the definition of Eq. (14) in Methods section] and 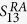 [see the definition of Eq. (15) in Methods section] are the same in Fig. 3(a–c). Yet, the value of
[see the definition of Eq. (15) in Methods section] are the same in Fig. 3(a–c). Yet, the value of 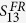 in Fig. 3(c) is larger than Fig. 3(b), and is further larger than Fig. 3(a).
in Fig. 3(c) is larger than Fig. 3(b), and is further larger than Fig. 3(a).
Performance of the FR model
The comparison of FR index with CN, AA and RA indices in twelve networks is summarized in Table 2. As shown in Table 2, FR index in general outperforms the other three indices in link prediction, regardless of AUC or Precision [see definitions in Methods section]. The highest accuracy in each line is emphasized in bold. Furthermore, Precision as a function of L in six networks is presented in Fig. S1 in Supplementary Information, which also confirms the good performance of FR index.
Table 2. Comparison of S FR with S CN , S AA and S RA in 12 networks, including AUC and Precision.
| Network | Metric | CN | AA | RA | FR |
|---|---|---|---|---|---|
| C. elegans | AUC | 0.8501 | 0.8663 | 0.8701 | 0.8756 |
| Precision | 0.1306 | 0.1374 | 0.1315 | 0.1504 | |
| NS | AUC | 0.9913 | 0.9916 | 0.9917 | 0.9916 |
| Precision | 0.8707 | 0.9731 | 0.9712 | 0.9832 | |
| FWEW | AUC | 0.6868 | 0.6939 | 0.7017 | 0.7595 |
| Precision | 0.1415 | 0.1551 | 0.1664 | 0.2763 | |
| FWFW | AUC | 0.6074 | 0.6097 | 0.6142 | 0.6623 |
| Precision | 0.0837 | 0.0853 | 0.082 | 0.1798 | |
| USAir | AUC | 0.9558 | 0.9676 | 0.9736 | 0.9752 |
| Precision | 0.606 | 0.6218 | 0.6337 | 0.6586 | |
| Jazz | AUC | 0.9563 | 0.963 | 0.9717 | 0.9714 |
| Precision | 0.8247 | 0.8401 | 0.8192 | 0.8406 | |
| Tap | AUC | 0.9538 | 0.9545 | 0.9548 | 0.955 |
| Precision | 0.7594 | 0.78 | 0.7818 | 0.8659 | |
| Power | AUC | 0.6249 | 0.6251 | 0.6245 | 0.6248 |
| Precision | 0.1215 | 0.0952 | 0.0801 | 0.1275 | |
| Metabolic | AUC | 0.9248 | 0.9565 | 0.9612 | 0.9623 |
| Precision | 0.2026 | 0.2579 | 0.3219 | 0.3302 | |
| Yeast | AUC | 0.9158 | 0.9161 | 0.9167 | 0.9172 |
| Precision | 0.6821 | 0.6958 | 0.4988 | 0.8041 | |
| Router | AUC | 0.6519 | 0.6523 | 0.652 | 0.6519 |
| Precision | 0.1144 | 0.1104 | 0.0881 | 0.0592 | |
| PB | AUC | 0.9239 | 0.9275 | 0.9286 | 0.9309 |
| Precision | 0.4205 | 0.3782 | 0.2509 | 0.3454 |
The highest value in each row is marked in bold.
Moreover, the correlation of ranking values between FR index and RA index is given in Fig. 5, where the percentage values in x or y axis is the top percentage of ranking values based on Precision. As a result, a small percentage value means a higher ranking value. Figure 5 indicates that a high RA ranking value of links gives rise to a high FR ranking value. However, a high FR ranking value of links may induce a low RA ranking value of links. Take Tap and Yeast networks as examples, based on FR index, some links have higher ranking values, however their corresponding ranking values based on RA index may be very small (see the regions marked by pink dash boundary in Fig. 5(g,j)).
By analyzing a typical case in the Yeast network (see Fig. 6), where two nodes A and B are the neighbors of introducer C (in fact, there has a link connecting A and B in the Yeast network). Since links {A, C} and {B, C} are strong-tie links. When using FR index, the similarity 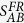 is rather large, which can predict the existence of link {A, B}. However, for RA index, since the large degree value of introducer C, the similarity
is rather large, which can predict the existence of link {A, B}. However, for RA index, since the large degree value of introducer C, the similarity 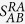 is very small, such an existing link {A, B} cannot be accurately predicted by RA index.
is very small, such an existing link {A, B} cannot be accurately predicted by RA index.
Figure 6. A typical case in the Yeast network is considered to emphasize the difference between FR index and RA index, where nodes A, B and C are the node 1175, 421 and 205 in the Yeast network.
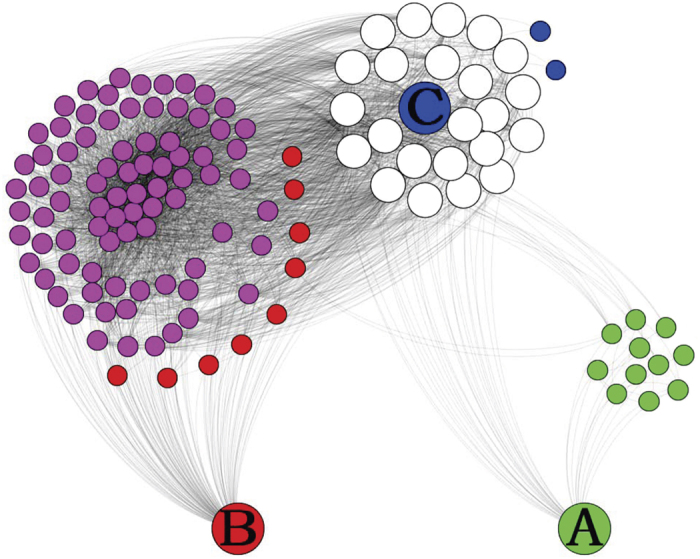
Two links {A, C} and {B, C} share a common endpoint C, and both of them are strong-tie links. Therefore, the similarity 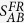 is rather large. However, when using RA index, the ranking number of
is rather large. However, when using RA index, the ranking number of 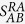 is very low owing to the large degree value of node C, causing the failure of RA index in predicting such an existing link. Red nodes, green nodes and blue nodes are the neighbors of A, B and C (including themselves), respectively. Purple nodes are the common neighbors of A and C; white nodes are the common neighbors of A, B and C.
is very low owing to the large degree value of node C, causing the failure of RA index in predicting such an existing link. Red nodes, green nodes and blue nodes are the neighbors of A, B and C (including themselves), respectively. Purple nodes are the common neighbors of A and C; white nodes are the common neighbors of A, B and C.
Role of PWCS
We have validated that the FR index based on PWCS phenomenon can improve the performance of link prediction, and the reasons were also analyzed. Here we want to know how the strength of PWCS affects the performance of link prediction. For this purpose, we propose a generalized friend recommendation (GFR) index, which is given as:
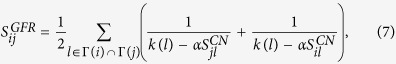 |
where parameter 0 ≤ α ≤ 1 is used to uncover the role of PWCS in link prediction. As α = 0, Eq. (7) returns to RA index, that is, 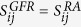 . When α = 1, the difference between FR method and GFR method is the absence of 1 in the denominators of Eq. (7), therefore, we can simply view GFR index is the same as FR index when α = 1. As a result, with the increasing of α from zero to one, SSFR index can comprehensively investigate the role of PWCS in the RA index and FR index.
. When α = 1, the difference between FR method and GFR method is the absence of 1 in the denominators of Eq. (7), therefore, we can simply view GFR index is the same as FR index when α = 1. As a result, with the increasing of α from zero to one, SSFR index can comprehensively investigate the role of PWCS in the RA index and FR index.
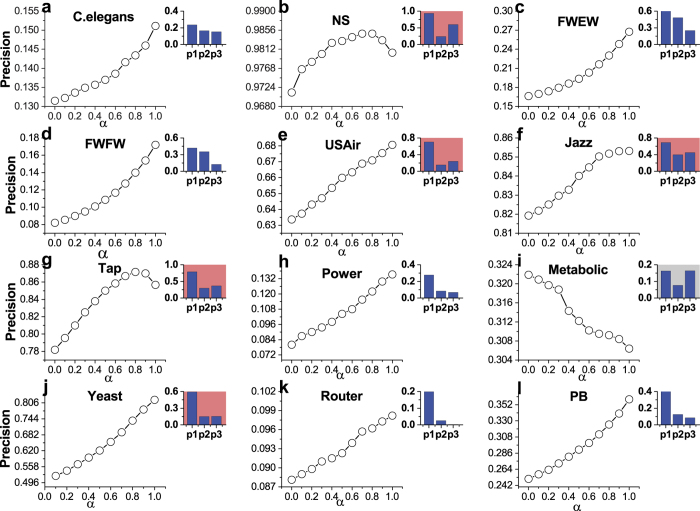
The effect of α on the Precision in all twelve networks is plotted in Fig. 7. As illustrated in Fig. 7, several interesting phenomena and meaningful conclusions can be summarized: First, except for Metabolic network, the Precision for the case of α > 0 is far larger than the case of α = 0 (i.e., RA index) in all other 11 networks. Since P1 > P2 and P1 > P3 in these 11 networks, which indicates that PWCS phenomenon in networks can ensure the higher accuracy of FR index (i.e., α = 1) in link prediction; Second, Metabolic network has non-PWCS phenomenon since P1 > P2 and P2 < P3, and Fig. 7(i) suggests that Precision decreases with the value of α. In other words, FR index is invalid in network with non-PWCS phenomenon, which again emphasizes the importance of PWCS in link prediction; At last, by systematically comparing the subfigures in Fig. 7, one can see that, when the networks with weak PWCS P1 > P3 ≥ P2 (i.e., the insets are light red background, see Fig. 7(b,e–g,j)), Precision increases with α at first and then decreases when α is further increased (except Fig. 7(e)). However, when P1 > P2 > P3 (i.e., networks with significant PWCS, the insets are white background, see Fig. 7(a,c,d,h,k,l)), Precision always increases with the value of α even when α = 1.0.
Figure 7. Effects of α in Eq. (8) on Precision are plotted in 12 networks.
Inset in each subfigure is to show the values of P1, P2 and P3. The background of inset is white color when P1 > P2 > P3; the background of inset is light red color when P1 > P3 ≥ P2. Otherwise, the background of inset for Metabolic is gray color.
In view of this observation, we can conjecture the role of PWCS can be further explored when the PWCS phenomenon is significant. Unfortunately, the maximal value α in Eq. (7) is one, the denominator may be negative if α > 1. So we design a new index to further explore the role of significant PWCS.
Since Eq. (7) can be rewritten as
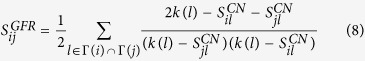 |
when α = 1. To further play the role of PWCS, another similarity index, called strong friend recommendation (labelled as SFR) index, is given in following
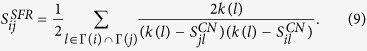 |
Combing Eq. (8) with Eq. (9), we can find that two subtrahends 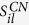 and
and 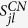 in the numerator of Eq. (8) are removed. So Eq. (9) can better play the role of PWCS.
in the numerator of Eq. (8) are removed. So Eq. (9) can better play the role of PWCS.
We conjecture that the performance of SFR index is better than GFR index when P1 > P2 > P3 (i.e., significant PWCS), and worse than that of GFR index when P1 < P2 and P1 < P3 (i.e., non-PWCS). However, it is difficult to distinguish which one has better performance when P1 > P3 ≥ P2 (i.e., weak PWCS). As presented in Table 3, Precision in 12 networks validates our conjecture.
Table 3. The comparison of Precision between SFR index and GFR index (α = 1) in 12 networks.
| Index |
P1 > P2 > P3 |
|||||
|---|---|---|---|---|---|---|
| C. elegans | FWEW | FWFW | Power | Router | PB | |
| GFR (α = 1) | 0.1511 | 0.2676 | 0.172 | 0.1354 | 0.0982 | 0.3595 |
| SFR | 0.1577 | 0.2912 | 0.2057 | 0.1658 | 0.112 | 0.4353 |
| P1 > P3 ≥ P2 | P1 < P3, P2 < P3 | |||||
| Index | NS | USAir | Jazz | Tap | Yeast | Metabolic |
| GFR (α = 1) | 0.9804 | 0.6807 | 0.8532 | 0.8568 | 0.8178 | 0.3064 |
| SFR | 0.9744 | 0.6866 | 0.8739 | 0.8485 | 0.8587 | 0.2912 |
The results suggest that the accuracy of link prediction can be further improved by SFR index when the networks have significant PWCS. The highest value in each case is marked as bold.
Synthesizing the above results, we can find that the ranking of P1, P2 and P3 has a determinant effect on the performance of the proposed index. Inspired by this clue, we may design a universal indicator to do link prediction based on the values of P1, P2 and P3 in different networks. To this end, we design a mixed friend recommendation (labelled MFR) index:
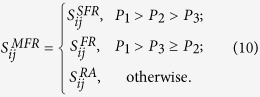 |
Table 4 lists the results of MFR index and FR index in 7 networks (since MFR index is the same to FR index when P1 > P3 ≥ P2, in this case, it is unnecessary to compare the two indices). The results in Table 4 indicate that, compared with FR index, MFR index can further improve the accuracy of link prediction.
Table 4. Comparison of Precision between FR index and MFR index in 7 networks.
| Metric | Index | C.elegans | FWEW | FWFW | Power | Router | PB | Metabolic |
|---|---|---|---|---|---|---|---|---|
| AUC | FR | 0.8756 | 0.7595 | 0.6623 | 0.6248 | 0.6519 | 0.9309 | 0.9623 |
| MFR | 0.8771 | 0.7771 | 0.6878 | 0.6247 | 0.6516 | 0.9314 | 0.9612 | |
| Precision | FR | 0.1504 | 0.2763 | 0.1798 | 0.1275 | 0.0592 | 0.3454 | 0.3302 |
| MFR | 0.1577 | 0.2912 | 0.2057 | 0.1658 | 0.112 | 0.4353 | 0.3219 |
The highest value in each case is given in bold.
Conclusion
In summary, by analyzing the structural properties in real networks, we have found that there exists a typical phenomenon: nodes are preferentially linked to the nodes with weak clique structure. Then we have proposed a friend recommendation model to better predict the missing links based on the observed phenomenon. Through the detailed analysis and experimental results, we have shown that FR index has several typical characteristics: First, FR index is based on the information of common neighbors, which is a local similarity index. Thus, the algorithm is simple and has low complexity; Second, the common neighbors with small degrees have greater contributions than the common neighbors with larger degrees; Third, FR index can take full advantage of the PWCS phenomenon, and so forth.
Furthermore, we have also proposed an SFR index to further improve the accuracy of link prediction when networks have significant PWCS phenomenon. At last, by judging whether the networks have significant PWCS, weak PWCS or non-PWCS phenomenon, we have also proposed a mixed friend recommendation index which can increase the accuracy of link prediction in different networks. In this work, we mainly applied FR index to unweighed and undirected networks, and how to generalize our FR index to weighted21,22 or directed networks23 is our further purpose.
Methods
Link prediction algorithm
Considering an undirected and unweighed network G(V, E), where V is the set of nodes and E is the set of links. The multiple links and self-connections are not allowed. For a network with size N, the universal set of all possible links, is denoted by U, consisting of 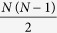 pairs of links. For each pair of nodes, x, y ∈ V, we assign a score, Sxy, according to a defined similarity measure. Higher score means higher similarity between x and y, and vice versa. Since G is undirected, the score is supposed to be symmetry, that is Sxy = Syx. All the nonexistent links are sorted in a descending order according to their scores, and the links at the top are most likely to exist14,15. To test the prediction accuracy of each index, we adopt the approach used in ref. 14. The link set E is randomly divided into two sets E = ET ∪ EP with ET ∩ EP = ∅. Where set ET is the training set and is supposed to be known information, and EP is the testing set for the purpose of testing and no information therein is allowed to be used for prediction. As in previous literatures, the training set ET always contains 90% of links in this work, and the remaining 10% of links constitute the testing set.
pairs of links. For each pair of nodes, x, y ∈ V, we assign a score, Sxy, according to a defined similarity measure. Higher score means higher similarity between x and y, and vice versa. Since G is undirected, the score is supposed to be symmetry, that is Sxy = Syx. All the nonexistent links are sorted in a descending order according to their scores, and the links at the top are most likely to exist14,15. To test the prediction accuracy of each index, we adopt the approach used in ref. 14. The link set E is randomly divided into two sets E = ET ∪ EP with ET ∩ EP = ∅. Where set ET is the training set and is supposed to be known information, and EP is the testing set for the purpose of testing and no information therein is allowed to be used for prediction. As in previous literatures, the training set ET always contains 90% of links in this work, and the remaining 10% of links constitute the testing set.
Evaluation metrics
Two standard metrics are used to quantify the accuracy of prediction algorithms: area under the receiver operating characteristic curve (AUC) and Precision5.
Area under curve (AUC) can be interpreted as the probability that a randomly chosen missing link (a link in EP) is given a higher score than a randomly chosen nonexistent link (a link in U − EP). When implementing, among n independent comparisons, if there are n′ times the missing link having a higher score and n′′ times they are of the same score, AUC can be read as follow5:
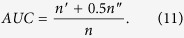 |
If all the scores generated from independent and identical distribution, the accuracy should be about 0.5. Therefore, the degree to which the accuracy exceeds 0.5 indicates how much the algorithm performs better than pure chance.
Precision is the ratio of the number of missing links predicted correctly within those top-L ranked links to L, and L = 100 in this paper [Precision as a function of L is compared in Fig. S1 in Supplementary Information, which confirms that our FR index is better than other three indices for a large range of L]. If m links are correctly predicted, then Precision can be calculated as5:
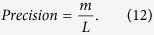 |
Benchmarks
We mainly compare three local similarity indices for link prediction, including (1) Common Neighbors(CN)24; (2) Adamic-Adar (AA) index25; (3) Resource Allocation (RA) index14. Among which, CN index is the simplest index. AA index and RA index have the similar form, and they both depress the contribution of the high-degree common neighbors, however, Zhou et al. have shown that the performance of RA index is generally better than AA index.
Let Γ(i) be the neighbor set of node i, |.| be the cardinality of the set, and k(i) be the degree of node i. Then CN index, AA index and RA index are defined as
CN index
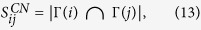 |
AA index
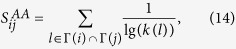 |
RA index
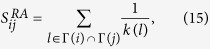 |
respectively.
Data Set
In this paper, we choose twelve representative networks drawn from disparate fields: including: (1) C. elegans-The neural network of the nematode worm C. elegans26; (2) NS-A coauthorship network of scientists working on network theory and experiment27; (3) FWEW-A 66 component budget of the carbon exchanges occurring during the wet and dry seasons in the graminoid ecosystem of South Florid28; (4) FWFW-A food web in Florida Bay during the rainy season28; (5) USAir-The US Air transportation system5; (6) Jazz-A collaboration network of jazz musicians29; (7) TAP-yeast protein-protein binding network generated by tandem affinity purification experiments30; (8) Power-An electrical power grid of the western US26; (9) Metabolic-A metabolic network of C. elegans31; (10) Yeast-A protein-protein interaction network in budding yeast32; (11) Router-A symmetrized snapshot of the structure of the Internet at the level of autonomous systems33; (12) PB-A network of the US political blogs34. Topological features of these networks are summarized in Table 5.
Table 5. The basic topological features of twelve example networks.
| Network | N | M | C | r | H |
|---|---|---|---|---|---|
| C. elegans | 297 | 2148 | 0.308 | −0.163 | 1.801 |
| NS | 1589 | 2742 | 0.791 | 0.462 | 2.011 |
| FWEW | 69 | 880 | 0.552 | −0.298 | 1.275 |
| FWFW | 128 | 2075 | 0.335 | −0.112 | 1.237 |
| USAir | 332 | 2126 | 0.749 | −0.208 | 3.464 |
| Jazz | 198 | 2742 | 0.633 | 0.02 | 1.395 |
| Tap | 1373 | 6833 | 0.557 | 0.579 | 1.644 |
| Power | 4941 | 6594 | 0.107 | 0.003 | 1.45 |
| Metabolic | 453 | 2025 | 0.655 | −0.226 | 4.485 |
| Yeast | 2375 | 11693 | 0.388 | 0.454 | 3,476 |
| Router | 5022 | 6258 | 0.033 | −0.138 | 5.503 |
| PB | 1222 | 16724 | 0.36 | −0.221 | 2.971 |
N and M are the total numbers of nodes and links, respectively. C and r are clustering coefficient and assortative coefficient, respectively. H is the degree heterogeneity, defined as 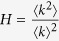 , where 〈k〉 denotes the average degree19.
, where 〈k〉 denotes the average degree19.
Additional Information
How to cite this article: Ma, C. et al. Playing the role of weak clique property in link prediction: A friend recommendation model. Sci. Rep. 6, 30098; doi: 10.1038/srep30098 (2016).
Supplementary Material
Acknowledgments
This work is supported by the National Natural Science Foundation of China (Grant No. 61473001, 61433014), and partially supported by open fund of Key Laboratory of Computer Network and Information Integration (Southeast University), Ministry of Education (No. K93-9-2015-03B).
Footnotes
Author Contributions C.M., T.Z. and H.-F.Z. devised the research project. C.M. and H.-F.Z. implemented experiments. C.M., T.Z. and H.-F.Z. analyzed the results. C.M., T.Z. and H.-F.Z. wrote the paper.
References
- Getoor L. & Diehl C. P. Link mining: a survey. ACM SIGKDD Explorations Newsletter 7, 3–12 (2005). [Google Scholar]
- Scellato S., Noulas A. & Mascolo C. Exploiting place features in link prediction on location-based social networks. In Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining, 1046–1054 (ACM, 2011).
- Cannistraci C. V., Alanis-Lobato G. & Ravasi T. From link-prediction in brain connectomes and protein interactomes to the local-community-paradigm in complex networks. Scientific Reports 3, 1613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimerà R. & Sales-Pardo M. Missing and spurious interactions and the reconstruction of complex networks. Proceedings of the National Academy of Sciences 106, 22073–22078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü L. & Zhou T. Link prediction in complex networks: A survey. Physica A: Statistical Mechanics and its Applications 390, 1150–1170 (2011). [Google Scholar]
- Lü L., Pan L., Zhou T., Zhang Y.-C. & Stanley H. E. Toward link predictability of complex networks. Proceedings of the National Academy of Sciences 112, 2325–2330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü L., Jin C.-H. & Zhou T. Similarity index based on local paths for link prediction of complex networks. Physical Review E 80, 046122 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang P., Wang X., Wang F., Zeng A. & Xiao J. Measuring the robustness of link prediction algorithms under noisy environment. Scientific Reports 6, 18881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarukkai R. R. Link prediction and path analysis using markov chains. Computer Networks 33, 377–386 (2000). [Google Scholar]
- Zhu J., Hong J. & Hughes J. G. Using markov chains for link prediction in adaptive web sites. In Soft-Ware 2002: Computing in an Imperfect World , 60–73 (Springer, 2002). [Google Scholar]
- Popescul A. & Ungar L. H. Statistical relational learning for link prediction. In IJCAI workshop on learning statistical models from relational data , vol. 2003 (Citeseer, 2003). [Google Scholar]
- Liben-Nowell D. & Kleinberg J. The link-prediction problem for social networks. Journal of the American Society for Information Science and Technology 58, 1019–1031 (2007). [Google Scholar]
- Clauset A., Moore C. & Newman M. E. Hierarchical structure and the prediction of missing links in networks. Nature 453, 98–101 (2008). [DOI] [PubMed] [Google Scholar]
- Zhou T., Lü L. & Zhang Y.-C. Predicting missing links via local information. The European Physical Journal B 71, 623–630 (2009). [Google Scholar]
- Liu W. & Lü L. Link prediction based on local random walk. EPL (Europhysics Letters) 89, 58007 (2010). [Google Scholar]
- Liu Z., Dong W. & Fu Y. Local degree blocking model for link prediction in complex networks. Chaos: An Interdisciplinary Journal of Nonlinear Science 25, 013115 (2015). [DOI] [PubMed] [Google Scholar]
- Feng X., Zhao J. & Xu K. Link prediction in complex networks: a clustering perspective. The European Physical Journal B 85, 1–9 (2012). [Google Scholar]
- Zhu B. & Xia Y. An information-theoretic model for link prediction in complex networks. Scientific Reports 5, 13707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. E. J. Networks: an introduction (Oxford University Press, 2010). [Google Scholar]
- Lü L. & Zhou T. Link prediction in weighted networks: The role of weak ties. EPL (Europhysics Letters) 89, 18001 (2010). [Google Scholar]
- Zhao J. et al. Prediction of links and weights in networks by reliable routes. Scientific Reports 5, 12261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aicher C., Jacobs A. Z. & Clauset A. Learning latent block structure in weighted networks. Journal of Complex Networks 3, 221–248 (2015). [Google Scholar]
- Guo F., Yang Z. & Zhou T. Predicting link directions via a recursive subgraph-based ranking. Physica A: Statistical Mechanics and its Applications 392, 3402–3408 (2013). [Google Scholar]
- Newman M. E. Clustering and preferential attachment in growing networks. Physical Review E 64, 025102 (2001). [DOI] [PubMed] [Google Scholar]
- Adamic L. A. & Adar E. Friends and neighbors on the web. Social Networks 25, 211–230 (2003). [Google Scholar]
- Watts D. J. & Strogatz S. H. Collective dynamics of small-worldnetworks. Nature 393, 440–442 (1998). [DOI] [PubMed] [Google Scholar]
- Von Mering C. et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403 (2002). [DOI] [PubMed] [Google Scholar]
- Ulanowicz R., Bondavalli C. & Egnotovich M. Network analysis of trophic dynamics in south florida ecosystem, fy 97: The florida bay ecosystem. Annual Report to the United States Geological Service Biological Resources Division Ref. No. [UMCES] CBL 98–123 (1998). [Google Scholar]
- Gleiser P. M. & Danon L. Community structure in jazz. Advances in Complex Systems 6, 565–573 (2003). [Google Scholar]
- Gavin A.-C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002). [DOI] [PubMed] [Google Scholar]
- Duch J. & Arenas A. Community detection in complex networks using extremal optimization. Physical Review E 72, 027104 (2005). [DOI] [PubMed] [Google Scholar]
- Bu D. et al. Topological structure analysis of the protein-protein interaction network in budding yeast. Nucleic Acids Research 31, 2443–2450 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring N., Mahajan R., Wetherall D. & Anderson T. Measuring isp topologies with rocketfuel. Networking, IEEE/ACM Transactions on 12, 2–16 (2004). [Google Scholar]
- Reese S. D., Rutigliano L., Hyun K. & Jeong J. Mapping the blogosphere professional and citizen-based media in the global news arena. Journalism 8, 235–261 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.