Summary
Cytomegalovirus (CMV) infection markedly expands NKG2C+/NKG2A− NK cells, which are potent killers of infected cells expressing human leucocyte antigen (HLA)‐E. As HLA‐E is also over‐expressed in several haematological malignancies and CMV has been linked to a reduced risk of leukaemic relapse, we determined the impact of latent CMV infection on NK cell cytotoxicity against four tumour target cell lines with varying levels of HLA‐E expression. NK cell cytotoxicity against K562 (leukaemia origin) and U266 (multiple myeloma origin) target cells was strikingly greater in healthy CMV‐seropositive donors than seronegative donors and was associated strongly with target cell HLA‐E and NK cell NKG2C expression. NK cell cytotoxicity against HLA‐E transfected lymphoma target cells (221.AEH) was ∼threefold higher with CMV, while NK cell cytotoxicity against non‐transfected 721.221 cells was identical between the CMV groups. NK cell degranulation (CD107a+) and interferon (IFN)‐γ production to 221.AEH cells was localized almost exclusively to the NKG2C subset, and antibody blocking of NKG2C completely eliminated the effect of CMV on NK cell cytotoxicity against 221.AEH cells. Moreover, 221.AEH feeder cells and interleukin (IL)−15 were found to expand NKG2C+/NKG2A– NK cells preferentially from CMV‐seronegative donors and increase NK cell cytotoxicity against HLA‐E+ tumour cell lines. We conclude that latent CMV infection enhances NK cell cytotoxicity through accumulation of NKG2C+ NK cells, which may be beneficial in preventing the initiation and progression of haematological malignancies characterized by high HLA‐E expression.
Keywords: 221 AEH, CD57, CD158, K562, leukaemia, lymphoma, multiple myeloma, NKG2A, U266
Introduction
Cytomegalovirus (CMV) is a prevalent β‐herpesvirus that infects 50–80% of all adults in the United States 1. While CMV is often well controlled in immunocompetent hosts, it has been linked to an increased risk of cancer‐related mortality 2, and exposure to CMV in immunocompromised organ transplant recipients is associated with increased post‐transplant mortality 3. CMV drives both T cell and natural killer (NK) cell differentiation, underscoring the importance of these cell types in curtailing CMV. Cytokine‐driven NK cell responses to CMV typically precede the antigen‐driven T cell response, with more effective NK cell responses requiring less vigorous T cell responses to exert viral control 4, 5. To overcome these rheostat‐like effects of NK cells on the regulation of persistent infections, CMV has evolved a myriad of immunoevasive strategies to evade detection 6, 7. For example, several CMV genes have been identified which down‐regulate ligands for NK cell activating receptors (such as NKG2D, DNAM‐1 and NKp46) that are critical to the recognition of CMV‐infected cells 8, 9 and the maintenance of NK cell effector functions in the host 9, 10, 11. CMV also induces expression of human leucocyte antigen (HLA) class I homologues by infected cells, which allows for inhibition of NK cells via ligation with inhibitory receptors, such as LIR‐1 12, 13.
NK cells are inherently plastic, and have evolved countermeasures to minimize viral escape 14, 15. NKG2C is an NK cell activating receptor that ligates with HLA‐E 16, a non‐classical HLA molecule expressed by CMV‐infected cells 17. Both acute and latent CMV infections are associated with a marked expansion of NKG2C+ (Ly49H+ in mice) NK cells in both mice and humans 15, 18, 19, 20, thus facilitating the recognition and destruction of CMV‐infected cells through NKG2C/HLA‐E interactions. A portion of these ‘CMV‐specific’ cells remain as long‐lived ‘memory’ NK cells capable of generating recall responses 21, and are still elevated markedly in transplant patients 250 days after resolution of viraemia 19. However, the high expression of the putative terminal differentiation marker CD57 indicates that NKG2C+ NK cells may have undergone clonal exhaustion 22. Despite this, it is evident that NKG2C+ NK cells help protect against CMV, as an increased proportion of these cells is associated with a lower risk of acute CMV infection in patients undergoing solid organ transplantation 23, and NKG2C+ NK cells taken from CMV‐infected donors show enhanced expansion in response to CMV reactivation in haematopoietic cell transplant recipients 24. Moreover, NKG2C+ NK cells can be expanded in vitro with interleukin (IL)−15 and transfected lymphoma target (221.AEH) cells (HLA‐Ehigh lymphoma), implicating the up‐regulation of HLA‐E in the clonal‐like response of NK cells to CMV infection 25, 26. It remains to be seen, however, how preferential expansion of NKG2C+ NK cells affects anti‐tumour cytotoxicity.
CMV is often considered to be an immunological burden within the T cell compartment that exerts mainly negative effects on immune status and overall health 27; however, it has been suggested recently that latent herpesviruses may play a pivotal role in ‘arming’ NK cells to destroy target cells adequately 28. Mice with latent murid herpesvirus 4 infection show increased granzyme B protein expression, interferon (IFN)‐γ production and NK cell cytotoxicity, which can protect against a lethal lymphoma challenge 28. However, it is not yet known, if the expansion of NKG2C+ NK cells with CMV infection impacts anti‐tumour immunity in humans. Given that many haematological malignancies and solid tumours are associated with an over‐expression of HLA‐E 29, cancer patients with a latent CMV infection, or who experience a mild but controllable CMV reactivation after solid organ or haematopoietic stem cell transplantation, could be at an advantage due to the CMV‐induced expansion of NKG2C+ NK cells in vivo. For example, donor CMV seropositivity 30 and CMV reactivation 31, 32 are associated with a decreased risk of relapse in acute myeloid leukaemia (AML) patients, although the mechanisms underpinning this beneficial CMV effect remain to be elucidated.
The aims of this study were twofold. First, we wanted to determine the effect of latent CMV infection and the proportion of NKG2C+ NK cells on cytotoxicity against four tumour cell lines with varying degrees of HLA‐E expression. Secondly, we determined the effect of HLA‐E+ feeder cells on NKG2C+/NKG2A– NK cell expansion and cytotoxicity in CMV‐seronegative subjects. In this report, we show that latent CMV infection is associated with an NKG2C‐dependent increase in NK cell cytotoxicity against HLA‐E‐expressing tumour cell lines. We also show that highly cytotoxic NKG2C+/NKG2A– NK cells can be expanded preferentially from CMV‐seronegative subjects using 221.AEH (HLA‐Ehigh lymphoma) feeder cells. Overall, our findings suggest that the enrichment of the NKG2C+ NK cell fraction may serve as a simple strategy for enhancing the anti‐tumour cytotoxicity of NK cells for immunotherapy.
Materials and methods
Subjects
Thirty healthy adults (aged 32.3 ± 4.8 years) participated voluntarily in this study. Subjects were between the ages of 18 and 50 years and not taking any immunomodulatory medication. Potential subjects were excluded if they had used tobacco products within the previous 6 months; had a body mass index (BMI) > 30 kg/m2; used any medication known to affect the immune system; were pregnant; had chronic/debilitating arthritis; had diabetes; were bedridden in the past 3 months; had a common illness (i.e. colds) within the past 6 weeks; had a central or peripheral nervous disorder; or had any autoimmune disease or chronic infectious disease (i.e. hepatitis or HIV). Abstinence from alcohol, caffeine and physical activity 24 h prior to trials as well as elimination of vitamin/mineral supplementation at least 4 weeks prior to taking part in the study was required and confirmed verbally with the subjects on their arrival to the laboratory. All subjects provided written informed consent prior to participating in the study and the Committee for the Protection of Human Subjects at the University of Houston approved the protocol. Physical characteristics of the subjects are presented in Table 1.
Table 1.
Physical characteristics of the participants (n CMV+ = 15; n CMV– = 15)
| Characteristics | CMV+ (NKG2Chigh) (n = 7) | CMV+ (NKG2Clow) (n = 8) | CMV– (n = 15) | One‐way anova F‐statistic (P‐value) |
|---|---|---|---|---|
| Gender | (2F, 5M) | (2F, 6M) | (4F, 11M) | |
| Age (years) | 34·9 ± 4·5 | 32·8 ± 5·6 | 31·0 ± 4·7 | 0·41 (0·67) |
| BMI (kg/m−2) | 25·3 ± 3·8 | 24·5 ± 2·4 | 24·9 ± 3·3 | 0·23 (0·79) |
| Physical activity Rating (0–7)* | 5·8 ± 1·9 | 6·0 ± 1·8 | 5·9 ± 1·4 | 0·11 (0·90) |
Data are mean ± standard deviation. There was no effect of CMV/NKG2C category on any of the physical characteristics (P > 0.05). *Jackson Physical Activity Rating (PA‐R) 33. CMV = cytomegalovirus (CMV); anova = analysis of variance; M = male; F = female; BMI = body mass index.
Blood processing
All blood samples were collected between 6:00 and 10:00 a.m. Fasting serum samples were frozen at −80° C until measurement of CMV immunoglobulin (Ig)G antibodies, which were analysed in duplicate using commercially available enzyme‐linked immunosorbent assay (ELISA) kits (BioCheck, Foster City, CA, USA) and a 96‐well microplate reader (Molecular Devices, Sunnyvale, CA, USA), in accordance with the manufacturer's instructions. The cut‐off for being defined as seropositive was an IgG index of 1, which corresponded to an antibody titre of 1·2 (IU/ml). Fasting whole blood samples were processed immediately for phenotypical and functional analyses of NK cells using flow cytometric techniques. Ethylenediamine tetraacetic acid (EDTA) blood tubes (Becton Dickinson, Franklin Lakes, NJ, USA) were used for the NK cell phenotypical analysis and ACD blood tubes (Becton Dickinson) were used for the NK cell functional assays.
Labelling with monoclonal antibodies against surface antigens
A four‐colour direct immunofluorescence procedure was used to label whole blood with the following monoclonal antibodies: peridinin chlorophyll (PerCP)‐eFluor710‐conjugated anti‐CD56 [IgG1, clone CMSSB (FL3)]; Alexa488‐conjugated anti‐KLRG1 (clone 13F12F2) 34, anti‐NKG2C (IgG1, clone 134591) or fluorescein isothiocyanate (FITC)‐conjugated anti‐CD3 [IgG1, clone SK7 (FL1)]; phycoerythrin (PE)‐conjugated anti‐CD57 (IgM, clone TB01), anti‐NKG2A (IgG2b, clone Z199), anti‐CD158a/h (IgG2b, clone HP‐MA4), anti‐CD158b1/b2/j (IgG1, clone GL183) or anti‐CD158e1/e2 [IgG1, clone Z27.3.7) (FL2)]; and either an allophycocyanin (APC)‐conjugated anti‐CD3 (IgG2a, clone UCHT1) or anti‐NKG2A [IgG2b, clone Z199 (FL4)]. Aliquots of 50 μl of whole blood were incubated with 5·0 μl of each monoclonal antibody (mAb) [1 : 1 dilution with phosphate‐buffered saline (PBS)] for 30 min at room temperature. The blood was then incubated with 500 μl of red blood cell (RBC) lysis buffer (eBioscience, San Diego, CA, USA) for 20 min at room temperature, washed with PBS and resuspended in 250 μl of PBS prior to flow cytometry analysis. The anti‐CD56, anti‐CD3, anti‐CD57 and anti‐CD158a/h antibodies were purchased from eBioscience; the anti‐NKG2A, anti‐CD158b1/b2/j and anti‐CD158e1/e2 antibodies were purchased from Beckman Coulter (Brea, CA, USA); the anti‐NKG2C antibody was purchased from R&D Systems (Minneapolis, MN, USA); and the anti‐KLRG1 antibody was generously provided by Dr Hanspeter Pircher.
Flow cytometry
NK cell phenotypes were assessed on a BD Accuri C6 flow cytometer (BD Accuri, Ann Arbor, MI, USA). The lymphocytes were identified and gated electronically using the forward and side light‐scatter mode using Accuri C6 (CFlow® software version 2). Side‐scatter against CD3 was then used to identify and gate the CD3– cells and the CD56+ population was identified in the CD3– population. Co‐expression of surface markers was then assessed on the CD3–/CD56+ NK cells in order to identify individual NK cell subsets by four‐colour flow cytometry. The antibody panel used in this study is described in Table 2. Single‐colour compensation control tubes were used to establish the compensation scheme for all flow cytometry‐based assays. Data were analysed directly using BD Accuri's CFlow Plus software. The percentages of all CD3–/CD56+ NK cells expressing the cell surface markers of interest were tabulated for statistical analysis. Total cell numbers of each NK cell subset were determined by multiplying the percentage of all lymphocytes expressing the surface markers of interest by the total lymphocyte count. The lymphocyte count was determined using a whole blood flow cytometric procedure 35 that was validated internally against a Mindray BC‐3200 Auto Hematology Analyzer (Nanshan, Shenzhen, China).
Table 2.
Antibody panel
| FL1 | FL2 | FL3 | FL4 |
|---|---|---|---|
| NKG2C | NKG2A | CD56 | CD3 |
| NKG2C | CD57 | CD56 | CD3 |
| NKG2C | CD158a | CD56 | CD3 |
| NKG2C | CD158b | CD56 | CD3 |
| NKG2C | CD158e | CD56 | CD3 |
| KLRG1 | CD57 | CD56 | CD3 |
| CD3 | CD158a | CD56 | NKG2A |
| CD3 | CD158b | CD56 | NKG2A |
| CD3 | CD158e | CD56 | NKG2A |
NK cell cytotoxicity assay and blocking experiment
This study employed the following target cell lines: 721.221, U266, K562 and 221.AEH. 721.221 is an HLA‐deficient/HLA‐E negative (HLA‐Eneg) lymphoma cell line; U266 is a multiple myeloma cell line that expresses classical HLA molecules [Group 1 HLA‐C (*0304 and *0702) and HLA‐Bw6] and dimly expresses HLA‐E (HLA‐Elow); K562 is an HLA‐E+ leukaemia cell line (HLA‐Emid) that lacks classical HLA molecules; and 221.AEH is a transfectant derived from the 721.221 cell line that highly expresses HLA‐E (HLA‐Ehigh) 36. As we have described previously 37, monocyte‐depleted lymphocytes (purity: 99 ± 1% lymphocytes) were co‐cultured with CD71‐labelled target cells (1·0 × 105 cells) at 1 : 1, 2 × 5 : 1, 5 : 1 and 10 : 1 lymphocyte : target cell ratios in a final volume of 2 × 2 ml of 10% fetal bovine serum (FBS)‐RPMI‐1640. Monocytes were depleted magnetically using CD14 Microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). After a 4‐h incubation at 37°C, the cells were washed and stained with anti‐CD3 and CD56 antibodies to quantify the number of NK cells in each tube. After a final wash, propidium iodide (PI) was added and the numbers of NK cells, live target cells and dead target cells were resolved using four‐colour flow cytometry. NK cell cytotoxicity was quantified as the percentage of specific lysis (% total lysis – % spontaneous cell death). Spontaneous cell death was less than 10% for the U266 and K562 cell lines and less than 20% for the 721.221 and 221.AEH cell lines for each assay. All antibodies and PI were purchased from eBioscience.
For the NKG2C/NKG2A blocking experiment, NK cells were treated with either 10 μl of media, isotype control, anti‐NKG2C (IgG1, clone 134591; R&D Systems), anti‐NKG2A (IgG2b, clone Z199; Beckman Coulter) or anti‐NKG2C and anti‐NKG2A. NK cells were then washed and resuspended in media.
NK cell degranulation and intracellular staining assay
Peripheral blood mononuclear cells (PBMCs) and 221.AEH cells (HLA‐Ehigh lymphoma) were washed and resuspended in 10% FBS‐RPMI‐1640 at a final concentration of 2·5 × 106 and 5·0 × 105 cells/ml, respectively. As described previously 37, the PBMC solution was plated out in a flat‐bottomed 96‐well plate (100 μl per well) under three conditions: negative control, 5 : 1 PBMC to target cell ratio and positive control. Five μl of 10% monensin (BD GolgiStopTM), 2 μl of anti‐CD107a PE‐CF594 (IgG1, clone H4A3) and 10 μl of 10% brefeldin A (Sigma‐Aldrich, St Louis, MO, USA) were added to each well.
Following a 4‐h incubation, the cells were harvested from the plate and pipetted into fluorescence activated cell sorter (FACS) tubes. The cells were washed and resuspended in 200 μl of PBS. The cells in each tube were then stained with anti‐CD56 BV605 (IgG1, clone HCD56), anti‐CD3 APC‐cyanin 7 (Cy7 (IgG1, SK7) and anti‐NKG2C FITC (IgG1, clone 134591). The cells were then lysed, permeabilized and stained with anti‐IFN‐γ VG‐450 (IgG1, clone B27), as described previously 38. The anti‐CD3, anti‐CD107a and anti‐IFN‐γ antibodies were purchased from BD Biosciences; the anti‐CD56 antibody from Biolegend (San Diego, CA, USA); and the anti‐NKG2C antibody from R&D Systems. The proportions of NKG2C+ NK cells expressing CD107a and IFN‐γ were resolved by five‐colour flow cytometry on a BD LSRFortessa flow cytometer (BD Biosciences).
NKG2C+ NK cell expansion assay
NKG2C+ NK cells were expanded preferentially from magnetically enriched CD3–/CD56+ NK cells (purity: 96 ± 1% NK cells) obtained from CMV‐seronegative subjects. First, PBMCs were sorted negatively using CD3 MicroBeads (Miltenyi Biotec) and then the CD3‐depleted cells were sorted positively using CD56 MicroBeads (Miltenyi Biotec). NK cells were cultured for 14 days with 30 ng/ml IL‐15 (eBioscience) and either 721.221 (HLA‐Eneg lymphoma) or 221.AEH (HLA‐Ehigh lymphoma) target cells at a 10 : 1 NK cell : target cell ratio (37°C). NK cell numbers were determined every 3‐4 days when the medium was changed. The phenotype, function and receptor specificity of the expanded NK cell lines was determined before (d0) and after expansion (d14) by flow cytometry (as described above). NK cell cytotoxicity was measured against the 721.221 (HLA‐Eneg lymphoma), U266 (HLA‐Elow myeloma), K562 (HLA‐Emid leukaemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines.
Statistical analysis
Data were analysed statistically using the Predictive Analytics SoftWare (PASW version 22·0) statistics computer program. To examine the effect of CMV status on NK cell cytotoxicity, a maximum likelihood linear mixed model (LMM) was built that included main effects for CMV status and dose (×1, ×2·5, ×5 or ×10) as well as an interaction effect of CMV status × dose. To examine the effect of NKG2C+ NK cell proportion on NK cell cytotoxicity in CMV‐seropositive subjects, an LMM was built that included main effects for NKG2C proportion (high or low) and dose as well as an interaction effect of NKG2C proportion × dose. Subjects were categorized as NKG2Chigh if their proportion of NKG2C+ NK cells was greater than the upper limit of the 95% confidence interval (CI) for CMV‐seronegative subjects (13·4% of total NK cells). Those subjects with an NKG2C+ NK cell proportion below 95% CI were defined as NKG2Clow. Bonferroni post‐hoc analysis was performed to determine the precise location of any significant effects for dose. To determine the effect of NKG2C/NKG2A blockade on NK cell killing of 221.AEH cells (HLA‐Ehigh lymphoma), a LMM was built that included main effects for CMV status, dose and condition (media only, isotype control, anti‐NKG2C, anti‐NKG2A or anti‐NKG2C + NKG2A) as well as interaction effects for CMV status × dose and CMV status × condition. Bonferroni post‐hoc analysis was again performed to determine the location of the significant effects for dose and condition. To determine the effect of HLA‐E on NK cell expansion, phenotype and function, a LMM was built that included main effects for culture conditions [baseline and 14 days co‐incubation with 721.221 (HLA‐Eneg lymphoma) or 221.AEH (HLA‐Ehigh lymphoma) cells] and NK cell dose (for the NK cell assay), as well as an interaction effect for culture condition × dose. The correlation between the proportion of NKG2C+ NK cells and cytotoxicity was determined by calculating the R 2 value for the appropriate best‐fitting line. The impact of CMV infection on NK cell phenotype was determined by using independent‐sample t‐tests based on CMV status. The effect of NKG2C expression on NK cell degranulation/IFN‐γ expression was determined using paired‐sample t‐tests. The upper bound of the 95% CI was defined as the mean + (1·96 × standard deviation). Statistical significance was accepted at P < 0·05.
Results
Latent CMV infection drives accumulation of NKG2C+ NK cells with a highly differentiated phenotype
The effect of latent CMV infection on NK cell phenotype is described in Fig. 1a,b. CMV seropositivity was associated with a marked increase in the percentage of NKG2C+ NK cells (P < 0·05). Specifically, the percentage of NKG2C+/CD57+, NKG2C+/NKG2A–, NKG2C+/CD158a+, NKG2C+/CD158b+ and NKG2C+/CD158e– NK cells was elevated in CMV‐seropositive subjects (n = 15) compared to CMV‐seronegative subjects (n = 15) (P < 0·05). Of the 15 CMV‐seropositive subjects, seven were NKG2Chigh (NKG2C+ proportion > upper bound of 95% CI for CMV‐seronegative subjects) and eight were NKG2Clow (NKG2C+ proportion < upper bound of 95% CI for CMV‐seronegative subjects). Further, the percentage of NK cells expressing CD158e was lower in those infected with CMV relative to CMV‐seronegative subjects (P < 0·05). The functions of individual NK cell receptors are described in Table 3. CMV‐induced changes in the number of NK cell subsets are presented in Table 4 (non‐significant findings are presented in Supporting information, Table S1). Representative flow cytometry dot‐plots that illustrate the effect of CMV infection on NK cell phenotype are shown in Fig. 1c.
Figure 1.
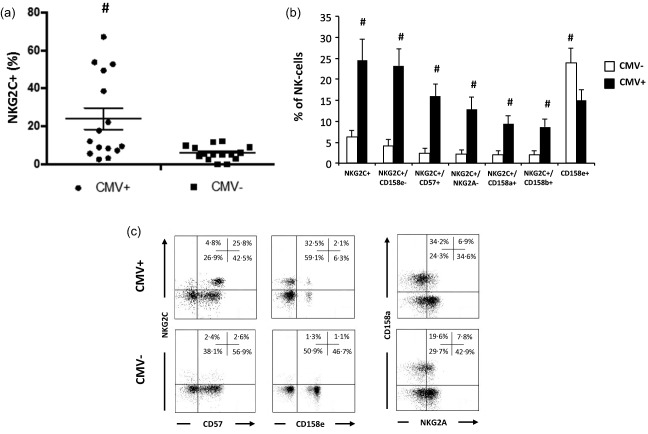
Latent cytomegalovirus (CMV) infection is associated with marked alterations in natural killer (NK) cell phenotype. (a) Whisker dot‐plot for the proportion of NKG2C+ NK cells based on latent CMV infection (n CMV+ = 15; n CMV– = 15). (b) Proportions of peripheral blood CD3–/CD56+ NK cell subsets based on latent CMV infection. Values are mean ± standard error. Statistically significant differences are indicated by #P < 0·05. (c) Representative flow cytometry dot‐plots for the co‐expression of NKG2C with CD57 and CD158e, and the co‐expression of CD158a with NKG2A on NK cells based on CMV status.
Table 3.
Natural killer (NK) cell surface markers
| Cell surface marker | Function |
|---|---|
| NKG2C | NK cell activating receptor that interacts with HLA‐E 15 |
| NKG2A | NK cell inhibitory receptor that interacts with HLA‐E 15 |
| CD57 | Differentiation marker associated with reduced replicative potential 22 |
| CD158a | NK cell inhibitory KIR that interacts with HLA‐C group 2 antigens 39 |
| CD158b | NK cell inhibitory KIR that interacts with HLA‐C group 1 antigens 39 |
| CD158e | NK cell inhibitory KIR that interacts with HLA‐B antigens 39 |
| KLRG1 | NK cell inhibitory receptor that interacts with E‐cadherin 34 |
HLA = human leucocyte antigen; KIR = killer cell immunoglobulin‐like receptor.
Table 4.
Natural killer (NK) cell subset numbers in healthy adults (n CMV+ = 15; n CMV– = 15) contrasted by cytomegalovirus (CMV) status
| Cell subset (cells/μl) | CMV‐seropositive | CMV‐seronegative |
|---|---|---|
| NKG2C+/NKG2A– | 23·8 ± 22·9* | 4·0 ± 3·6 |
| NKG2C+/CD57+ | 29·5 ± 28·2* | 4·3 ± 3·6 |
| NKG2C+/CD158a+ | 17·3 ± 17·2* | 3·8 ± 3·6 |
| NKG2C+/CD158b+ | 16·0 ± 13·9* | 3·8 ± 3·5 |
| NKG2C+/CD158e– | 42·8 ± 42·3* | 7·8 ± 6·6 |
Data are mean ± standard deviation. Statistically significant differences are indicated by *P < 0·05.
NK cell cytotoxicity against HLA‐E‐expressing target cells is enhanced in CMV‐seropositive individuals in association with an increased proportion of NKG2C+ NK cells
The effect of CMV infection on NK cell cytotoxicity against the 721.221 (HLA‐Eneg lymphoma), U266 (HLA‐Elow myeloma), K562 (HLA‐Emid leukaemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines is described in Fig. 2a. NK cell cytotoxicity (%) increased with increasing effector : target cell ratios for all four cell lines (P < 0·001) and all four doses were distinct from each other (P < 0·05). The dose response for the NK cell assay is presented in Supporting information, Table S2. To determine the effect of CMV status on NK cell cytotoxicity, a LMM was built that included main effects for CMV status and dose as well as an interaction effect of CMV status × dose. CMV seropositivity was associated with increased NK cell cytotoxicity (main effect) against the U266 (HLA‐Elow myeloma) (F (1, 88) = 7.909, P < 0·01), K562 (HLA‐Emid leukaemia) (F (1, 120) = 12·560, P < 0·01) and 221.AEH (HLA‐Ehigh lymphoma) (F (1, 80) = 58·554, P < 0·001) cell lines, but not the 721.221 (HLA‐Eneg lymphoma) cell line (F (1, 80) = 0·019, P = 0·89). The effect of CMV status on NK cell cytotoxicity increased with effector cell dose (interaction effect) for the 221.AEH (HLA‐Ehigh lymphoma) cell line (F (3, 80) = 5·507, P < 0·01), but not the U266 (HLA‐Elow myeloma) (F (3, 88) = 0·890, P = 0·45) and K562 (HLA‐Emid leukaemia) (F (3, 120) = 1·317, P = 0·27) cell lines. In summary, CMV‐seropositive individuals had higher NK cell cytotoxicity against HLA‐E+ target cell lines, and this effect was greater at higher doses for the 221.AEH cell line (HLA‐Ehigh lymphoma).
Figure 2.
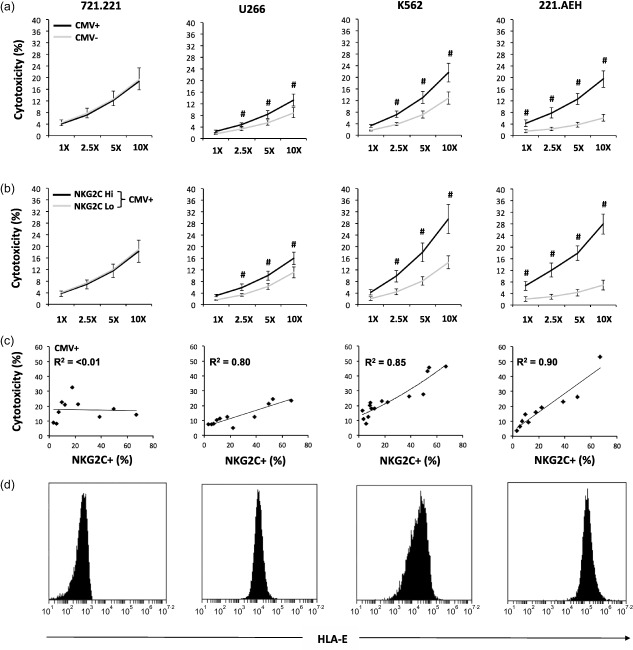
Latent cytomegalovirus (CMV) infection enhances natural killer (NK) cell cytotoxicity against human leucocyte antigen (HLA)‐E‐expressing target cells in association with an increased proportion of NKG2C+ NK cells. (a) NK cell cytotoxicity (%) against 721.221 (HLA‐Eneg lymphoma) (n CMV+ = 10; n CMV– = 10), U266 (HLA‐Elow myeloma) (n CMV+ = 11; n CMV– = 11), K562 (HLA‐Emid leukaemia) (n CMV+ = 15; n CMV– = 15) and 221.AEH (HLA‐Ehigh lymphoma) cells (n CMV+ = 10; n CMV– = 10) at 1 : 1, 2·5 : 1, 5 : 1 and 10 : 1 effector : target cell ratios based on latent CMV infection. (b) NK cell cytotoxicity (%) against 721·221 (n NKG2Chi = 5; n NKG2Clo = 5), U266 (n NKG2Chi = 5; n NKG2Clo = 6), K562 (n NKG2Chi = 7; n NKG2Clo = 8) and 221.AEH cells (n NKG2Chi = 5; n NKG2Clo = 5) at 1 : 1, 2·5 : 1, 5 : 1 and 10 : 1 effector : target cell ratios based on the proportion of NKG2C+ NK cells in CMV‐seropositive subjects. Values are mean ± standard error. Statistically significant differences are indicated by #P < 0·05. (c) Correlation between the proportion of NKG2C+ NK cells and NK cell cytotoxicity (%) against 721·221 (n = 10), U266 (n = 11), K562 (n = 15) and 221.AEH cells (n = 10) at a 10 : 1 effector : target cell ratio in CMV‐seropositive subjects. (d) Flow cytometry histograms for the expression of HLA‐E by 721·221, U266, K562 and 221.AEH cells.
The effect of NKG2C+ NK cell proportion on cytotoxicity against the 721.221 (HLA‐Eneg lymphoma), U266 (HLA‐Elow myeloma), K562 (HLA‐Emid leukaemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines in CMV‐seropositive subjects is described in Fig. 2b. To determine the effect of NKG2C+ NK cell proportion on cytotoxicity in CMV‐seropositive subjects, an LMM was built that included main effects for NKG2C proportion (high or low) and dose as well as an interaction effect of NKG2C proportion × dose. In CMV‐seropositive subjects, a high proportion of NKG2C+ NK cells (NKG2Chigh) was associated with increased cytotoxicity (main effect) against the U266 (HLA‐Elow myeloma) (F (1, 44) = 8·724, P < 0·01), K562 (HLA‐Emid leukaemia) (F (1, 60) = 42·947, P < 0·001) and 221.AEH (HLA‐Ehigh lymphoma) (F (1, 40) = 54·047, P < 0·001) cell lines, but not the 721.221 (HLA‐Eneg lymphoma) (F (1, 40) = 0·409, P = 0·53) cell line. The effect of NKG2C+ NK cell proportion increased with effector cell dose (interaction effect) for the K562 (HLA‐Emid leukaemia) (F (3, 60) = 5·106, P < 0·01) and 221.AEH (HLA‐Ehigh lymphoma) (F (3, 40) = 4·840, P < 0·01) cell lines, but was independent of dose (interaction effect) for the U266 (HLA‐Elow myeloma) cell line (F (3, 44) = 0·380, P = 0·77). In summary, CMV‐seropositive individuals with a high proportion of NKG2C+ NK cells had higher cytotoxicity against HLA‐E+ target cell lines and this effect was greater at higher doses for the K562 (HLA‐Emid leukaemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines.
The correlation between the proportion of NKG2C+ NK cells and cytotoxicity against the 721.221 (HLA‐Eneg lymphoma), U266 (HLA‐Elow myeloma), K562 (HLA‐Emid leukaemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines in CMV‐seropositive subjects is described in Fig. 2c. The proportion of NKG2C+ NK cells was correlated strongly (in CMV‐seropositive subjects) with cytotoxicity against the U266 (HLA‐Elow myeloma) (R 2 = 0·80), K562 (HLA‐Emid leukaemia) (R 2 = 0·85) and 221.AEH (HLA‐Ehigh lymphoma) (R 2 = 0·90) cell lines, but not the 721.221 (HLA‐Eneg lymphoma) cell line (R 2 < 0·01). The correlation between the proportion of NKG2C+ NK cells and cytotoxicity in CMV‐seronegative subjects was R 2 < 0·1 for all four cell lines. Flow cytometry histograms for the expression of HLA‐E by the 721.221 (HLA‐Eneg lymphoma), U266 (HLA‐Elow myeloma), K562 (HLA‐Emid leukaemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines are shown in Fig. 2d. In summary, the proportion of NKG2C+ NK cells correlates strongly with cytotoxicity against HLA‐E+ target cell lines (in CMV‐seropositive subjects).
NK cell cytotoxicity against 221.AEH (HLA‐Ehigh lymphoma) cells is enhanced in CMV‐seropositive individuals in an NKG2C‐dependent manner
The effects of NKG2C and NKG2A blockade on NK cell cytotoxicity against the 221.AEH (HLA‐Ehigh lymphoma) cell line are described in Fig. 3a,b. Three CMV‐seropositive (aged 31·0 ± 2·6 years; gender: two male, 1 female; physical activity rating: 7; BMI: 23·0 ± 2·0) and three CMV‐seronegative subjects (aged 30·0 ± 2·0 years; gender: two male, 1 female; physical activity rating: 6·3 ± 0·5; BMI: 24·0 ± 1·0) participated in this blocking experiment. To determine the effect of NKG2C/NKG2A blockade on NK cell killing of 221.AEH cells (HLA‐Ehigh lymphoma), a LMM was built that included main effects for CMV status, dose and culture condition (media only, isotype control, anti‐NKG2C, anti‐NKG2A or anti‐NKG2C + NKG2A) as well as interaction effects for CMV status × dose and CMV status × culture condition. CMV seropositivity was associated with increased NK cell cytotoxicity (main effect) against the 221.AEH (HLA‐Ehigh lymphoma) cell line (F (1, 120) = 84·179, P < 0·001). The effect of CMV status on NK cell cytotoxicity against the 221.AEH (HLA‐Ehigh lymphoma) cell line was eliminated when NKG2C was blocked (P < 0·001) even if NKG2A was blocked at the same time (P < 0·001). Treatment with anti‐NKG2A or isotype control (IgG1) did not alter the effect of CMV on NK cell cytotoxicity against the 221.AEH (HLA‐Ehigh lymphoma) cell line (P > 0·05). The percentages of degranulating (CD107a+) and IFN‐γ expressing NKG2C+ and NKG2C– NK cells in response to the 221.AEH (HLA‐Ehigh lymphoma) cell line are described in Fig. 3c. Expression of NKG2C was associated with higher 221.AEH‐induced degranulation and IFN‐γ expression by NK cells (P < 0·05). Representative flow cytometry dot‐plots that illustrate the effect of NKG2C expression on NK cell degranulation (CD107a+) and IFN‐γ expression in response to the 221.AEH (HLA‐Ehigh lymphoma) cell line are shown in Fig. 3d. In summary, NKG2C+ NK cells are responsible for the increased cytotoxicity against 221.AEH cells (HLA‐Ehigh lymphoma) in CMV‐seropositive individuals.
Figure 3.

Latent cytomegalovirus (CMV) infection enhances natural killer (NK) cell cytotoxicity against human leucocyte antigen (HLA)‐E‐expressing target cells in an NKG2C‐dependent manner. (a) NK cell cytotoxicity (%) against 221.AEH (HLA‐Ehigh lymphoma) cells at 1 : 1, 2·5 : 1, 5 : 1 and 10 : 1 effector : target cell ratios based on latent CMV infection and anti‐NKG2C treatment (n CMV+ = 3; n CMV– = 3). (b) NK cell cytotoxicity (%) against 221.AEH cells at a 10 : 1 effector : target cell ratio under five different treatments: media, isotype control [immunoglobulin (Ig)G1], anti‐NKG2C, anti‐NKG2A and anti‐NKG2C+ NKG2A (n CMV+ = 3; n CMV– = 3). (c) Percentage of NKG2C+ and NKG2C– NK cells expressing the degranulation marker CD107a, interferon (IFN)‐γ and both CD107a and IFN‐γ in response to 221.AEH cells (n = 6). Values are mean ± standard error. Statistically significant differences are indicated by #P < 0·05. (d) Representative flow cytometry dot‐plots for the degranulation assay.
Preferential expansion of NKG2C+/NKG2A– NK cells from CMV‐seronegative individuals enhances NK cell cytotoxicity against HLA‐E‐expressing tumour cell lines
The effect of culturing NK cells with 221.AEH (HLA‐Ehigh lymphoma) feeder cells for 14 days on the proportion of NKG2C+ and NKG2C+/NKG2A– NK cells is described in Fig. 4a. Four CMV‐seronegative subjects participated in this expansion experiment (aged 31·3 ± 2·9 years; gender: three male, one female; physical activity rating: 6·5 ± 0·5; BMI: 24·0 ± 0·8). To determine the effect of HLA‐E on NK cell expansion, phenotype and function, a LMM was built that included main effects for culture conditions [baseline and 14 days co‐incubation with 721.221 (HLA‐Eneg lymphoma) or 221.AEH cells (HLA‐Ehigh lymphoma)] and NK cell dose (for the NK cell assay), as well as an interaction effect for culture condition × dose. There was a main effect of culture condition on the proportion of NKG2C+ (F (2, 12) = 50·4, P < 0·001) and NKG2C2+/NKG2A– NK cells (F (2, 12) = 123·0, P < 0·001) that was driven by a greater proportion of these subsets after 14 days culture with 221.AEH cells (HLA‐Ehigh lymphoma) (P < 0·05). The effect of 221.AEH‐driven NKG2C+/NKG2A– NK cell expansion on cytotoxicity against the 721.221 (HLA‐Eneg lymphoma), U266 (HLA‐Elow myeloma), K562 (HLA‐Emid leukaemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines is described in Fig. 4b,c. There was a main effect of culture condition on NK cell cytotoxicity against 721.221 (HLA‐Eneg lymphoma) (F (2, 48) = 32·0, P < 0·001), U266 (HLA‐Elow myeloma) (F (2, 48) = 80·7, P < 0·001), K562 (HLA‐Emid leukaemia) (F (2, 48) = 68·1, P < 0·001) and 221.AEH cells (HLA‐Ehigh lymphoma) (F (2, 48) = 50.4, P < 0·001) that was driven by increased cytotoxicity after 14 days culture with 221.AEH (HLA‐Ehigh lymphoma) cells (P < 0·001). The increase in NK cell cytotoxicity against the 221.AEH (HLA‐Ehigh lymphoma) cell line was greater than for the other three cell lines (P < 0·05). The effect of 14 days of co‐culture with 221.AEH cells on NKG2C+ NK cell numbers is shown in Fig. 4d. There was a main effect of culture condition on the number of NKG2C+ (F (2, 12) = 4.1, P < 0·5) and NKG2C+/NKG2A– NK cells (F (2, 12) = 4·0, P < 0·05) that was driven by a greater number of these subsets after 14 days culture with 221.AEH cells (HLA‐Ehigh lymphoma) (P < 0·05). In summary, co‐culture of NK cells with HLA‐E+ feeder cells (221.AEH) preferentially expands NKG2C+ NK cells and increases cytotoxicity against HLA‐E+ target cell lines.
Figure 4.
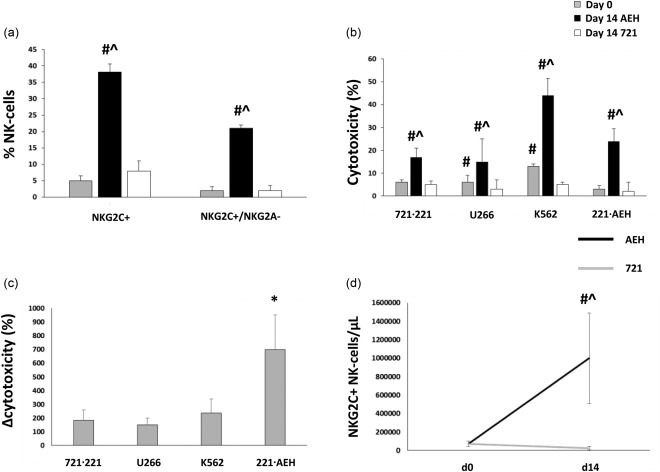
Isolated natural killer (NK) cells from four cytomegalovirus (CMV) seronegative donors were expanded over 14 days in the presence of interleukin (IL)−15 and human leucocyte antigen (HLA)‐E transfected (221.AEH) or non‐transfected (721.221) feeder cells. Panel (a) shows the proportion of NKG2C+ and NKG2C+/NKG2A– NK cells relative to culture conditions. (b) NK cell cytotoxicity (%) against the 721.221 (HLA‐Eneg lymphoma), U266 (HLA‐Elow myeloma), K562 (HLA‐Emid leukemia) and 221.AEH (HLA‐Ehigh lymphoma) cell lines at a 1 : 1 NK : target cell ratio relative to culture conditions. (c) The change in NK cell cytotoxicity (Δcytotoxicity) against the 721.221, U266, K562 and 221.AEH cell lines at a 1 : 1 NK : target cell ratio after 14 days of co‐culture with the 221.AEH cell line. (d) Number of NKG2C+ NK cells before and after 14 days of co‐culture with 221.AEH or 721.221 cells. Values are mean ± standard error. Statistically significant differences from baseline and 721.221 expanded NK cells are indicated by ^P < 0·05 and #P < 0·05, respectively. A statistically significant difference in Δcytotoxicity is indicated by *P < 0·05.
Discussion
The current literature on CMV infection and NK cell function is tilted overwhelmingly towards studies of solid organ and haematopoietic cell transplantation where patients are severely immunocompromised or elderly 23, 32, 39, 40. In this study, we investigated how latent CMV infection in otherwise healthy non‐elderly adults (≤ 50 years) affects NK cell function and linked these functional changes to CMV‐induced phenotypical alterations. The phenotypical imprint of CMV infection is a marked increase in the proportion of NKG2C+ NK cells expressing the terminal differentiation marker CD57, lacking the HLA‐E‐specific inhibitory receptor NKG2A, and showing a skewed killer‐cell immunoglobulin‐like receptor (KIR) repertoire (CD158a+/CD158b+/CD158e–). We show here that latent CMV infection is associated with increased NK cell cytotoxicity against leukaemia, lymphoma and multiple myeloma target cells, and have implicated target cell HLA‐E expression and NK cell NKG2C expression in this effect. Signalling through the activating receptor NKG2C can drive NK cell cytotoxicity in CMV‐infected individuals, as evidenced by enhanced NK cell cytotoxicity against lymphoma cells constitutively expressing the NKG2C ligand HLA‐E (221.AEH), but not those lacking HLA‐E expression (721.221). This CMV‐induced increase in NK cell cytotoxicity against the 221.AEH cell line (HLA‐Ehigh lymphoma) is abrogated completely by antibody blockade of the NKG2C receptor. In addition, we show that cytotoxicity against HLA‐E+ tumour cell lines can be enhanced in CMV‐seronegative individuals by expanding NKG2C+ NK cells via co‐culture of NK cells with 221.AEH (HLA‐Ehigh lymphoma) feeder cells. Collectively, these data suggest that CMV infection might prime NK cells to recognize and destroy malignant target cells through the accumulation of highly functional NKG2C+ NK cells.
We have shown previously that latent CMV infection is associated with increased NK cell cytotoxicity against leukaemia and multiple myeloma target cell lines 37. We show here that this CMV effect was correlated strongly with the proportion of NKG2C+ NK cells in CMV‐infected individuals. The beneficial effect of a high NKG2C+ NK cell proportion on cytotoxicity against the K562 (HLA‐Emid leukaemia) cell line was much larger than with cytotoxicity against U266 (HLA‐Elow myeloma) cells (+18.2 versus +4.0%), which was in line with the higher HLA‐E expression of K562 cells relative to U266 cells. The expansion of NKG2C+ NK cells is a hallmark of CMV infection and the magnitude of expansion is highly variable between individuals 18, 19, 20. It has been shown that NKG2C+ NK cells are expanded selectively in response to CMV‐infected cells due to the interaction of NKG2C with HLA‐E expressed on the surface of CMV‐infected cells 25, 26. NKG2C+ NK cells are capable of generating recall responses during active CMV infection and a higher percentage of donor NKG2C+ NK cells is associated with a reduced risk of CMV reactivation during allogeneic haematopoietic cell transplantation 23, 24. Our work builds on a previous murine study, which showed that latent herpesvirus infection ‘arms’ NK cells and can protect against lymphoma challenge 28, suggesting that CMV‐expanded NKG2C+ NK cells are not just effective mediators of anti‐viral immunity, but are also superior killers of tumour cells. Future studies should determine how CMV affects anti‐tumour NK cell cytotoxicity in older donors as multiple myeloma and AML have their highest prevalence in patients over 50 years of age 41, 42 and recent evidence suggests that tumour immunosurveillance decreases with increasing age in CMV‐seropositive individuals 43. It could be that age (or duration of infection) contributes to the accumulation of NKG2C+ NK cells in a similar manner to that seen with CMV‐specific T cells.
It has been reported that CMV reactivation is associated with a marked reduction in the risk of relapse in AML patients receiving a haematopoietic cell transplant 31, 32. The mechanism behind this reduced relapse risk is currently unknown; however, it has been hypothesized that it may be the result of CMV‐mediated alterations in the composition of NK cell subsets 32. In this study, we show that the accumulation of NKG2C+ NK cells with latent CMV infection is associated with a strong anti‐leukaemia and anti‐myeloma effect in vitro that is proportionate to the HLA‐E expression of the target cell lines. Many tumour cells express HLA‐E, the ligand for NKG2C 29; thus, we hypothesized that the increased anti‐tumour cytotoxicity of NK cells in CMV‐infected individuals was the result of an increased proportion of NKG2C+ NK cells. HLA‐E can signal through either the activating receptor NKG2C or the inhibitory receptor NKG2A, both of which form a complex with the signal transduction protein CD94 44, 45. Signalling through the inhibitory receptor NKG2A is dominant, thus only NKG2C+/NKG2A– NK cells are able to lyse HLA‐E‐expressing target cells effectively 46. Using the HLA‐E transfected 221.AEH (HLA‐Ehigh lymphoma) cell line, we were able to demonstrate that the CMV effect on NK cell cytotoxicity was restricted to HLA‐E+ target cells, as cytotoxicity was increased markedly against 221.AEH (HLA‐Ehigh lymphoma) cells, but not non‐transfected, HLA‐E‐negative 721.221 cells (HLA‐Eneg lymphoma). The effects of CMV and NKG2C+ NK cell proportion on NK cell cytotoxicity against HLA‐E‐expressing target cells increases with effector cell dose, due probably to the fact that at lower doses some of the NK cells never come into contact with target cells, while at higher doses most (or all) NK cells have the opportunity to interact with target cells (thus allowing for greater NKG2C/HLA‐E interaction). Interestingly, this increased NK cell cytotoxicity against the 221.AEH (HLA‐Ehigh lymphoma) cell line was abrogated completely by antibody blocking of NKG2C, but remained intact when NKG2A was blocked. Thus, the increased percentage of NKG2C+/NKG2A– NK cells appears to be directly responsible for the increased anti‐tumour cytotoxicity observed in CMV‐seropositive individuals. Further, it is the NKG2C+ NK cells specifically that degranulate and produce IFN‐γ in response to 221.AEH cells (HLA‐Ehigh lymphoma), thus linking these cells directly to NK cell‐mediated cytotoxicity. It is possible that our results may help to explain the anti‐leukaemia effect of CMV reactivation in AML patients receiving an allogeneic haematopoietic cell transplant 31, 47, as NKG2C+ NK cells expand during reactivation of CMV 24 and AML blasts are known to express high levels of HLA‐E 48. Further, we show that the CMV effect on NK cell cytotoxicity can be mimicked in CMV‐seronegative individuals through 221.AEH‐driven expansion of NKG2C+/NKG2A– NK cells. It remains to be determined if an already large NKG2C+ NK cell pool taken from a CMV‐seropositive individual can be expanded further using 221.AEH cells. Overall, our present findings suggest that CMV‐induced accumulation of NKG2C+ NK cells drives increased cytotoxicity against blood cancer target cell lines that express HLA‐E. Future studies should seek to determine how CMV infection affects NK cell cytotoxicity against solid tumours, as CMV seropositivity and elevated NKG2C+ NK cell proportion are associated with an increased risk of head/neck and colorectal tumours in liver transplant patients 49, which suggests that CMV may have divergent effects on NK cell cytotoxicity depending on the category of tumour. It is also important to determine how CMV infection and the proportion of NKG2C+ NK cells affect prognosis/relapse risk in patients with a haematological malignancy as well as cytotoxicity against primary tumours. While it has been reported that donor CMV seropositivity 30 and CMV reactivation 31, 32 are associated with a decreased risk of relapse in AML patients, this beneficial effect is more than nullified by increased non‐relapse mortality 30, 50. Our findings suggest that the beneficial effect of CMV on AML relapse risk may be attributable to an increased proportion of NKG2C+ NK cells that are capable of recognizing and killing HLA‐E+ AML blasts 48. If this is the case, infusion of ex‐vivo expanded NKG2C+ NK cells may mimic the effect of CMV on preventing AML relapse, while at the same time reducing the risk of CMV reactivation 24, 40 and consequently reducing the risk of non‐relapse mortality.
Our results also demonstrate that the accumulation of NKG2C+ NK cells with CMV infection favours KIR specific for HLA‐C (CD158a and CD158b) over those specific for HLA‐B (CD158e). The literature concerning the effects of CMV infection on KIR expression is ambiguous, as some studies report that ‘unlicensed’ NK cells expand during CMV infection and suppress viraemia 19, 51, while other studies report that NKG2C+ NK cells that expand during CMV infection are licensed 40. Our data support the notion that NKG2C+ NK cells that expand with CMV infection are licensed relative to HLA‐C antigens as the percentages of NKG2C+/CD158a+ and NKG2C+/CD158b+ NK cells were markedly increased, but this was not the case with NKG2C+/CD158e+ NK cells. Previous studies have reported a marked preferential expansion of NKG2C+/CD158b+ NK cells relative to NKG2C+ NK cells expressing other KIR 40, 52, 53. Our results, however, show that NKG2C+ NK cells express similar levels of CD158a and CD158b, while expression of CD158e is clearly reduced in CMV‐infected individuals, the latter being consistent with previous reports 19. Hence, it appears that NKG2C+ NK cells are licensed relative to some HLA antigens (HLA‐C groups 1 and 2) and unlicensed relative to others (HLA‐B). The difference between our findings and those reported in the literature are due probably to differences in CMV status (active versus latent infection) and subject pool (healthy versus immunocompromised) between the earlier studies and our own.
In conclusion, our results show for the first time that latent CMV infection enhances NK cell cytotoxicity against HLA‐E expressing tumour target cell lines through selective accumulation of NKG2C+ NK cells. The beneficial effect of CMV on NK cell cytotoxicity can be abrogated completely by blockade of NKG2C and preferential expansion of NKG2C+/NKG2A– NK cells can enhance cytotoxicity in CMV‐seronegative individuals. Collectively, our data suggest that the CMV‐mediated increase in the proportion of NKG2C+ NK cells may prime NK cell cytotoxicity and could be beneficial in preventing the progression and development of haematological malignancies characterized by high HLA‐E expression, although future studies using primary tumour target cells will be required to support this assertion. Moreover, enrichment of the NKG2C+ NK cell fraction may serve as a simple strategy for enhancing the anti‐tumour cytotoxicity of NK cells for immunotherapeutic applications.
Disclosure
None of the authors have any disclosures to declare.
Author contributions
A. B. B., T. S., N. B., M. P., N. A. and H. K. performed the experiments; A. B. B., K. R., N. S., C. M. B., D. P. O'C. and R. J. S. designed the study; A. B. B., R. J. S., K. R., C. M. B. and N. S. interpreted data; A. B. B. and R. J. S. wrote the paper.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. Natural killer (NK) cell subset numbers in healthy adults (n CMV+ = 15; n CMV– = 15) contrasted by cytomegalovirus (CMV) status. Data are mean ± standard deviation. All cell counts for which there was no significant CMV effect are reported here (P > 0·05)
Table S2. Cytotoxicity against natural killer (NK) cell targets in healthy adults. Doses are defined as effector : target cell ratios (E : T). Data are mean ± standard deviation. Main effects and interactions among the nominal variables (cytomegalovirus status and dose) are shown with significance indicated by *P < 0·05
Acknowledgements
This work was supported by National Space Biomedical Research Institute through NASA NCC 9‐58 to A. B. B. and NASA grants NNX12AB48G and NNJ14ZSA001N‐FLAGSHIP to R. J. S.. The authors thank Dr Hanspeter Pircher for providing the Alexa488‐conjugated anti‐KLRG1 (clone 13F12F2) monoclonal antibody and Dr Dan Geraghty for providing the 721·221 and 221·AEH cells.
References
- 1. Bate SL, Dollard SC, Cannon MJ. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988‐2004. Clin Infect Dis 2010; 50:1439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gkrania‐Klotsas E, Langenberg C, Sharp SJ, Luben R, Khaw KT, Wareham NJ. Seropositivity and higher immunoglobulin g antibody levels against cytomegalovirus are associated with mortality in the population‐based European prospective investigation of Cancer – Norfolk cohort. Clin Infect Dis 2013; 56:1421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desai R, Collett D, Watson CJ, Johnson PJ, Moss P, Neuberger J. Impact of cytomegalovirus on long‐term mortality and cancer risk after organ transplantation. Transplantation 2015; 99:1989–94. [DOI] [PubMed] [Google Scholar]
- 4. Bigley AB, Spielmann G, Agha N, O'Connor DP, Simpson RJ. Dichotomous effects of latent CMV infection on the phenotype and functional properties of CD8+ T‐cells and NK‐cells. Cell Immunol 2016; 300:26–32. [DOI] [PubMed] [Google Scholar]
- 5. Orange JS, Biron CA. Characterization of early IL‐12, IFN‐alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol 1996; 156:4746–56. [PubMed] [Google Scholar]
- 6. Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol 2008; 8:259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lopez‐Botet M, Angulo A, Guma M. Natural killer cell receptors for major histocompatibility complex class I and related molecules in cytomegalovirus infection. Tissue Antigens 2004; 63:195–203. [DOI] [PubMed] [Google Scholar]
- 8. Cosman D, Mullberg J, Sutherland CL et al ULBPs, novel MHC class I‐related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 2001; 14:123–33. [DOI] [PubMed] [Google Scholar]
- 9. Magri G, Muntasell A, Romo N et al NKp46 and DNAM‐1 NK‐cell receptors drive the response to human cytomegalovirus‐infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood 2011; 117:848–56. [DOI] [PubMed] [Google Scholar]
- 10. Bennett NJ, Ashiru O, Morgan FJ et al Intracellular sequestration of the NKG2D ligand ULBP3 by human cytomegalovirus. J Immunol 2010; 185:1093–102. [DOI] [PubMed] [Google Scholar]
- 11. Welte SA, Sinzger C, Lutz SZ et al Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol 2003; 33:194–203. [DOI] [PubMed] [Google Scholar]
- 12. Beck S, Barrell BG. Human cytomegalovirus encodes a glycoprotein homologous to MHC class‐I antigens. Nature 1988; 331:269–72. [DOI] [PubMed] [Google Scholar]
- 13. Prod'homme V, Griffin C, Aicheler RJ et al The human cytomegalovirus MHC class I homolog UL18 inhibits LIR‐1+ but activates LIR‐1‐ NK cells. J Immunol 2007; 178:4473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bigley AB, Spielmann G, LaVoy EC, Simpson RJ. Can exercise‐related improvements in immunity influence cancer prevention and prognosis in the elderly? Maturitas 2013; 76:51–6. [DOI] [PubMed] [Google Scholar]
- 15. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature 2009; 457:557–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vales‐Gomez M, Reyburn HT, Erskine RA, Lopez‐Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2‐A and the activating receptor CD94/NKG2‐C to HLA‐E. EMBO J 1999; 18:4250–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tomasec P, Braud VM, Rickards C et al Surface expression of HLA‐E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000; 287:1031 [DOI] [PubMed] [Google Scholar]
- 18. Guma M, Angulo A, Vilches C, Gomez‐Lozano N, Malats N, Lopez‐Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004; 104:3664–71. [DOI] [PubMed] [Google Scholar]
- 19. Lopez‐Verges S, Milush JM, Schwartz BS et al Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA 2011; 108:14725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Monsivais‐Urenda A, Noyola‐Cherpitel D, Hernandez‐Salinas A et al Influence of human cytomegalovirus infection on the NK cell receptor repertoire in children. Eur J Immunol 2010; 40:1418–27. [DOI] [PubMed] [Google Scholar]
- 21. Sun JC, Lopez‐Verges S, Kim CC, DeRisi JL, Lanier LL. NK cells and immune ‘memory’. J Immunol 2011; 186:1891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez‐Verges S, Milush JM, Pandey S et al CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK‐cell subset. Blood 2010; 116:3865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hadaya K, de Rham C, Bandelier C et al Natural killer cell receptor repertoire and their ligands, and the risk of CMV infection after kidney transplantation. Am J Transplant 2008; 8:2674–83. [DOI] [PubMed] [Google Scholar]
- 24. Foley B, Cooley S, Verneris MR et al Human cytomegalovirus (CMV)‐induced memory‐like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J Immunol 2012; 189:5082–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beziat V, Liu LL, Malmberg JA et al NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013; 121:2678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guma M, Budt M, Saez A et al Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus‐infected fibroblasts. Blood 2006; 107:3624–31. [DOI] [PubMed] [Google Scholar]
- 27. Sansoni P, Vescovini R, Fagnoni FF et al New advances in CMV and immunosenescence. Exp Gerontol 2014; 55:54–62. [DOI] [PubMed] [Google Scholar]
- 28. White DW, Keppel CR, Schneider SE et al Latent herpesvirus infection arms NK cells. Blood 2010; 115:4377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lo Monaco E, Tremante E, Cerboni C et al Human leukocyte antigen E contributes to protect tumor cells from lysis by natural killer cells. Neoplasia 2011; 13:822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nachbaur D, Clausen J, Kircher B. Donor cytomegalovirus seropositivity and the risk of leukemic relapse after reduced‐intensity transplants. Eur J Haematol 2006; 76:414–9. [DOI] [PubMed] [Google Scholar]
- 31. Elmaagacli AH, Steckel NK, Koldehoff M et al Early human cytomegalovirus replication after transplantation is associated with a decreased relapse risk: evidence for a putative virus‐versus‐leukemia effect in acute myeloid leukemia patients. Blood 2011; 118:1402–12. [DOI] [PubMed] [Google Scholar]
- 32. Green ML, Leisenring WM, Xie H et al CMV reactivation after allogeneic HCT and relapse risk: evidence for early protection in acute myeloid leukemia. Blood 2013; 122:1316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jackson AS, Blair SN, Mahar MT, Wier LT, Ross RM, Stuteville JE. Prediction of functional aerobic capacity without exercise testing. Med and Sci Sports Exer 1990; 22:863–70. [DOI] [PubMed] [Google Scholar]
- 34. Marcolino I, Przybylski GK, Koschella M et al Frequent expression of the natural killer cell receptor KLRG1 in human cord blood T cells: correlation with replicative history. Eur J Immunol 2004; 34:2672–80. [DOI] [PubMed] [Google Scholar]
- 35. Bigley AB, Lowder TW, Spielmann G et al NK‐cells have an impaired response to acute exercise and a lower expression of the inhibitory receptors KLRG1 and CD158a in humans with latent cytomegalovirus infection. Brain Behav Immun 2012; 26:177–86. [DOI] [PubMed] [Google Scholar]
- 36. Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA‐E surface expression depends on binding of TAP‐dependent peptides derived from certain HLA class I signal sequences. J Immunol 1998; 160:4951–60. [PubMed] [Google Scholar]
- 37. Bigley AB, Rezvani K, Chew C et al Acute exercise preferentially redeploys NK‐cells with a highly‐differentiated phenotype and augments cytotoxicity against lymphoma and multiple myeloma target cells. Brain Behav Immun 2014; 39:160–71. [DOI] [PubMed] [Google Scholar]
- 38. de Lavallade H, Khoder A, Hart M et al Tyrosine kinase inhibitors impair B‐cell immune responses in CML through off‐target inhibition of kinases important for cell signaling. Blood 2013; 122:227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Duin D, Avery RK, Hemachandra S et al KIR and HLA interactions are associated with control of primary CMV infection in solid organ transplant recipients. Am J Transplant 2014; 14:156–62. [DOI] [PubMed] [Google Scholar]
- 40. Foley B, Cooley S, Verneris MR et al Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 2012; 119:2665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deschler B, Lubbert M. Acute myeloid leukemia: epidemiology and etiology. Cancer 2006; 107:2099–107. [DOI] [PubMed] [Google Scholar]
- 42. Turesson I, Velez R, Kristinsson SY, Landgren O. Patterns of multiple myeloma during the past 5 decades: stable incidence rates for all age groups in the population but rapidly changing age distribution in the clinic. Mayo Clin Proc 2010; 85:225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cramer DW, Finn OJ. Epidemiologic perspective on immune‐surveillance in cancer. Curr Opin Immunol 2011; 23:265–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res 2006; 35:263–78. [DOI] [PubMed] [Google Scholar]
- 45. Colonna M, Moretta A, Vely F, Vivier E. A high‐resolution view of NK‐cell receptors: structure and function. Immunol Today 2000; 21:428–31. [DOI] [PubMed] [Google Scholar]
- 46. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008; 9:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beck JC, Wagner JE, DeFor TE et al Impact of cytomegalovirus (CMV) reactivation after umbilical cord blood transplantation. Biol Blood Marrow Transplant 2010; 16:215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen S, Dhedin N, Vernant JP et al NK‐cell reconstitution after haploidentical hematopoietic stem‐cell transplantations: immaturity of NK cells and inhibitory effect of NKG2A override GvL effect. Blood 2005; 105:4135–42. [DOI] [PubMed] [Google Scholar]
- 49. Achour A, Baychelier F, Besson C et al Expansion of CMV‐mediated NKG2C+ NK cells associates with the development of specific de novo malignancies in liver‐transplanted patients. J Immunol 2014; 192:503–11. [DOI] [PubMed] [Google Scholar]
- 50. Takenaka K, Nishida T, Asano‐Mori Y et al Cytomegalovirus reactivation after allogeneic hematopoietic stem cell transplantation is associated with a reduced risk of relapse in patients with acute myeloid leukemia who survived to day 100 after transplantation: the Japan Society for Hematopoietic Cell Transplantation Transplantation‐Related Complication Working Group. Biol Blood Marrow Transplant 2015; 21:2008–16. [DOI] [PubMed] [Google Scholar]
- 51. Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol 2010; 11:321–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bjorkstrom NK, Lindgren T, Stoltz M et al Rapid expansion and long‐term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med 2011; 208:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petitdemange C, Becquart P, Wauquier N et al Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog 2011; 7:e1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. Natural killer (NK) cell subset numbers in healthy adults (n CMV+ = 15; n CMV– = 15) contrasted by cytomegalovirus (CMV) status. Data are mean ± standard deviation. All cell counts for which there was no significant CMV effect are reported here (P > 0·05)
Table S2. Cytotoxicity against natural killer (NK) cell targets in healthy adults. Doses are defined as effector : target cell ratios (E : T). Data are mean ± standard deviation. Main effects and interactions among the nominal variables (cytomegalovirus status and dose) are shown with significance indicated by *P < 0·05


