Abstract
Phosphorus (P) is an essential macronutrient for plant growth and development. Inorganic phosphate (Pi) is the major form of P taken up from the soil by plant roots. It is well established that under Pi deficiency condition, plant roots undergo striking morphological changes; mainly a reduction in primary root length while increase in lateral root length as well as root hair length and density. This typical phenotypic change reflects complex interactions with other nutrients such as iron, and involves the activity of a large spectrum of plant hormones. Although, several key proteins involved in the regulation of root growth under Pi-deficiency have been identified in Arabidopsis, how plants adapt roots system architecture in response to Pi availability remains an open question. In the current post-genomic era, state of the art technologies like high-throughput phenotyping and sequencing platforms,“omics” methods, together with the widespread use of system biology and genome-wide association studies will help to elucidate the genetic architectures of root growth on different Pi regimes. It is clear that the large-scale characterization of molecular systems will improve our understanding of nutrient stress phenotype and biology. Herein, we summarize the recent advances and future directions towards a better understanding of Arabidopsis root developmental programs functional under Pi deficiency. Such a progress is necessary to devise strategies to improve the Pi use efficiency in plants that is an important issue for agriculture.
Keywords: Phosphate, Root, Genes, Signaling, Crosstalk
INTRODUCTION
Phosphorus (P) is the 11th most abundant element of the earth’s crust; while simultaneously the most immobile nutrients in the soils resulting in in its poorly availability for plants [1]. The major available form of P for plants in the soils is inorganic P (Pi). To overcome Pi deficiency, a massive Pi supply is chosen as an immediate remedy, a practice that is neither ecologically sustainable nor economically viable. Interestingly, crops take up only 15–30% of the applied Pi fertilizer within the year of its application [2], whereas rest of the applied fertilizer is lost in form of leaching and polluting the water bodies. Thus improving the ability of crops to use the available Pi is necessary to reduce a plant dependency on Pi-fertilizers while maintaining an optimum yield. Such an objective requires a better understanding on mechanisms regulating both the Pi transport system in plants and the root growth capacity in response to fluctuating Pi concentrations in soil. During the last few decades, our knowledge on the transport of Pi and its accumulation in plants has been considerably advanced, mainly using Arabidopsis as a reference model plant [3, 4]. For an extensive review of the Pi transporter gene family, readers are referred to [5, 6]. The physiological and molecular aspects of root growth and development in response to Pi deficiency has been also investigated in Arabidopsis ecotype Columbia (Col) [7]. The current knowledge on theses mechanisms is summarized below.
Primary and lateral root developmental response to Pi availability
Low Pi conditions have been shown to affect the course of root development in plants, for instance on low Pi media, a reduction in primary root length as well as increase in lateral root length have been reported [8-11]. These morphological changes have been opined to increase the root surface, thus enabling plants for the better exploration of the top layer of soil to improve the acquisition of the poorly mobile Pi.
Number of key genes involved in the primary root growth inhibition upon Pi deficiency has been identified in Arabidopsis through classical genetic (reverse and forward) studies. Based on detailed mutant analysis we can distinguish the following three categories, 1) mutants hypersensitive to low Pi such as the phosphate deficiency response 2 mutant (pdr2, P5-Type ATPase, At5g23630); the siz1 mutant (SUMO E3 ligase, At5g60410) and the prd mutant (DNA binding protein, At1g79700); 2) mutants able to maintain primary root growth in low Pi, such as the low phosphate root lpr mutants (lpr1, At1g23010; lpr2, At1g71040; lpr3) [11] and; 3) mutants that are low phosphorus insensitive, namely lpi1, lpi2, lpi3 and lpi4, which are characterized by a long primary root despite of low Pi in the growth media [12]. In general, low Pi causes a redistribution of root growth from the primary root to the lateral roots and the later becoming denser [8, 13]. Although this observation is overstated in some mutants such as the siz1 mutant displaying an increase of lateral root number, or the pdr2 and the ribonuclease polynucleotide phosphorylase mutant (pnp, At3g03710) that presents highly branched lateral roots.
Substantial natural variation of root developmental response to Pi deficiency can be easily observed using hundreds of available accessions of Arabidopsis genus [14]. Numerous initiatives in the development of high-throughput plant phenotyping platforms using robotic-assisted imaging and computer vision-assisted analysis tools are engaged [15, 16].
The availability of the complete Arabidopsis genome sequence has dramatically accelerated traditional genetic research on root biology, and has also enabled entirely new experimental strategies to be applied [17]. The availability of genome sequences of various plant species coupled with root phenotyping tools have allowed the emergence of the genome-wide association studies (GWAS) as an excellent strategy to dissect the genetic basis of many plant traits in responses to abiotic stresses. GWAS combined with expression analyses, allows the identification of genomic regions and causal genes, associated with biological processes such as root development. For instance, [15] reports a cost-efficient phenotyping system for Arabidopsis roots that enables scalable image acquisition and processing, as well as storing of positional information of plant genotypes and automated annotation of multiple genotypes per plate. The setup and evaluation of the performance of this system to produce and process a large data set as well as its robustness toward different growth conditions was discussed [15]. Recently, this system was used and allowed the identification of a new F-box gene, KUK, involved in the regulation of root meristem and cell length [18]. The availability of nucleotide and protein sequences allowed the identification of the polymorphisms in the coding sequences as the major causes of KUK (F-box) allele–dependent natural variation in root development [18]. Therefore, GWAS strategy has proved its reliability to explore the genetic determinants underlying the plasticity of root growth in response to Pi availability.
Pi starvation activates a large-scale change at the transcriptome and proteome levels in plant shoots and roots [19, 20]. Gene expression profiles (microarrays) of a high-resolution set of developmental time points within a single Arabidopsis root and a comprehensive map of nearly all root cell types has been reported [21]. These data revealed complex programs that define Arabidopsis root development in both space and time. It will very interesting to combines cell sorting with microarray analysis to generate the global expression pattern for every cell type in the root under Pi deficiency conditions. If this information could be obtained for every cell type and every developmental stage of the root grown under limited Pi condition, it would provide an all-encompassing picture of the regulatory networks controlling root development. From this dataset all transcription factors that are expressed in a tissue-specific pattern can be identified. Localizing these transcription factors and determining their immediate targets will be instrumental for a better understanding of complex biological systems such as root development.
In conclusion, combination of the above mentioned innovative approaches will certainly complete the current understanding on genes and their regulatory network involved in the regulation of primary root development, but also others root traits in response to Pi availability.
Role of root tip in sensing and responding to PI starvation
Recent works have shown that the external, but not the internal, low-Pi content conditions the primary root growth arrest; this is a typical local response to low-Pi [22, 23]. Accumulating evidence indicates that the root tip plays a critical role for sensing and responding to Pi starvation. The physical contact of the root tip with low-Pi medium is necessary and sufficient to severely reduce the root growth capacity [24]. The conditional short root phenotype of pdr2 under Pi limitation can be rescued by supplementing the Pi-limiting media with phosphite (Phi), which leads to resume the root meristem activity [25, 26]. The PDR2 gene (P5-type ATPase) appears to function in the endoplasmic reticulum (ER) and act as a Pi sensitive checkpoint in root development in low Pi media through the maintenance of the stem-cell fate [27]. PDR2 in Pi-deprived roots is required for proper expression of a key regulator gene of root patterning, SCARECROW (SCR), and affects SHORT-ROOT (SHR) movement towards the endodermis [27]. It was also observed that PRD, an Arabidopsis AINTEGUMENTA-like gene, was involved in root architectural changes in response to Pi starvation by controlling primary and lateral root elongation [28]. The Low Phosphate Root1 (LPR1, ferroxidase), as its paralog LPR2, are expressed in the root tip, comprising the meristem and root cap [24]. It is noteworthy that PDR2 and LPR1 expression domains overlap in the stem-cell niche and distal root meristem [27], which strengthens the hypothesis that these two genes function together in an ER-resident pathway that adjusts root meristem activity in response to external Pi. Taken together, these data attest the critical role of the root tip in Pi starvation response.
Fortuitously, multiple sets of Arabidopsis microarray data have been assembled in various available databases, as well as data mining and analysis tool boxes such as TAIR [29], NASCArrays [30], the Stanford Microarray Database [31] and GENEVESTIGATOR [32]. These data sources can be combined with several meta-analysis tools using a guilt-by-association principle to help visualise correlated gene expression, such as ATTED-II [33], and CressExpress [34]. The functional annotations in the gene's co-expression neighbourhood can then be used to hypothesise a biologically relevant relationship. Using ATTED ver7.1 (http://atted.jp/), the co-expression relationship of AtLPR1 (At1g23010) and AtLPR2 (At1g71040) and AtPDR2 (AT5G23630) can be investigated. Interestingly, such an analysis revealed that LPR1, LPR2, and PDR2 genes constitute a different set of co-expressed genes, which illustrate the existence of co-expression clusters that correspond to different functional modules involved in regulation of root development. A comprehensive protein–protein interaction map would be another valuable resource that would help identifying new network connections.
Primary root growth under PI starvation: interaction with other nutrient availability
Variation in the experimental design and the composition of the growth media impacts the morphology of the roots of Pi-deficient plants [10, 11, 35]. Concentrations of iron (Fe) and other microelement contaminants in gelling agents cause significant variations in the morphological responses to Pi deprivation [35]. Therefore, the root phenotype could not be the only “placemark” to define status of Pi in plants [36]. For example, similarly to low Pi, Fe deprivation induces an increase in root hair length and density [37-39]. We have learned recently that the morphological changes in root architecture upon Pi starvation are most likely an outcome of the complex interactions between Pi and the Fe [24, 35]. Indeed, the Pi-deprived plants over-accumulate Fe, resulting supposedly to a Fe toxicity effects in roots. In line with this statement, the reduction of Fe concentration in the medium, in spite of low concentrations of Pi, leads to the recovery of the primary root elongation and the ability of plant to uptake Pi [35]. Therefore, Pi deficiency-induced root growth inhibition depends on external Fe presence and is initiated by accelerated differentiation of elongating cells, followed by a decline in meristematic cells [40]. The molecular basis and the nature of the cross-talks between Pi and Fe homeostasis start to emerge. Very recently, [40] reported that the two functionally interacting genes, LPR1 and PDR2, facilitate cell-specific apoplastic Fe and callose deposition in the meristem and elongation zone of primary roots upon Pi limitation. This work highlights the importance of callose-regulated symplastic communication in root meristems for the perception of Pi availability, which likely depends on Fe redox cycling. However, taken into account that the inhibitory effect on the growth of Pi-deprived pdr2 mutant roots was shown to be mostly independent of Fe concentration in the media [27], it is thus clear that plants have evolved distinct Pi-signalling pathways that are dependent and independent of metal ions availability. In addition to Fe, zinc (Zn) availability in the growth media has also been shown to inversely impact the Pi uptake and accumulation on plants [41-43]. It will be interesting to decipher the root phenotype under conditions of Pi availability linked with Zn excess or deficiency conditions. To further address the question of a regulatory connection between ions signalling pathways and root growth capacity in Arabidopsis thaliana, combination of bioinformatics, system biology, molecular genetics and genomics experimental approaches should be more effective at unravelling complex cross-talk mechanisms compared to previous, single approach, studies.
Root hairs development in response to PI availability
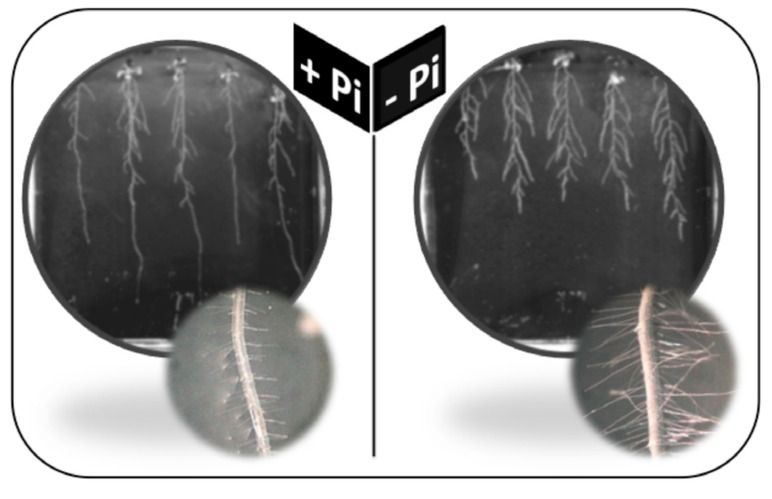
The role of root hair in anchoring the plant to the soil is proposed but not fully established. The ability of Arabidopsis mutants affected in root hair development to adhere to the growth medium is compromised in the case of actin double mutant actin2actin7 (act2act7; AT3G18780, AT5G09810), but not in the case of the ROOT Hair Defective 2 (rhd2-1; AT5G51060) mutant, which is defective in NADPH oxidase and hence ROS production [44]. Nevertheless, the implication of root hair in the acquisition of nutrients from the soil solution is well established. Pi deficiency affects the root hairs growth (Fig. 1).
Fig. (1).
Changes in primary root length and root hairs in Arabidopsis thaliana grown either in presence (1 mM) or in absence of inorganic phosphate (Pi) for two weeks.
In case of Pi, the importance of root hairs in the absorption of this element has been proven earlier using the Arabidopsis mutants affected in root hair elongation (rdh2, AT5G51060) or reduced root hair density (rhd6, At1g66470) [45-48]. It is a proven fact that a plant increases its root hair length and density under Pi deficiency. These morphological changes can be seen as a strategy to further empower the capacity of a root system to explore more soil surface, to penetrate into the finest structures of the soil and to enhance the acquisition of available Pi. For instance, Pi-deficient plants increase root hair density by the formation of shorter cells, resulting in a higher frequency of hairs per unit root length, and additional trichoblast cell fate assignment via increased expression of the ENHANCER OF TRY AND CPC (ETC1, At1g01380) gene [49]. There is a growing list of Arabidopsis genes involved in root hair proliferation in response to low Pi condition. Mutants in these genes can promote root hair formation such as the SUMO E3 ligase gene SIZ1 (At5g60410) [50], the transcription factor genes (WRKY75, At5g13080; bHLH32, At3g25710) [51], the transcription factor phl1/phr1(At5g29000/At4g28610) [52]. Other genes have been also identified including the F-Box protein with WD40 domain (fbx2, At5g21040), inositol polyphosphate kinase (ipk, At5g42810), Raf like Kinase (hsp2, At5g03730) [53], loss of heat shock protein (HSP2, At5g03730), or the loss of the F-box gene BX2 (At5g21040) [54]. All these gene mutants have higher root hair density in low Pi conditions. Mutants that also reduce and or inhibit root hair growth have been described for e.g. the ubiquitin protease (ubp14/per1, At3g20630) [55], gibberellin biosynthesis (ga1-3, At4g02780), and the transcription factor (rsl4, At1g27740). A recent study on the natural variation of the root hair responses to local scarcity of Pi in a large panel of A. thaliana accessions (166), using GWA mapping, allowed the identification of some accessions showing no root hair under Pi deficiency or in the opposite longer/denser root hair formation under this condition. GWAS analysis revealed new genes involved in the response of root hairs to scarce local Pi level including (CYR1, At1g32360 and RLP48, AT4G13880) [56].
In the post genomic area, root hair cells can be seen as a system biology model to investigate Pi uptake, and response to Pi deficiency at a plant cell level. A system model for root hairs should include gene regulatory and signal transduction networks, and metabolic pathways related to various aspects of root hair function [57]. To generate this knowledge, there is a need of methodological development and improvement. In method development, efficient ways to isolate and manipulate the fragile root hair cells is the first crucial step to generate the “omics” data. To date, the transcriptome data on isolated root hair cells have been reported only on limited number of plant species such as Soybean [58]. Analysis of transcriptome of root hair in presence or absence of Pi, using the DNA microarray hybridization and high-throughput sequencing technologies will help in obtaining the repertoire of gene expression which can be used for the identification of Pi- specific regulatory genes. At the microscopic level, the observation of root hair, with minimal interference from surrounding cells, is feasible and enables the analysis of different biological processes related to root hair elongation under different Pi regimes. Nevertheless, further method improvement is also needed to measure and visualize intercellular Pi dynamics and Pi signalling molecules accurately. Integration of “omics” data to generate a comprehensive model remains particularly challenging. Bioinformatics and systems biology are promising approaches to help integrating these large biological data sets.
The combination of traditional forward genetics, genomics and systems biology approaches emerge as a powerful strategy to uncover key players in ion signalling pathways crosstalk in plant. These approaches were successfully used to predict network modeling of the high-resolution dynamic plant transcriptome in response to nitrate [59, 60]. Such approaches, implemented as part of the adaptive response of the root system to the Pi deficiency, should emerge original crosstalk between Pi and plant hormone signalling pathways.
Phytohormones influence root development in response to PI availability
Hormonal activities have been implicated in different developmental aspects of root growth under Pi deficiency, with either promotive or repressive effects [61]. Nevertheless, how Pi signal interacts with those of others nutrients at the molecular level, is a largely unanswered question in plants.
The responses of the Arabidopsis root system architecture to Pi deficiency involve both auxin-dependent and auxin-independent mechanisms [8, 10, 13, 39, 62, 63]. In a similar manner to wild type plants, all auxin-related mutants (aux1 and eir1, axr1, axr2 and axr4) showed a decrease in primary root length as well as an increase in lateral root number and density in response to Pi deficiency. The only exception is the auxin response mutant iaa28 which is severely defective in lateral root formation in plants grown in either Pi-sufficient or Pi-deficient conditions [13]. Lateral root formation in Pi starvation appeared to require a class of transcriptional regulators that mediate growth and developmental responses to auxins AUXIN RESPONSE FACTOR 19 (ARF19, AT1G19220) and which involves an SCFTIR1-dependent signalling mechanism [64]. Like wild type, the root hair elongation in the hairless auxin-resistant mutant axr1 and axr2, as well as in the auxin-insensitive mutant aux1, is stimulated in low Pi condition [65].
Recent experimental evidences indicate that auxin could affect root architecture in Pi-starved plants via modulation of the status of other phytohormones. For example, it has been proposed that auxin may regulate root growth by modulating gibberellic acid (GA) signalling [66]. In line with this report, Jiang et al., (2007) showed that low Pi in plants resulted in a reduction in bioactive GA levels and accumulation of DELLA proteins. Worth noting that DELLA-mediated signalling contributes only to certain aspects of Pi-deficiency response in roots, typically are suppression of the primary root growth and promotion of root hairs [67].
The gaseous phytohormone, ethylene, has been shown to play an integral local role in the root hair formation, the lateral root elongation, and the reduction of primary root elongation upon Pi starvation [67]. The root system of all ethylene mutants, such as the ethylene-insensitive mutants etr1, ein2, ein3, and hls, the ethylene-overproducing mutant eto1 and the ethylene constitutive response mutant ctr1, respond to Pi deprivation by a decrease in primary root growth and an increase in lateral root formation, although the response of the ctr1 and eto1 mutants was reduced compared to wild type [8, 13]. Some indication for the role of ethylene in systemic Pi signalling through modulation a number of systemically controlled Pi starvation responses start to emerge [68].
Recently role of strigolactones (SLs) in plant responses to Pi growth conditions have been reported [69]. Pi-deficiency has been shown to increase SLs biosynthesis in Arabidopsis roots and transport through the xylem to the shoot [70]. SLs act as long-distance shoot-branching inhibitors [70, 71]. SLs have also been suggested to have a positive effect on root-hair elongation, mediated via the MAX2 F-box. The SLs' ability to regulate root development may be executed by induction of the ethylene pathway in conjunction with regulation of auxin transport [72, 73]. SLs were suggested to negatively regulate lateral root (LR) formation in Arabidopsis, under conditions of sufficient Pi nutrition [74].
The brassinosteroid (BR) signaling pathway regulates numerous physiological and developmental processes in plants. In roots, BRs have both promoting and inhibitory effects on growth, depending on the intensity of the signal [75]. [61] reported that two homologous BR transcriptional effectors, namely BRASSINAZOLE-RESISTANT 1 (BZR1) and BRASSINAZOLE-RESISTANT 2 (BZR2), block these responses (namely exhaustion of the primary meristem, impaired unidirectional cell expansion and elevated density of lateral roots), consequently maintaining normal root development under low Pi conditions. The roots of the bzr1-D mutants remain longer at the very low Pi concentration compared to wild type.
CONCLUSION
Over the past few decades, many research works aimed to elucidate the molecular mechanisms controlling Pi homeostasis in plants have been performed using Arabidopsis thaliana as a model system. A combination of genetics, molecular biology and genomics led to the identification and characterization of a number of genes that regulate growth and development of the root in Arabidopsis in response to Pi deficiency. Nevertheless, it is clear that much remains to be discovered to fully appreciate the molecular processes that govern the adaptation of plants root system to Pi availability. Achieving this objective becomes possible thanks to the available resources in the current post-genomic era including completion of the Arabidopsis genome, developing genome-wide profiling technology, the system biology methods and an arrays of experimental tools including phenotyping platforms. Orthologous of the Arabidopsis regulatory genes in crop plants could be then targeted for biotechnological and agronomical applications.
ACKNOWLEDGEMENTS
The authors wish to thank Dr Sikander Pal and Dr Aida Rouached for his constructive comments on the review. This work was supported, in part, by the “Institut National de la Recherche Agronomique” to H.R and P.D and from the Tunisian government to N.B. The authors apologize to any colleagues whose relevant work has not been mentioned.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Neset T.S., Cordell D. Global phosphorus scarcity: identifying synergies for a sustainable future. J. Sci. Food Agric. 2012;92(1):2–6. doi: 10.1002/jsfa.4650. [DOI] [PubMed] [Google Scholar]
- 2.Syers J.K., Johnston A.E., Curtin D.C. Efficiency of Soil and Fertilizer Phosphorus Use. FAO Fertilizer and Plant Nutrition Bulletin; 2008. p. 18. [Google Scholar]
- 3.Rouached H., Arpat A.B., Poirier Y. Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol. Plant. 2010;3(2):288–299. doi: 10.1093/mp/ssp120. [DOI] [PubMed] [Google Scholar]
- 4.Briat J.F., Rouached H., Tissot N., Gaymard F., Dubos C. Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front. Plant Sci. 2015;6:290. doi: 10.3389/fpls.2015.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Ribot C., Rezzonico E., Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 2004;135(1):400–411. doi: 10.1104/pp.103.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nussaume L., Kanno S., Javot H., Marin E., Pochon N., Ayadi A., Nakanishi T.M., Thibaud M.C. Phosphate Import in Plants: Focus on the PHT1 Transporters. Front. Plant Sci. 2011;2:83. doi: 10.3389/fpls.2011.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peret B., Desnos T., Jost R., Kanno S., Berkowitz O., Nussaume L. Root architecture responses: in search of phosphate. Plant Physiol. 2014;166(4):1713–1723. doi: 10.1104/pp.114.244541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williamson L.C., Ribrioux S.P., Fitter A.H., Leyser H.M. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 2001;126(2):875–882. doi: 10.1104/pp.126.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linkohr B.I., Williamson L.C., Fitter A.H., Leyser H.M. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002;29(6):751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- 10.Al-Ghazi Y., Muller B., Pinloche S., Tranbarger T.J., Nacry P., Rossignol M., Tardieu F., Doumas P. Temporal responses of Arabidopsis root architecture to phosphate starvation: Evidence for the involvement of auxin signalling. Plant Cell Environ. 2003;26:1053–1066. [Google Scholar]
- 11.Reymond M., Svistoonoff S., Loudet O., Nussaume L., Desnos T. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ. 2006;29(1):115–125. doi: 10.1111/j.1365-3040.2005.01405.x. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez-Calderon L., Lopez-Bucio J., Chacon-Lopez A., Gutierrez-Ortega A., Hernandez-Abreu E., Herrera-Estrella L. Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol. 2006;140(3):879–889. doi: 10.1104/pp.105.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Bucio J., Hernandez-Abreu E., Sanchez-Calderon L., Perez-Torres A., Rampey R.A., Bartel B., Herrera-Estrella L. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 2005;137(2):681–691. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chevalier F., Pata M., Nacry P., Doumas P., Rossignol M. Effects of phosphate availability on the root system architecture: large-scale analysis of the natural variation between Arabidopsis accessions. Plant Cell Physiol. 2003;26(11):1839–1850. [Google Scholar]
- 15.Slovak R., Goschl C., Su X., Shimotani K., Shiina T., Busch W. A Scalable Open-Source Pipeline for Large-Scale Root Phenotyping of Arabidopsis. Plant Cell. 2014;26(6):2390–2403. doi: 10.1105/tpc.114.124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahlgren N., Gehan M.A., Baxter I. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015;24:93–99. doi: 10.1016/j.pbi.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Benfey P.N., Bennett M., Schiefelbein J. Getting to the root of plant biology: impact of the Arabidopsis genome sequence on root research. Plant J. 2010;61(6):992–1000. doi: 10.1111/j.1365-313X.2010.04129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijon M., Satbhai S.B., Tsuchimatsu T., Busch W. Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat. Genet. 2014;46(1):77–81. doi: 10.1038/ng.2824. [DOI] [PubMed] [Google Scholar]
- 19.Hammond J.P., Bennett M.J., Bowen H.C., Broadley M.R., Eastwood D.C., May S.T., Rahn C., Swarup R., Woolaway K.E., White P.J. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003;132(2):578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong S.E., Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Cell Biol. 2005;1(5):252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 21.Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 22.Thibaud M.C., Arrighi J.F., Bayle V., Chiarenza S., Creff A., Bustos R., Paz-Ares J., Poirier Y., Nussaume L. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010;64(5):775–789. doi: 10.1111/j.1365-313X.2010.04375.x. [DOI] [PubMed] [Google Scholar]
- 23.Peret B., Clement M., Nussaume L., Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 2011;16(8):442–450. doi: 10.1016/j.tplants.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Svistoonoff S., Creff A., Reymond M., Sigoillot-Claude C., Ricaud L., Blanchet A., Nussaume L., Desnos T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 2007;39(6):792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- 25.Ticconi C.A., Abel S. Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci. 2004;9(11):548–555. doi: 10.1016/j.tplants.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Ticconi C.A., Delatorre C.A., Lahner B., Salt D.E., Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 2004;37(6):801–814. doi: 10.1111/j.1365-313x.2004.02005.x. [DOI] [PubMed] [Google Scholar]
- 27.Ticconi C.A., Lucero R.D., Sakhonwasee S., Adamson A.W., Creff A., Nussaume L., Desnos T., Abel S. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. USA. 2009;106(33):14174–14179. doi: 10.1073/pnas.0901778106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho-Cristobal J.J., Rexach J., Conejero G., Al-Ghazi Y., Nacry P., Doumas P. PRD, an Arabidopsis AINTEGUMENTA-like gene, is involved in root architectural changes in response to phosphate starvation. Planta. 2008;228(3):511–522. doi: 10.1007/s00425-008-0754-9. [DOI] [PubMed] [Google Scholar]
- 29.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., Miller N., Mueller L.A., Mundodi S., Reiser L., Tacklind J., Weems D.C., Wu Y., Xu I., Yoo D., Yoon J., Zhang P. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31(1):224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32(Database issue):D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ball C.A., Awad I.A., Demeter J., Gollub J., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Brown P.O., Sherlock G. The Stanford Microarray Database accommodates additional microarray platforms and data formats. Nucleic Acids Res. 2005;33(Database issue):D580–D582. doi: 10.1093/nar/gki006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136(1):2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35(Database issue):D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasasainagendra V., Page G.P., Mehta T., Coulibaly I., Loraine A.E. CressExpress: a tool for large-scale mining of expression data from Arabidopsis. Plant Physiol. 2008;147(3):1004–1016. doi: 10.1104/pp.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward J.T., Lahner B., Yakubova E., Salt D.E., Raghothama K.G. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 2008;147(3):1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouached H., Secco D., Arpat B.A. Regulation of ion homeostasis in plants: current approaches and future challenges. Plant Signal. Behav. 2010;5(5):501–502. doi: 10.4161/psb.11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schikora A., Schmidt W. Acclimative changes in root epidermal cell fate in response to Fe and P deficiency: a specific role for auxin? Protoplasma. 2001;218(1-2):67–75. doi: 10.1007/BF01288362. [DOI] [PubMed] [Google Scholar]
- 38.Schikora A., Schmidt W. Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol. 2001;125(4):1679–1687. doi: 10.1104/pp.125.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt W., Schikora A. Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 2001;125(4):2078–2084. doi: 10.1104/pp.125.4.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller J., Toev T., Heisters M., Teller J., Moore K.L., Hause G., Dinesh D.C., Burstenbinder K., Abel S. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell. 2015;33(2):216–230. doi: 10.1016/j.devcel.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 41.Khan G.A., Bouraine S., Wege S., Li Y., de Carbonnel M., Berthomieu P., Poirier Y., Rouached H. Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 2014;65(3):871–884. doi: 10.1093/jxb/ert444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouain N., Kisko M., Rouached A., Dauzat M., Lacombe B., Belgaroui N., Ghnaya T., Davidian J.C., Berthomieu P., Abdelly C., Rouached H. Phosphate/zinc interaction analysis in two lettuce varieties reveals contrasting effects on biomass, photosynthesis, and dynamics of Pi transport. BioMed Res. Int. 2014;2014:548254. doi: 10.1155/2014/548254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouain N., Shahzad Z., Rouached A., Khan G.A., Berthomieu P., Abdelly C., Poirier Y., Rouached H. Phosphate and zinc transport and signalling in plants: toward a better understanding of their homeostasis interaction. J. Exp. Bot. 2014;65(20):5725–5741. doi: 10.1093/jxb/eru314. [DOI] [PubMed] [Google Scholar]
- 44.Gilliland L.U., Kandasamy M.K., Pawloski L.C., Meagher R.B. Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol. 2002;130(4):2199–2209. doi: 10.1104/pp.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bates T.R., Lynch J.P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19:529–538. [Google Scholar]
- 46.Bates T.R., Lynch J.P. Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 2000;87:958–963. [PubMed] [Google Scholar]
- 47.Bates T.R., Lynch J.P. The efficiency of Arabidopsis thaliana (Brassicaceae) root hairs in phosphorus acquisition. Am. J. Bot. 2000;87:964–970. [PubMed] [Google Scholar]
- 48.Bates T.R., Lynch J.P. Root hairs confer a competitive advantage under low phosphorus availability. Plant Soil. 2001;236:243–250. [Google Scholar]
- 49.Savage N., Yang T.J., Chen C.Y., Lin K.L., Monk N.A., Schmidt W. Positional signaling and expression of ENHANCER OF TRY AND CPC1 are tuned to increase root hair density in response to phosphate deficiency in Arabidopsis thaliana. PLoS One. 2013;8(10):e75452. doi: 10.1371/journal.pone.0075452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A.S., Raghothama K.G., Baek D., Koo Y.D., Jin J.B., Bressan R.A., Yun D.J., Hasegawa P.M. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA. 2005;102(21):7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z.H., Nimmo G.A., Jenkins G.I., Nimmo H.G. BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem. J. 2007;405:191–198. doi: 10.1042/BJ20070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Perez-Perez J., Solano R., Leyva A., Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6(9):e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei M., Zhu C., Liu Y., Karthikeyan A.S., Bressan R.A., Raghothama K.G., Liu D. Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol. 2011;189(4):1084–1095. doi: 10.1111/j.1469-8137.2010.03555.x. [DOI] [PubMed] [Google Scholar]
- 54.Chen Z.H., Jenkins G.I., Nimmo H.G. Identification of an F-box protein that negatively regulates P(i) starvation responses. Plant Cell Physiol. 2008;49(12):1902–1906. doi: 10.1093/pcp/pcn157. [DOI] [PubMed] [Google Scholar]
- 55.Li W.F., Perry P.J., Prafulla N.N., Schmidt W. Ubiquitin-specific protease 14 (UBP14) is involved in root responses to phosphate deficiency in Arabidopsis. Mol. Plant. 2010;3(1):212–223. doi: 10.1093/mp/ssp086. [DOI] [PubMed] [Google Scholar]
- 56.Stetter M.G., Schmid K., Ludewig U. Uncovering genes and ploidy involved in the high diversity in root hair density, length and response to local scarce phosphate in Arabidopsis thaliana. PLoS One. 2015;10(3):e0120604. doi: 10.1371/journal.pone.0120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Libault M., Brechenmacher L., Cheng J., Xu D., Stacey G. Root hair systems biology. Trends Plant Sci. 2010;15(11):641–650. doi: 10.1016/j.tplants.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 58.Libault M., Farmer A., Brechenmacher L., May G.D., Stacey G. Soybean root hairs: a valuable system to investigate plant biology at the cellular level. Plant Signal. Behav. 2010;5(4):419–421. doi: 10.4161/psb.5.4.11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krouk G., Lacombe B., Bielach A., Perrine-Walker F., Malinska K., Mounier E., Hoyerova K., Tillard P., Leon S., Ljung K., Zazimalova E., Benkova E., Nacry P., Gojon A. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell. 2010;18(6):927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Krouk G., Mirowski P., LeCun Y., Shasha D.E., Coruzzi G.M. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010;11(12):R123. doi: 10.1186/gb-2010-11-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh A.P., Savaldi-Goldstein S. Growth control: brassinosteroid activity gets context. J. Exp. Bot. 2015;66(4):1123–1132. doi: 10.1093/jxb/erv026. [DOI] [PubMed] [Google Scholar]
- 62.Nacry P., Canivenc G., Muller B., Azmi A., Van Onckelen H., Rossignol M., Doumas P. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005;138(4):2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopez-Bucio J., Hernandez-Abreu E., Sanchez-Calderon L., Nieto-Jacobo M.F., Simpson J., Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002;129(1):244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez-Torres C.A., Lopez-Bucio J., Cruz-Ramirez A., Ibarra-Laclette E., Dharmasiri S., Estelle M., Herrera-Estrella L. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008;20(12):3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma Z., Bielenberg D.G., Brown K.M., Lynch J.P. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 2001;24:459–467. [Google Scholar]
- 66.Fu X., Harberd N.P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421(6924):740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- 67.Jiang C., Gao X., Liao L., Harberd N.P., Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145(4):1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagarajan V.K., Smith A. Ethylene's role in phosphate starvation signaling: more than just a root growth regulator. Plant Cell Physiol. 2012;53(2):277–286. doi: 10.1093/pcp/pcr186. [DOI] [PubMed] [Google Scholar]
- 69.Kumar M., Pandya-Kumar N., Dam A., Haor H., Mayzlish-Gati E., Belausov E., Wininger S., Abu-Abied M., McErlean C.S., Bromhead L.J., Prandi C., Kapulnik Y., Koltai H. Arabidopsis response to low-phosphate conditions includes active changes in actin filaments and PIN2 polarization and is dependent on strigolactone signalling. J. Exp. Bot. 2015;66(5):1499–1510. doi: 10.1093/jxb/eru513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M.A., Beguerie S., Verstappen F., Leyser O., Bouwmeester H., Ruyter-Spira C. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011;155(2):974–987. doi: 10.1104/pp.110.164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gomez-Roldan V., Fermas S., Brewer P.B., Puech-Pages V., Dun E.A., Pillot J.P., Letisse F., Matusova R., Danoun S., Portais J.C., Bouwmeester H., Becard G., Beveridge C.A., Rameau C., Rochange S.F. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 72.Kapulnik Y., Delaux P.M., Resnick N., Mayzlish-Gati E., Wininger S., Bhattacharya C., Sejalon-Delmas N., Combier J.P., Becard G., Belausov E., Beeckman T., Dor E., Hershenhorn J., Koltai H. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta. 2011;233(1):209–216. doi: 10.1007/s00425-010-1310-y. [DOI] [PubMed] [Google Scholar]
- 73.Kapulnik Y., Resnick N., Mayzlish-Gati E., Kaplan Y., Wininger S., Hershenhorn J., Koltai H. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 2011;62(8):2915–2924. doi: 10.1093/jxb/erq464. [DOI] [PubMed] [Google Scholar]
- 74.Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., Cardoso C., Lopez-Raez J.A., Matusova R., Bours R., Verstappen F., Bouwmeester H. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 2011;155(2):721–734. doi: 10.1104/pp.110.166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fridman Y., Savaldi-Goldstein S. Brassinosteroids in growth control: how, when and where. Plant Sci. 2013;209:24–31. doi: 10.1016/j.plantsci.2013.04.002. [DOI] [PubMed] [Google Scholar]