Abstract
MicroRNAs (miRNAs) are well-conserved noncoding RNAs that broadly regulate gene expression through posttranscriptional silencing of coding genes. Dysregulated miRNA expression in prostate and other cancers implicates their role in cancer biology. Moreover, functional studies provide support for the contribution of miRNAs to several key pathways in cancer initiation and progression. Comparative analyses of miRNA gene expression between malignant and nonmalignant prostate tissues, healthy controls and prostate cancer (PCa) patients, as well as less aggressive versus more aggressive disease indicate that miRNAs may be future diagnostic or prognostic biomarkers in tumor tissue, blood, or urine. Further, miRNAs may be future therapeutics or therapeutic targets. In this review, we examine the miRNAs most commonly observed to be de-regulated in PCa gene expression analyses and review the potential contribution of these miRNAs to important pathways in PCa initiation and progression.
Keywords: miRNA, prostate cancer, noncoding RNA
INTRODUCTION
Prostate cancer (PCa) is the second most commonly diagnosed cancer in men worldwide and it is the fifth leading cause of cancer deaths.1 Historically, PCa has provided important knowledge to the areas of early diagnosis and treatment of cancer, from highly-sensitive PSA screening and the modern radical prostatectomy, to the current recognition of PCa over-diagnosis and over-treatment of indolent disease. PCa was one of the first solid tumors with a targeted treatment for metastatic disease through androgen deprivation therapy (ADT), which has provided important clinical, molecular, and genetic knowledge of pathways that contribute to cancer progression and tumor cell therapeutic resistance.
The PCa field is again paving new paths in cancer management through the active surveillance of low-risk disease, as opposed to more radical treatments, and through the development and approval of several novel targeted drugs for metastatic disease and the metastatic microenvironment.2,3,4 As more is discovered about PCa biology and as PCa management evolves, our knowledge of microRNA (miRNA) biology and expression has grown. Here, we review the recent discoveries of numerous miRNAs reported to be associated with PCa, and we consider the potential contributions of these miRNAs to PCa biology and cancer management.
PROSTATE CANCER PROGRESSION AND MANAGEMENT
To understand in what ways miRNAs might contribute to PCa biology and how they might be helpful in its management, it is important to understand the natural progression of PCa, how it is managed at each stage, and the potential sources for miRNA analysis (Figure 1). PCa is thought to arise from focally damaged epithelium that is characterized as proliferative inflammatory atrophy (PIA) and/or prostatic intraepithelial neoplasia (PIN).5,6 These lesions are considered precancerous and are of insufficient risk alone to warrant treatment. Cancer is suspected upon detection of an elevated serum PSA and/or an abnormal digital rectal exam; it is then confirmed by pathologic diagnosis of a biopsy specimen. Most often, the tumors diagnosed by PSA screening are too small to be clinically imaged. Therefore, individual biopsies are not targeted to a specific lesion; rather, multiple systematic biopsies are used to screen regions suspected to have cancer. There is a significant risk that biopsy needles may miss the tumor and result in a false negative diagnosis. In light of this, there are opportunities to discovery new miRNA biomarkers that may improve the specificity of PCa diagnosis and reduce unnecessary repeat biopsy and/or patient anxiety.
Figure 1.
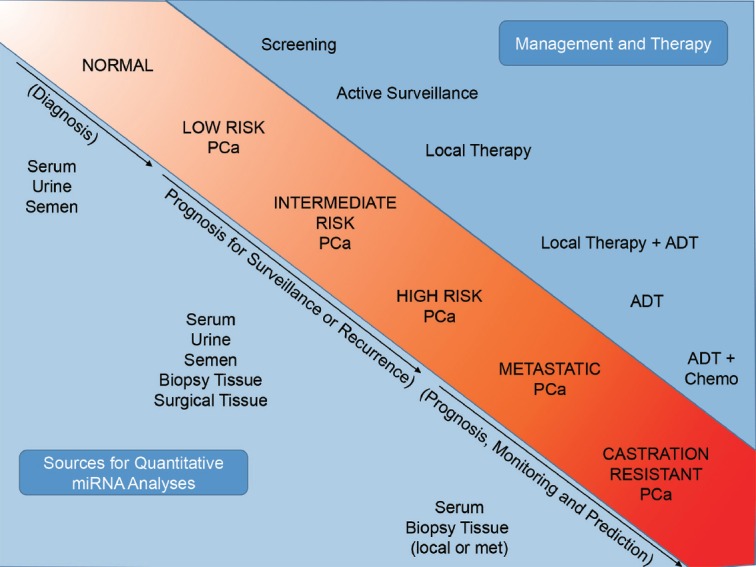
Opportunities and unmet needs in the progression of prostate cancer. The natural progression of prostate cancer from initiation to castration-resistant metastatic disease is highlighted. The upper right quadrant indicates the most common methods for management and therapy as the disease progresses. The lower left quadrant indicates resources for miRNA discovery and biomarker analyses and the potential areas for utility, such as diagnosis or active surveillance.
Once diagnosed, the risk of PCa progression is assessed pathologically by tumor stage and volume, serum PSA level, and Gleason score. Lower-risk patients (e.g., those with less than three cancer-positive biopsy cores, low PSA (<10 ng ml−1), and/or Gleason score ≤6) are given the option to enter active surveillance (AS) programs in which treatment is delayed until the disease shows the signs of increased risk upon repeat biopsy or elevated serum markers.7 Intermediate-risk PCa is characterized by intermediate stage (T2b, T2c) and grade (Gleason 7), and can be successfully treated with surgery or ionizing radiation therapy. High-risk PCa has an elevated stage (T3-T4), high serum PSA (>20 ng ml−1), and/or high grade (Gleason ≥8). Due to the elevated risk for local recurrence or metastasis, treatment includes both local therapies, such as surgery or radiation, as well as systemic ADT.8 One important consideration for the management of localized PCa is the multifocal nature of the disease, where a patient may be inaccurately labeled as having lower-risk disease because a higher-grade lesion was not detected by biopsy. The prevalence of upgrading after repeat biopsy or surgery is significant, as more than 30% of men initially diagnosed with low-risk disease are upgraded.9,10 Consequently, there are considerable unmet needs in the localized PCa patient population that could be addressed by the discovery of miRNA biomarkers that are associated with low-risk disease that does not require immediate treatment or high-risk disease that requires surgery or radiation therapy.
There is regrettably no cure for those PCa cases detected later in disease progression or that progress to metastasis after unsuccessful local treatment. Metastatic disease is initially treated with ADT through luteinizing hormone-releasing hormone (LHRH) analogs or anti-androgens, such as flutamide or bicalutamide. As resistance develops to these traditional ADT agents, second-generation agents such as abiraterone and enzalutamide are utilized, as well as bone-targeted radiotherapeutics and taxane-based chemotherapy. These new drugs have resulted in the development of a new type of therapeutically resistant PCa, which the field is just beginning to study.4 Here, miRNAs have a potential value as novel biomarkers of disease aggressiveness or therapeutic resistance, as well as therapeutic targets or as oligonucleotide therapeutics.
In summary, each step of PCa progression, from initiation to therapeutic resistance, is a potential source for miRNA discovery. MiRNA analysis in patients may improve the clinical decision making regarding cancer management or therapeutic outcome. Sources of miRNA may include the direct analysis of miRNA expression in biopsy or surgical tissue or noninvasive sources such as serum, urine, or semen (Figure 1). Finally, miRNAs may prove useful as novel therapeutic targets or as gene therapeutic agents in the future treatment of castration-resistant disease. Below, we review the approximately 10 years of miRNA research in PCa thus far.
miRNA BIOGENESIS AND PCA
The biogenesis of miRNAs in cancer has been recently reviewed elsewhere in detail.11 Briefly, miRNAs are initially transcribed by RNA polymerase II as a large primary miRNA (pri-miRNA) transcript (Figure 2). In light of this, miRNA expression can be altered in cancer through oncogenic or tumor suppressive pathways or through genetic or epigenetic aberrations commonly observed in cancer including gene amplification, rearrangement, deletion, or promoter silencing. The miRNA stem-loop structure is then recognized by the Microprocessor complex and the hairpin is released following cleavage by the RNAse III enzyme, DROSHA. The liberated pre-miRNA is then bound and exported into the cytoplasm by Exportin 5 (XPO5) and RanGTP. In the cytoplasm, the pre-miRNA is further cleaved by the RNAse III enzyme, DICER, as guided by the RNA-binding protein (TRBP). The mature miRNA is then loaded into the RNA-induced silencing complex (RISC), which includes the Argonaut proteins and GW182 family proteins. The miRNA and RISC then directly suppress target gene expression through complementary binding of the seed sequence (6–8 nucleotides) to target mRNAs, leading to the inhibition of translation and/or decreased mRNA stability.
Figure 2.
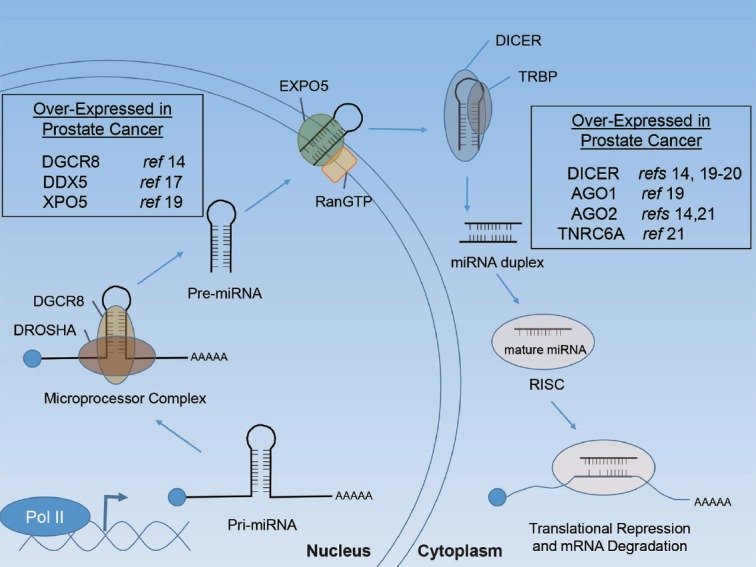
miRNA biogenesis. Schematic representation of miRNA biogenesis initiating with primary miRNA (pri-miRNA) transcription. Boxes indicate miRNA biogenesis components that have been reported to be deregulated in human PCa. References (ref) are indicated.
Various components of the miRNA biogenesis pathway have been found to be over-expressed, under-expressed, amplified, deleted, and/or mutated in human cancers.11 In both localized and metastatic PCa, genetic deletion or mutation of miRNA biogenesis machinery appears to be rare.12,13 Rather, the miRNA biogenesis machinery is more often overactive in PCa, suggesting a possible driver role of miRNAs. Early studies found the Microprocessor subunit, DGCR8, to be over-expressed in a large cohort of primary tumors.14 Moreover, knockout (KO) of DGCR8 in a transgenic PTEN KO model of PCa inhibited tumor progression.15 DDX5, the DEAD box RNA helicase p68 that associates with the Microprocessor complex, is also over-expressed in localized PCa and enhances androgen receptor (AR)-regulated gene expression.16,17 Further, like the TMPRSS2 gene, fusions of the DDX5 gene with ETV4 have been observed in PCa.18 Elevated expression of XPO5 has also been reported in metastatic versus organ-confined PCa.19 The RNAse III enzyme DICER has also been shown to be over-expressed in both localized and metastatic PCa.14,19,20 Notably, however, low DICER expression has been associated with PCa recurrence.20 This may be reflective of studies in PTEN KO transgenic mouse models in which complete Dicer KO blocked tumor growth and progression, whereas hemizygous loss of Dicer resulted in a more locally invasive phenotype.20 Components of the RISC complex have also been reported to be over-expressed in PCa. AGO1, encoded by the EIF2C1 gene, is upregulated in metastatic PCa.19 Similarly, AGO2 has been reported to be over-expressed at the RNA and protein level.14,21 The genetic loci of AGO2 are also located on chromosome 8q24, a region commonly amplified in PCa.22,23,24 TNRC6A, a component of GW-bodies that contributes to RISC-mediated silencing, has also been reported to be over-expressed in localized PCa.21 Collectively, these studies support that miRNA processing and activity may contribute to PCa biology and progression. These early results warrant additional research to define the upstream and downstream mechanisms of deregulated miRNA biogenesis machinery in PCa, their frequency in clinical specimens, and their potential contribution to PCa initiation and progression in cellular and animal models.
miRNA EXPRESSION IN HUMAN PCA
Approximately, 10 years ago, the first miRNA gene expression profiling studies were performed on clinical cancer samples, including prostate carcinoma.25 These studies found that miRNA expression profiles accurately reflected the developmental lineage as well as the differentiation state of tumors. This was further supported by two studies in 2006 and 2008, in which unsupervised clustering analysis of miRNA expression clearly separated different tissue types and several miRNAs were identified to be commonly de-regulated in multiple human cancers.26,27 These pioneering studies highlight the potential of miRNA profiling for cancer research, and suggest that miRNAs may be even better than mRNAs for tissue- and tumor-specific molecular classification. Below, we summarize miRNA gene expression studies conducted on both prostate tissues and bodily fluids.
miRNA gene expression profiles in prostate cancer tissues
Initial high-throughput miRNA expression studies sought to identify differences between nonmalignant prostate tissue and PCa. More recent studies have increased in complexity to include additional tumor-versus-tumor analyses for miRNAs associated with stage, grade, biochemical recurrence, metastases, or castration resistance. Many studies have also validated discovery through additional analysis of distinct tissue sample sets or different quantitative techniques. As new technology has been developed, such as RNA sequencing, additional high-throughput data have accumulated. Here, we focus this portion of the review on high-throughput studies to highlight those miRNAs that are most commonly reported to be differentially expressed in genome-wide expression analyses (Table 1). While there are numerous additional expression studies that focus on specific miRNAs, they are not, for the most part, included in this review.
Table 1.
Commonly reported miRNAs from gene expression analysis of prostate cancer tissue, serum, and exosomes
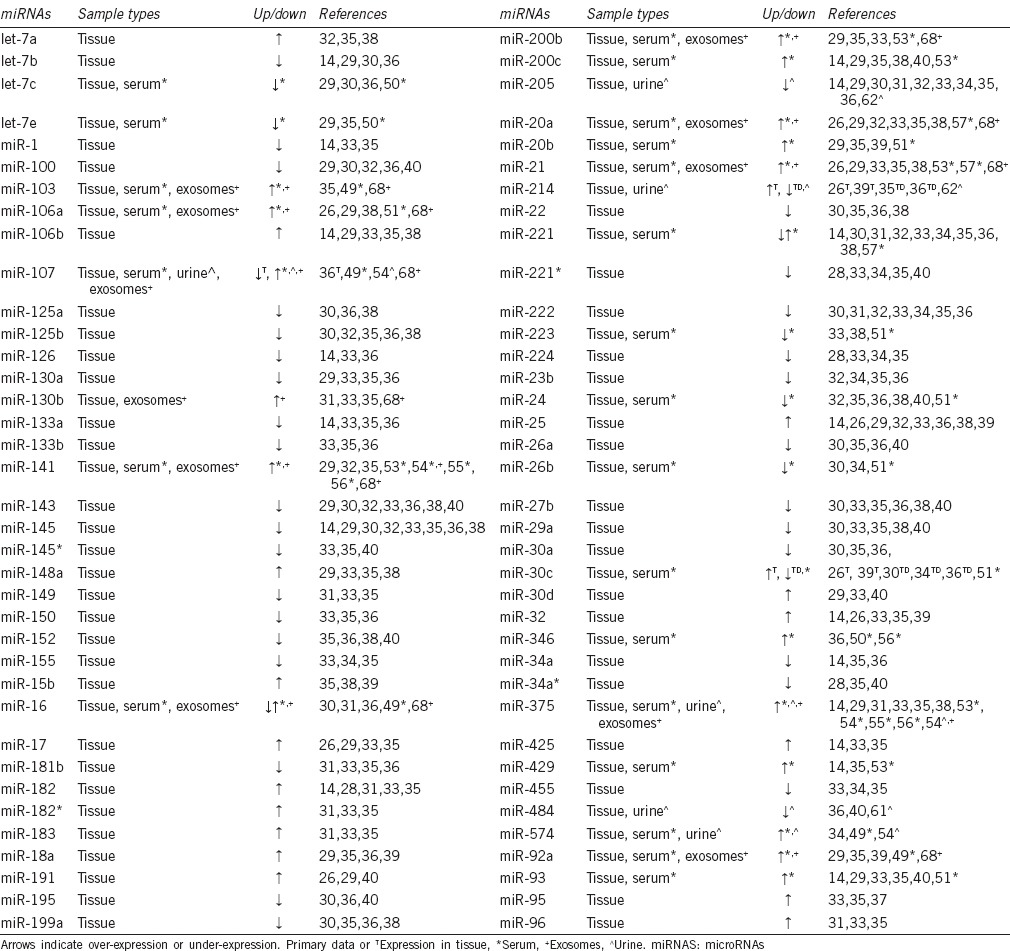
Several miRNAs have been commonly reported to be significantly over-expressed in PCa, when compared to nonmalignant tissue, across several tissue resources and analysis platforms (Table 1), including let-7a, miR-106a and miR-106b, miR-130b, miR-141, miR-148a, members of the miR-17-5p cluster, miR-182, miR-183, the miR-200 family, miR-21, miR-25, miR-32, miR-375, and miR-93.14,26,28,29,30,31,32,33,34,35,36,37,38,39 A considerably higher number of miRNAs have also been commonly observed to have significantly decreased expression in PCa (Table 1). Some of the most commonly reported under-expressed miRNAs are members of the miR-143/145 family;14,29,30,32,33,35,36,38,40 however, new research indicates that this may be reflective of the relative loss of stromal tissue in the tumor microenvironment.41 This phenomenon may influence the perceived levels of other miRNAs commonly observed to be over-expressed or under-expressed in tissue specimens. Other miRNAs commonly reported to be significantly down-regulated in PCa tissue include members of the let-7 family, miR-100, miR-125a, miR-125b, miR-130a, miR-133a, miR-199a, miR-205, miR-221, miR-222, miR-24, miR-26a, miR-27b, and miR-29a.14,28,29,30,31,32,33,34,35,36,37,38,40,42 The collective contribution of these miRNAs to PCa initiation and progression, as well as their potential as diagnostic and prognostic biomarkers, is still being evaluated.
Several analyses have further compared miRNA expression in higher-risk versus lower-risk tumor tissues, as defined by grade and stage. The most common approach for these studies was to first identify a series of miRNAs differentially expressed between normal tissue and tumor tissue, and then to compare the expression of this selected list of miRNAs between high-risk and low-risk tumors. Gleason score is a significant indicator of risk and is often a major determinant for treatment in newly diagnosed and AS patients. Several gene expression analyses have reported differences in miRNA expression between lower and higher Gleason score tumors.28,37,39,40 However, miRNA expression patterns in tissue that have the ability to differentiate between low and high Gleason score have not yet been identified.14,37,39 Several miRNAs have also been reported to be differentially expressed between nonmalignant prostate tissue and low-stage or high-stage PCa.28,29 Similarly, there does not appear to be a clear miRNA expression signature that can differentiate between lower- and higher-stage PCa. Further studies directly comparing tumor versus tumor miRNA expression may reveal new signatures associated with stage and/or grade.
Many gene expression studies have also evaluated miRNA expression in the biochemical recurrence (BCR) of PCa. The common approach has been to first identify miRNAs by comparison of nonmalignant prostate tissue to PCa, with later analysis for association with BCR.31,32 A common observation between these studies has been elevated miR-96 in samples with BCR. Two other studies have directly compared miRNA gene expression from the tumor tissue of recurrent and nonrecurrent PCa.43,44 Both found decreased miR-24-1* expression in recurrent prostate tumors. Down-regulated miR-154 expression has also been commonly found in tumors that biochemically progress.32,44
There have been very few miRNA gene expression studies in metastatic or castration-resistant PCa. Two studies have directly evaluated miRNA expression profiles in nonmalignant prostate tissues versus castration-resistant prostate tumors.30,35 Several miRNAs were found to be differentially expressed between normal and castration-resistant tissue, as well as normal and castration-sensitive tissue, though a gene expression signature of castration-sensitive versus castration-resistant was not apparent. In a separate study of metastasis, Peng et al. evaluated miRNA expression profiles in primary PCa versus bone metastasis samples.45 Martens-Uzunova et al. have similarly compared miRNA expression in primary versus lymph node metastatic PCa.33 Comparative review of these two studies indicates that upregulated miR-210 and decreased miR-125b, miR-27b, and miR-99a are more commonly observed in metastases when compared to primary tumors.
Collectively, these gene expression analyses provide an important insight into miRNA association with disease and disease progression. The last decade has seen several advances in miRNA gene expression analyses and interpretation, as well as improved understanding in tissue selection, processing, storage, and the importance of RNA stability and quality. For example, significant differences in miRNA expression could be expected for laser microdissected versus manually dissected tumors, for tissue isolated by trans-urethral resection of the prostate (TURP) versus radical prostatectomy specimens, or for frozen versus formalin-fixed and paraffin-embedded samples. Differential sensitivities and specificities may also be expected between miRNA hybridization arrays, qRT-PCR libraries, or RNA sequencing platforms. All of these features have contributed to the inconsistencies of miRNA gene expression studies seen thus far in PCa.46 Here, we have attempted to focus on the common observations, rather than the inconsistencies, to highlight those miRNAs that have been most commonly observed to be differentially expressed across different tissue sources and gene expression platforms. This does not signify that these miRNAs are more important or significant than others, as there have been numerous focused studies of miRNAs and miRNA biology that are independent of high-throughput gene expression analysis platforms. Nonetheless, we feel these common observations may be valuable and reflective of common biologic events in PCa.
Detection of aberrant miRNA expression in bodily fluids
The analysis of miRNAs in bodily fluids has uncovered their potential value as highly stable biomarkers. miRNAs can be detected in bodily fluids as free nucleic acids, within disseminated tumors cells or within exosomes or other microvesicles.47 Here, we review the current literature of circulating and secreted miRNAs in PCa. As above, we attempt to highlight those miRNAs most commonly observed to be de-regulated.
Circulating miRNAs
The discovery of circulating miRNAs uncovered their possible value as noninvasive biomarkers for a variety of indications. Mitchell et al. first described circulating miRNAs as blood-based markers for PCa, noting their surprisingly high stability.48 miR-141, a miRNA commonly observed to be over-expressed in PCa tissue (Table 1), was elevated in the serum of patients with PCa, indicating that circulating miRNA levels may be informative in PCa diagnosis.48 Independent miRNA gene expression analyses have also found elevated miR-141 levels in the serum of men with PCa, as well as elevated levels of several other miRNAs including miR-16, miR-92a, miR-103, miR-107, miR-197, miR-34b, miR-328, miR-485-3p, miR-486-5p, miR-92b, miR-574-3p, miR-636, miR-640, miR-766, and miR-885-5p.49 A separate miRNA expression analysis found five miRNAs, let-7c, let-7e, miR-30c, miR-622, and miR-1285, as potential serum diagnostic biomarkers for PCa.50 In another study, Moltzahn et al. found decreased miR-223, miR-26b, miR-30c, and miR-24 and increased miR-20b, miR-874, miR-1274a, miR-1207-5p, miR-93, and miR-106a levels in the sera of men with PCa.51 Circulating miRNA levels may also be indicative of disease aggressiveness. Differential levels of miR-93, miR-24, and miR-106a have been reported in low- and intermediate-risk PCa patients. RNA sequencing of exosomal miRNAs has also identified miR-1290 and miR-375 as potential prognostic makers for clinical outcome in castration-resistant PCa.52 Numerous studies focused on specific miRNAs such as miR-21, miR-221, miR-141, and miR-375 continue to shed light on the potential value of circulating miRNAs as diagnostic, prognostic, or predictive biomarkers.53,54,55,56,57,58,59
The continued study of circulating miRNAs has a great potential to uncover new insights into the function of miRNAs in cancer biology and to capitalize on their potential as biomarkers. As with studies of miRNAs in tissue, a variety of techniques and platforms likely account for the inconsistencies between studies. Nonetheless, several miRNAs have been consistently identified to be elevated in the serum of PCa patients, including miR-375, miR-141, and miR-21 (Table 1). Future unbiased analyses of circulating miRNAs in additional patient populations such as newly diagnosed low-risk versus high-risk patients or patients in AS programs may uncover new value for miRNAs as serum biomarkers in early disease management. Further studies of castration-resistant, advanced patients following second-generation ADT will provide further areas for discovery.
miRNAs as urine biomarkers
Urine represents a valuable resource for gene, protein, and RNA biomarkers in urological cancers. While there have been a limited number of miRNA studies in urine, results support that miRNAs are readily detected in urine and that they have promise as next-generation biomarkers (Table 1). Using a panel of miRNAs initially identified in the serum of patients with PCa, Bryant et al. found elevated miR-107 and miR-574-3p expression levels in the urine of men with PCa when compared to healthy controls.54 Importantly, the diagnostic value of these miRNAs appeared to be greater than that of PCA3, an established urine RNA biomarker for PCa diagnosis.60 Differential urine levels of miR-1825 and miR-484 have also shown diagnostic potential for men with PCa,61 as well as miR-205 and miR-214.62 The detection of miRNAs in the urine is not limited to cellular RNA. Elevated levels of miR-483-5p can be detected in the cell-free and nonexosome-enriched fraction of urine from patients with PCa.63 While these studies are promising, there is a clear need for stringent assay development and validation for miRNAs as urine biomarkers.64 Future studies will clarify their value and reliability. The analysis of miRNA biomarkers might be most helpful for the growing number of men diagnosed with low-risk disease who need accurate, noninvasive assays to detect or predict progression during AS, without the need for frequent invasive biopsies.
Exosomal miRNAs
Exosomes are small extracellular vesicles, 40–100 nm in diameter, that release cellular components including proteins and RNA into the extracellular milieu.65 These vesicles contain miRNAs and may be vehicles for transferring miRNAs between cells and tissues; however, it is unclear whether the levels of miRNA in patient exosomes are sufficiently high to be functional.66 Nonetheless, exosomes present a separate compartment for miRNA expression analysis in tissue, serum, and urine that may provide additional value beyond total, cellular, or cell-free miRNAs (Table 1). Serum-derived exosomes have been applied to miRNA expression analyses and elevated miR-141 and miR-375 have been observed in men with metastatic PCa.54 MiRNAs have also been found in larger extracellular vesicles (>200 nm), indicating that various microvessicles or microvessicle isolation platforms may be informative for miRNA studies.67 Indeed, several studies support that miRNAs can be readily detected in PCa-associated exosomes.52,68,69
THE FUNCTION OF miRNAS IN PROSTATE CANCER PROGRESSION
The progression of PCa can be described as a series of common histologic, genetic, and molecular events (Figure 3). There is accumulating evidence that miRNAs can act as upstream or downstream regulators of many of these events. Here, we review literature in which direct studies of miRNA function have been evaluated in PCa cells or models and identify areas for potential discovery.
Figure 3.
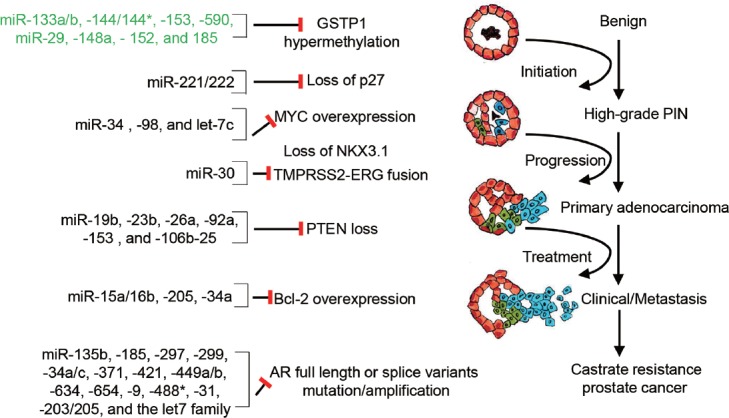
miRNAs in the molecular progression of prostate cancer. Schematic representation of key pathways in the molecular and genetic progression of PCa. miRNAs previously identified to directly regulate each molecule in cellular models of prostate cancer are provided. Those in green font indicate miRNAs that are implicated in regulation, but which have not yet been directly demonstrated in prostate models.
GSTP1
One of the earliest and most universal events in PCa progression is the loss of GSTP1 expression through CpG island methylation.70 The potential role of miRNAs in this early event, either through regulation of GSTP1 itself or the epigenetic regulation of its promoter, has not been well characterized. Aberrant DNA methylation can alter miRNA expression and, likewise, miRNA dysregulation can cause aberrant DNA methylation. Several miRNAs have been identified to regulate the key components of DNA methylation, including the miR-29 family, miR-148a, miR-152, and miR-185.71 Interestingly, many of these miRNAs are deregulated in PCa (Table 1). Several miRNAs have also been predicted to directly target GSTP1, including miR-133a/b, miR-144, miR-153, and miR-542. However, to our knowledge, there has not yet been direct evidence of miRNA regulation of GSTP1 expression or DNA methylation in prostatic models.72
CDKN1B/p27
Another early event in PCa is the loss of p27/CDKN1B expression, which progressively decreases with increasing tumor grade and stage.73 A well-characterized mechanism of p27 loss is through posttranslational proteasomal degradation,74 though decreased expression can occur through additional mechanisms.75 The only direct evidence for miRNA regulation of p27 in PCa involves miR-221 and miR-222, which directly suppress expression through interaction sites in the 3’ untranslated region (3’ UTR).76 While miR-221/222 has been commonly reported to be down-regulated in PCa (Table 1), they have also been observed to be upregulated in castration-resistant PCa cells and to enhance PCa cell proliferation.76,77 Further studies may uncover additional miRNAs capable of regulating p27 expression either through direct 3’ UTR interaction or through indirect regulation of p27 expression or posttranslational stability.
MYC
Another early event in human prostate carcinogenesis is the over-expression of the nuclear Myc protein.78 While the MYC gene is located on chromosome 8q24, a region commonly amplified in PCa, MYC protein over-expression does not necessarily correlate with the gain of 8q24.78 Thus, there may be alternative mechanisms for elevated MYC over-expression. Several miRNAs have been demonstrated to regulate MYC expression in PCa cells, including miR-34, miR-98, and let-7c,79,80,81 either directly or indirectly through upstream mechanisms. MYC is also known to directly regulate the transcription of several miRNAs commonly deregulated in PCa, including the miR-17-5p cluster.79 This cluster was one of the first miRNAs found to be directly regulated by an oncogenic transcription factor.82 Several other miRNAs, including miR-145, have been found to directly regulate MYC in other cancer models.83 Additional functional studies of MYC-regulated miRNAs and MYC-regulating miRNAs in PCa models may uncover additional contributions to PCa initiation or progression.
ETS gene fusions
One of the most common genomic alterations in PCa involves the fusion of TMPRSS2 and ERG.84 The role of miRNAs in TMPRSS2/ERG re-arrangement and downstream signaling has not yet been well defined. Members of the miR-30 family, which have been found to be both over- and under-expressed in PCa (Table 1), directly target ERG and ERG downstream function in PCa cells.85 Two known targets of the ERG transcription factor are EZH2 and MYC,86,87 which may further contribute to aberrant miRNA expression in PCa. Interestingly, the expression of miR-26a was found to be preferentially decreased in ERG rearrangement negative PCa, which was associated with hypermethylation of the miR-26 gene locus.88 EZH2 is a direct target of miR-26a; thus, this miRNA may contribute to EZH2 over-expression in ERG rearrangement negative PCa. Similarly, miR-221 has been observed to be down-regulated in TMPRSS2-ERG positive PCa.89 Further analyses may uncover new roles for miRNAs in these genetic rearrangements or ERG signaling.
PTEN
Genetic mutation and loss of PTEN is a frequent event in PCa, leading to aberrant PI3K/AKT signaling.90 miRNA-mediated suppression of PTEN may provide an alternate route for elevated AKT signaling in PCa. miR-22 and the miR-106b-25 cluster have been reported to be over-expressed in PCa and to directly target PTEN, potentiating cellular transformation.91 Similarly, miR-153 has been reported to be highly expressed in PCa and to directly target and down-regulate PTEN, leading to activated AKT signaling.92 miR-23b, miR-19b, miR-26a, and miR-92a have also been demonstrated to directly regulate PTEN in PCa cells.93 PTEN may also be regulated through competition between endogenous RNAs and miRNAs. PTENP1, a nonprotein-coding PTEN pseudogene, appears to compete with PTEN mRNA for miR-19b/20a binding.94 Accordingly, loss of PTENP1 enhances miR-19b/20a-mediated suppression of PTEN, thus promoting tumor development.
BCL-2
Another important pathway that is deregulated in advanced metastatic and castration-resistant PCa is the anti-apoptotic protein BCl-2,95 which is an established direct target of miR-15/16. Knockdown of miR-15/16 in prostate epithelial cells leads to enhanced survival, proliferation, and tumorigenesis in NOD-SCID mice.96 miR-205, whose expression is down-regulated in PCa, also regulates BCl-2 expression.97 Further characterization of miRNAs involved in apoptosis and survival may reveal additional mechanisms for the progression and survival of PCa cells.
AR
One of the most studied pathways in PCa progression and therapeutic resistance is the AR signaling pathway. Aberrant AR expression and activity can be attributed to numerous genetic, endocrine, and molecular mechanisms.98,99 In addition, several miRNAs have been identified as direct regulators of AR expression or AR activity. These include miR-135b, miR-34c, miR-488*, miR-31, miR-203/205, let-7, and others.100,101,102,103 Likewise, the expression of miRNAs can be directly regulated by AR transcriptional activity. Several miRNAs, including the commonly over-expressed miR-21 and miR-32 (Table 1), have been shown to be directly regulated by AR at the transcriptional level.35,104 Further characterization of AR-targeting and AR-targeted miRNAs, as well as miRNA-mediated regulation of hormone metabolism, AR cofactors, or mechanisms of aberrant AR splicing, may reveal additional roles for miRNAs in aberrant AR activity in PCa.
In summary, aberrant miRNA expression and activity are commonly observed in clinical PCa, and functional studies clearly implicate miRNAs in PCa initiation and progression. While some mechanisms of miRNA regulation have been relatively well established in PCa biology and several miRNAs have been routinely implicated in PCa development and progression, the field is just beginning to understand and appreciate the role of miRNAs. As the field of PCa research continues to evolve, as well as the study of miRNA expression and function, new areas for discovery will surely be revealed. Some of the most notable areas of unmet need in PCa include the need for diagnostic markers of clinically relevant PCa, biomarkers that can reduce the biopsy frequency of men undergoing AS, and new molecular biomarkers and targets for the management and treatment of locally aggressive and metastatic castration-resistant PCa. The next decade of research holds great promise for continued discovery and translation. The value of these efforts and data will be best realized with systematic and consistent research methods and validation across multiple cohorts and research platforms.
COMPETING INTERESTS
No conflicts of interest to disclose.
ACKNOWLEDGMENTS
Funding for this manuscript was supported by grants from the National Institutes of Health 5R01CA143299, 5P50CA058236, and the Patrick C Walsh Prostate Cancer Research Fund.
REFERENCES
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Roobol MJ, Carlsson SV. Risk stratification in prostate cancer screening. Nat Rev Urol. 2013;10:38–48. doi: 10.1038/nrurol.2012.225. [DOI] [PubMed] [Google Scholar]
- 3.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pienta KJ, Walia G, Simons JW, Soule HR. Beyond the androgen receptor: new approaches to treating metastatic prostate cancer. Report of the 2013 Prouts Neck Prostate Cancer Meeting. Prostate. 2014;74:314–20. doi: 10.1002/pros.22753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7:256–69. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van der Kwast TH, Roobol MJ. Defining the threshold for significant versus insignificant prostate cancer. Nat Rev Urol. 2013;10:473–82. doi: 10.1038/nrurol.2013.112. [DOI] [PubMed] [Google Scholar]
- 8.Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, et al. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 9.Trock BJ. Circulating biomarkers for discriminating indolent from aggressive disease in prostate cancer active surveillance. Curr Opin Urol. 2014;24:293–302. doi: 10.1097/MOU.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 10.Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol. 2012;61:1019–24. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belair CD, Paikari A, Moltzahn F, Shenoy A, Yau C, et al. DGCR8 is essential for tumor progression following PTEN loss in the prostate. EMBO Rep. 2015;16:1219–32. doi: 10.15252/embr.201439925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark EL, Hadjimichael C, Temperley R, Barnard A, Fuller-Pace FV, et al. p68/DdX5 supports beta-catenin and RNAP II during androgen receptor mediated transcription in prostate cancer. PLoS One. 2013;8:e54150. doi: 10.1371/journal.pone.0054150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–46. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han B, Mehra R, Dhanasekaran SM, Yu J, Menon A, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–37. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Chen H, Zhang L, Dakhova O, Zhang Y, et al. A dosage-dependent pleiotropic role of dicer in prostate cancer growth and metastasis. Oncogene. 2014;33:3099–108. doi: 10.1038/onc.2013.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo NJ, Hur SY, Kim MS, Lee JY, Lee SH. Immunohistochemical analysis of RNA-induced silencing complex-related proteins AGO2 and TNRC6A in prostate and esophageal cancers. APMIS. 2010;118:271–6. doi: 10.1111/j.1600-0463.2010.02588.x. [DOI] [PubMed] [Google Scholar]
- 22.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 23.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 28.Casanova-Salas I, Rubio-Briones J, Calatrava A, Mancarella C, Masia E, et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J Urol. 2014;192:252–9. doi: 10.1016/j.juro.2014.01.107. [DOI] [PubMed] [Google Scholar]
- 29.Hart M, Nolte E, Wach S, Szczyrba J, Taubert H, et al. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol Cancer Res. 2014;12:250–63. doi: 10.1158/1541-7786.MCR-13-0230. [DOI] [PubMed] [Google Scholar]
- 30.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 31.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–76. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 32.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–16. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 33.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–91. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 34.He HC, Han ZD, Dai QS, Ling XH, Fu X, et al. Global analysis of the differentially expressed miRNAs of prostate cancer in Chinese patients. BMC Genomics. 2013;14:757. doi: 10.1186/1471-2164-14-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31:4460–71. doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 36.Schubert M, Spahn M, Kneitz S, Scholz CJ, Joniau S, et al. Distinct microRNA expression profile in prostate cancer patients with early clinical failure and the impact of let-7 as prognostic marker in high-risk prostate cancer. PLoS One. 2013;8:e65064. doi: 10.1371/journal.pone.0065064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song C, Chen H, Wang T, Zhang W, Ru G, et al. Expression profile analysis of microRNAs in prostate cancer by next-generation sequencing. Prostate. 2015;75:500–16. doi: 10.1002/pros.22936. [DOI] [PubMed] [Google Scholar]
- 38.Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–38. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 39.Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA profiling of prostate cancer. J Cancer. 2013;4:350–7. doi: 10.7150/jca.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlsson J, Davidsson S, Helenius G, Karlsson M, Lubovac Z, et al. A miRNA expression signature that separates between normal and malignant prostate tissues. Cancer Cell Int. 2011;11:14. doi: 10.1186/1475-2867-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104–16. doi: 10.1016/j.cell.2014.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 43.Karatas OF, Guzel E, Suer I, Ekici ID, Caskurlu T, et al. miR-1 and miR-133b are differentially expressed in patients with recurrent prostate cancer. PLoS One. 2014;9:e98675. doi: 10.1371/journal.pone.0098675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mortensen MM, Hoyer S, Orntoft TF, Sorensen KD, Dyrskjot L, et al. High miR-449b expression in prostate cancer is associated with biochemical recurrence after radical prostatectomy. BMC Cancer. 2014;14:859. doi: 10.1186/1471-2407-14-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng X, Guo W, Liu T, Wang X, Tu X, et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS One. 2011;6:e20341. doi: 10.1371/journal.pone.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandellini P, Folini M, Zaffaroni N. Towards the definition of prostate cancer-related microRNAs: where are we now? Trends Mol Med. 2009;15:381–90. doi: 10.1016/j.molmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, et al. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen ZH, Zhang GL, Li HR, Luo JD, Li ZX, et al. A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate. 2012;72:1443–52. doi: 10.1002/pros.22495. [DOI] [PubMed] [Google Scholar]
- 51.Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, et al. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res. 2011;71:550–60. doi: 10.1158/0008-5472.CAN-10-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X, Yuan T, Liang M, Du M, Xia S, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brase JC, Johannes M, Schlomm T, Falth M, Haese A, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–16. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 54.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768–74. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen HC, Xie W, Yang M, Hsieh CL, Drouin S, et al. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate. 2013;73:346–54. doi: 10.1002/pros.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selth LA, Townley SL, Bert AG, Stricker PD, Sutherland PD, et al. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br J Cancer. 2013;109:641–50. doi: 10.1038/bjc.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen J, Hruby GW, McKiernan JM, Gurvich I, Lipsky MJ, et al. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate. 2012;72:1469–77. doi: 10.1002/pros.22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, et al. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011;32:583–8. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 59.Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326–31. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 60.Crawford ED, Rove KO, Trabulsi EJ, Qian J, Drewnowska KP, et al. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J Urol. 2012;188:1726–31. doi: 10.1016/j.juro.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 61.Haj-Ahmad TA, Abdalla MA, Haj-Ahmad Y. Potential urinary miRNA biomarker candidates for the accurate detection of prostate cancer among benign prostatic hyperplasia patients. J Cancer. 2014;5:182–91. doi: 10.7150/jca.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava A, Goldberger H, Dimtchev A, Ramalinga M, Chijioke J, et al. MicroRNA profiling in prostate cancer – The diagnostic potential of urinary miR-205 and miR-214. PLoS One. 2013;8:e76994. doi: 10.1371/journal.pone.0076994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korzeniewski N, Tosev G, Pahernik S, Hadaschik B, Hohenfellner M, et al. Identification of cell-free microRNAs in the urine of patients with prostate cancer. Urol Oncol. 2015;33:16–e17-22. doi: 10.1016/j.urolonc.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 64.Sapre N, Hong MK, Macintyre G, Lewis H, Kowalczyk A, et al. Curated microRNAs in urine and blood fail to validate as predictive biomarkers for high-risk prostate cancer. PLoS One. 2014;9:e91729. doi: 10.1371/journal.pone.0091729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–4. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–93. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morello M, Minciacchi VR, de Candia P, Yang J, Posadas E, et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12:3526–36. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hessvik NP, Phuyal S, Brech A, Sandvig K, Llorente A. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim Biophys Acta. 2012;1819:1154–63. doi: 10.1016/j.bbagrm.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 69.Hessvik NP, Sandvig K, Llorente A. Exosomal miRNAs as biomarkers for prostate cancer. Front Genet. 2013;4:36. doi: 10.3389/fgene.2013.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin X, Tascilar M, Lee WH, Vles WJ, Lee BH, et al. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. Am J Pathol. 2001;159:1815–26. doi: 10.1016/S0002-9440(10)63028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki H, Maruyama R, Yamamoto E, Kai M. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013;4:258. doi: 10.3389/fgene.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh S, Shukla GC, Gupta S. MicroRNA regulating glutathione S-transferase P1 in prostate cancer. Curr Pharmacol Rep. 2015;1:79–88. doi: 10.1007/s40495-014-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macri E, Loda M. Role of p27 in prostate carcinogenesis. Cancer Metastasis Rev. 1998;17:337–44. doi: 10.1023/a:1006133620914. [DOI] [PubMed] [Google Scholar]
- 74.Loda M, Cukor B, Tam SW, Lavin P, Fiorentino M, et al. Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas. Nat Med. 1997;3:231–4. doi: 10.1038/nm0297-231. [DOI] [PubMed] [Google Scholar]
- 75.le Sage C, Nagel R, Agami R. Diverse ways to control p27Kip1 function: miRNAs come into play. Cell Cycle. 2007;6:2742–9. doi: 10.4161/cc.6.22.4900. [DOI] [PubMed] [Google Scholar]
- 76.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–24. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 77.Sun T, Wang Q, Balk S, Brown M, Lee GS, et al. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–63. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–67. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benassi B, Flavin R, Marchionni L, Zanata S, Pan Y, et al. MYC is activated by USP2a-mediated modulation of microRNAs in prostate cancer. Cancer Discov. 2012;2:236–47. doi: 10.1158/2159-8290.CD-11-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamamura S, Saini S, Majid S, Hirata H, Ueno K, et al. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells. PLoS One. 2012;7:e29722. doi: 10.1371/journal.pone.0029722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nadiminty N, Tummala R, Lou W, Zhu Y, Zhang J, et al. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem. 2012;287:1527–37. doi: 10.1074/jbc.M111.278705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 83.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–12. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 85.Kao CJ, Martiniez A, Shi XB, Yang J, Evans CP, et al. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene. 2014;33:2495–503. doi: 10.1038/onc.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS One. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–53. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Borno ST, Fischer A, Kerick M, Falth M, Laible M, et al. Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation. Cancer Discov. 2012;2:1024–35. doi: 10.1158/2159-8290.CD-12-0041. [DOI] [PubMed] [Google Scholar]
- 89.Gordanpour A, Stanimirovic A, Nam RK, Moreno CS, Sherman C, et al. miR-221 Is down-regulated in TMPRSS2:ERG fusion-positive prostate cancer. Anticancer Res. 2011;31:403–10. [PMC free article] [PubMed] [Google Scholar]
- 90.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 91.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73:596–604. doi: 10.1002/pros.22600. [DOI] [PubMed] [Google Scholar]
- 93.Tian L, Fang YX, Xue JL, Chen JZ. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One. 2013;8:e75885. doi: 10.1371/journal.pone.0075885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Catz SD, Johnson JL. BCL-2 in prostate cancer: a minireview. Apoptosis. 2003;8:29–37. doi: 10.1023/a:1021692801278. [DOI] [PubMed] [Google Scholar]
- 96.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 97.Verdoodt B, Neid M, Vogt M, Kuhn V, Liffers ST, et al. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol. 2013;43:307–14. doi: 10.3892/ijo.2013.1915. [DOI] [PubMed] [Google Scholar]
- 98.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 99.Chan SC, Dehm SM. Constitutive activity of the androgen receptor. Adv Pharmacol. 2014;70:327–66. doi: 10.1016/B978-0-12-417197-8.00011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ostling P, Leivonen SK, Aakula A, Kohonen P, Makela R, et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–67. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 101.Sikand K, Slaibi JE, Singh R, Slane SD, Shukla GC. miR 488* inhibits androgen receptor expression in prostate carcinoma cells. Int J Cancer. 2011;129:810–9. doi: 10.1002/ijc.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013;73:1232–44. doi: 10.1158/0008-5472.CAN-12-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boll K, Reiche K, Kasack K, Morbt N, Kretzschmar AK, et al. miR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2013;32:277–85. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- 104.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–9. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]


