Abstract
In the postscreening era, physicians are in need of methods to discriminate aggressive from nonaggressive prostate cancer (PCa) to reduce overdiagnosis and overtreatment. However, studies have shown that prognoses (e.g., progression and mortality) differ even among individuals with similar clinical and pathological characteristics. Existing risk classifiers (TMN grading system, Gleason score, etc.) are not accurately enough to represent the biological features of PCa. Using new genomic technologies, novel biomarkers and classifiers have been developed and shown to add value to clinical or pathological risk factors for predicting aggressive disease. Among them, RNA testing (gene expression analysis) is useful because it can not only reflect genetic variations but also reflect epigenetic regulations. Commercially available RNA profiling tests (Oncotype Dx, Prolaris, and Decipher) have demonstrated strong abilities to discriminate PCa with poor prognosis from less aggressive diseases. For instance, these RNA profiling tests can predict disease progression in active surveillance patients or early recurrence after radical treatments. These tests may offer more dependable methods for PCa prognosis prediction to make more accurate and personal medical decisions.
Keywords: precision medicine, prostate cancer, RNA profiling
INTRODUCTION
Prostate cancer (PCa) has become the second leading cause of cancer-related death among men, with an estimated 914 000 new cases and 258 000 deaths worldwide every year.1 This makes PCa a major public health problem worldwide.
The introduction of prostate-specific antigen (PSA) testing has provided a method for early detection of PCa and has been associated with a decline in PCa mortality; however, it has also been associated with a widespread problem of overdiagnosis and overtreatment of the non-aggressive PCa.2 To solve this problem, active surveillance (AS) is recommended for patients with PCa cases that are at low-risk of progressing.3 However, studies have found that the accuracy of available risk assessment tools (based on clinical information, tests such as PSA, Gleason score of biopsy, etc.) should be challenged.4 For example, a proportion of up to 60% of patients with preoperational low-risk PCa were found to have higher grades of disease after surgery.5,6,7 This raises concerns of potential for missed-treatment using AS for patients with high-grade diseases in which curative treatment would be necessary. However, the prognosis of patients after radical treatments varies widely. For instance, studies have shown that approximately 70% of patients who undergo radical prostatectomy, who are at high-risk for aggressive disease (with a high Gleason score, extraprostatic extension, seminal vesicle invasion, or having positive lymph node) would not die of PCa after 15 years.8 In addition, several studies have suggested that patients with adverse pathology outcomes may be cured by surgery alone and that adjuvant therapy would not be necessary for all of them.9,10
To address the issue of being unable to accurately predict PCa prognosis, novel biomarkers have been shown to determine whether PCa is aggressive and to predict poor prognosis.11,12,13 In addition, new approaches that utilize new genomic technologies can assess to genetic alterations and epigenetic events. Among them, RNA testing (gene expression analysis) is considered highly useful, for reflecting not only genetic variations but also epigenetic regulations. Several RNA tests have been approved for clinical use in prostate cancer and have been found to add value to clinical and pathological risks for prostate cancer progression. In this review, we focus on the value of commercially available RNA profiling tests in precision medicine practice for PCa.
COMMERCIALLY AVAILABLE RNA PROFILING PANELS FOR PROSTATE CANCER
Oncotype Dx
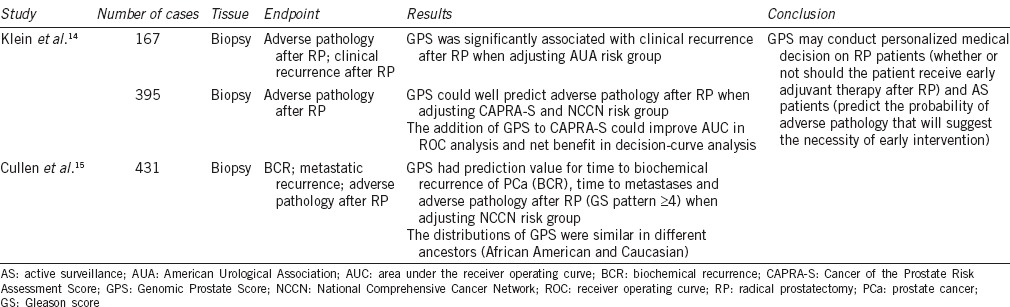
Oncotype Dx Prostate Cancer Assay is a multigene expression assay based on a real-time polymerase chain reaction (RT-PCR) technique developed by Genomic Health Inc., Redwood City, CA, USA. The assay measures expression of 17 genes using approximately 1 mm of fixed paraffin-embedded (FPE) prostate biopsy tissue. After assessing gene expression, a Genomic Prostate Score (GPS) is calculated. Among these 17 genes, 12 are cancer-related, representing a stromal response pathway (BGN, COL1A1, and SFRP4), an androgen signaling pathway (AZGP1, KLK2, SRD5A2, and FAM13C), a cellular organization pathway (FLNC, GSN, TPM2, and GSTM2) and a proliferation pathway (TPX2). The remaining five genes are housekeeping genes, including ARF1, ATP5E, CLTC, GPS1, and PGK1. Previous studies suggest that this assay could accurately predict PCa recurrence after radical prostatectomy (RP) or PCa progression in active surveillance (AS) patients, which could help make the decision regarding further treatment (e.g., adjuvant therapy after RP, radical treatment for patients undertaking AS).14,15
During discovery period, 727 genes were first evaluated in 441 patients (111 of whom had a clinical recurrence and 45 of whom died of PCa) who underwent RP from 1987 to 2004 at the Cleveland Clinic in Cleveland, Ohio, USA. The associations of these genes with Gleason score (GS) patterns and PCa recurrence after surgery were investigated. Eighty-one of 727 genes with the highest differential expression (P < 0.10) were included in further analysis under the following criteria: (1) added value to American Urological Association (AUA) risk stratification system and Cancer of the Prostate Risk Assessment Score (CAPRA-S); (2) were significantly associated with PCa death and adverse pathology at RP; (3) represented carcinogenesis pathways, or ER and AR. The associations of these 81 genes with GS and PCa recurrence were evaluated in a 167 biopsy population, confirming that 58 genes involved in six biological pathways were significantly associated with aggressive disease. Finally, 12 cancer-related genes and five housekeeping genes were used to build a scoring model of GPS based on consistency of the testing.14
A summarization of Oncotype Dx studies is shown in Table 1. Oncotype Dx has been shown to add value to the prediction of PCa recurrence after RP. In the 167 biopsy population, investigators found that GPS was an independent predictor when adjusted for AUA group. In the low-risk AUA group, the 10-year risk of clinical recurrence was 7.0% for patients with high GPS, which was 3 times higher than that of the patients with low GPS. In the AUA high-risk group, in which patients are considered to have a high probability of recurrence, the 10-year recurrence rates varied from 6.2% to 28.6% for patients with different levels of GPS.14 In a validation study that consisted of 395 patients who met AS criteria but underwent RP, GPS was able to discriminate high-grade from low-grade prostate cancer in various clinical risk groups including CAPRA-S and National Comprehensive Cancer Network (NCCN) risk groups.14 This indicates that the Oncotype Dx GPS might also be able to predict adverse pathology and high-risk prostate cancer in an AS population and may be able to supplement other clinical and pathological information to develop personalized AS plans for PCa. Further analysis showed that combining CAPRA-S and GPS might bring even more benefit, leading to fewer unnecessary treatments without increasing the number of high-risk PCa cases left untreated.14 Another validation study was performed with a median follow-up of 5.2 years by Cullen et al. The study indicated that GPS had prediction value for time to biochemical recurrence (BCR) of PCa, time to metastasis and adverse pathology after RP (GS pattern ≥4) when adjusting for NCCN risk group.15 In addition, the distributions of GPS were similar in different races such as African American and Caucasian.15 Therefore, researchers suggested that the Oncotype Dx GPS could predict cancer recurrence after RP and PCa progression for AS patients and could help further inform personalized medical decision making to RP patients and AS patients.
Table 1.
Summary of Oncotype Dx PCa assay studies
Prolaris
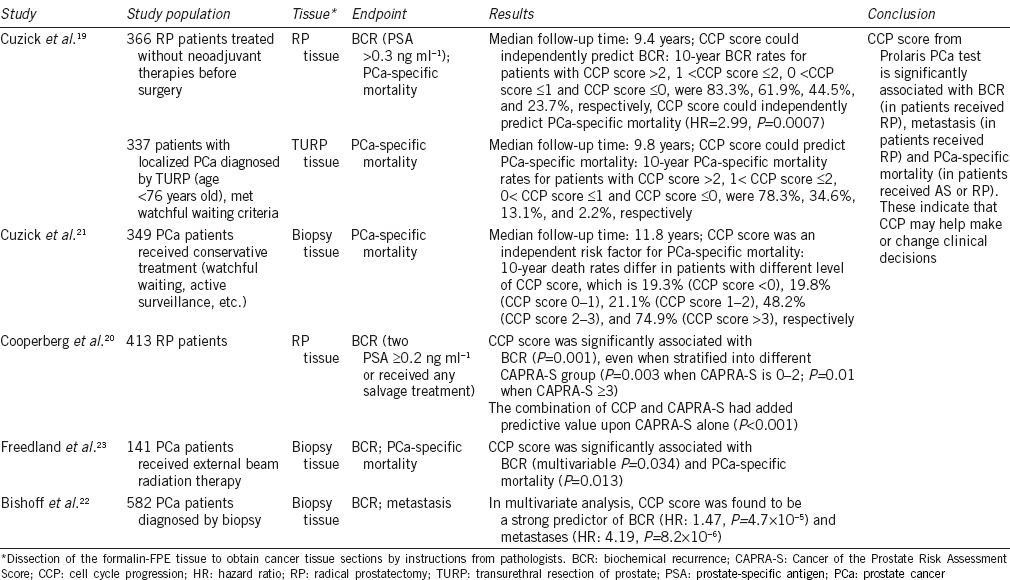
It has been shown that the expression of cell cycle progression (CCP) genes varies among different types of cells and reflects the pattern of mitosis.16 Cancer cells, especially aggressive cancer cells, will transcribe more CCP genes than normal cells due to continuous proliferation. Thus, CCP gene expression could reflect tumor biology (i.e., the more aggressive the tumor is, the more CCP genes are expressed), which may be useful for predicting the outcomes of cancers. This has been demonstrated in other types of malignancies, as well.17,18,19 The Prolaris PCa test (Myriad Genetics Inc., Salt Lake City, UT, USA) was designed based on this theory to test the expression of 31 CCP genes (FOXM1, CDC20, CDKN3, CDC2, KIF11, KIAA0101, NUSAP1, CENPF, ASPM, BUB1B, RRM2, DLGAP5, BIRC5, KIF20A, PLK1, TOP2A, TK1, PBK, ASF1B, C18orf24, RAD54L, PTTG1, CDCA3, MCM10, PRC1, DTL, CEP55, RAD51, CENPM, CDCA8, and ORC6L) and 15 housekeeping genes using quantitative RT-PCR. After testing, a CCP score is calculated for predicting cancer recurrence, metastases, PCa-specific mortality in RP patients, and PCa progression in AS patients.19,20,21,22,23
A total of 126 CCP genes chosen from the Gene Expression Omnibus database were evaluated. According to the database, genes with the highest differential expression (compared to the mean expressions of the 126 CCP genes) were selected, and further evaluated in multivariable analyses. Thirty-one of the 126 CCP genes were ultimately selected for having robust and independent abilities to measure levels of cell proliferation. In addition to these CCP genes, 15 housekeeping genes were included in the panel.19
Several studies have evaluated the clinical utility of Prolaris (Table 2). For patients that underwent RP, CCP score had better prediction abilities for BCR and PCa-specific mortality than any other clinical or pathological variables. One study found that 10-year BCR rates and PCa-specific mortality rates significantly increased if the patient had a higher CCP.19 For patients who received other radical therapies (e.g., external beam radiation therapy), CCP score was considered an independent risk factor of both BCR and PCa-specific mortality when adjusting to other clinical variables.20,23 The predictive value of Prolaris CCP scores was even further improved when combined with clinical variables, such as CAPRA-S risk group.20 In addition, the most recent study indicated that CCP could predict metastasis after RP (HR: 4.19, P = 8.2 × 10−6).22 CCP also predicted prognosis of AS (or “watchful waiting”) patients. In a watchful waiting cohort where patients were occasionally diagnosed with PCa via TURP, investigators found that CCP score (from TURP tissue) was able to predict PCa mortality after a median follow-up time of 9.8 years. They found that 10-year PCa-specific mortality rates for patients with CCP score >2, 1< CCP score ≤2, 0< CCP score ≤1 and CCP score ≤0, were 78.3%, 34.6%, 13.1% and 2.2%, respectively.19 A similar result was also observed in another cohort after a median follow-up time of 11.8 years, where patients were diagnosed with low clinical risk PCa via biopsy and received conservative treatments (e.g., watchful waiting, active surveillance, etc.).21 Thus, studies have suggested that CCP score from the Prolaris PCa test added predictive value for PCa prognosis for clinical and pathological risk factors in both RP patients and AS patients.
Table 2.
Summary of Prolaris studies
Decipher
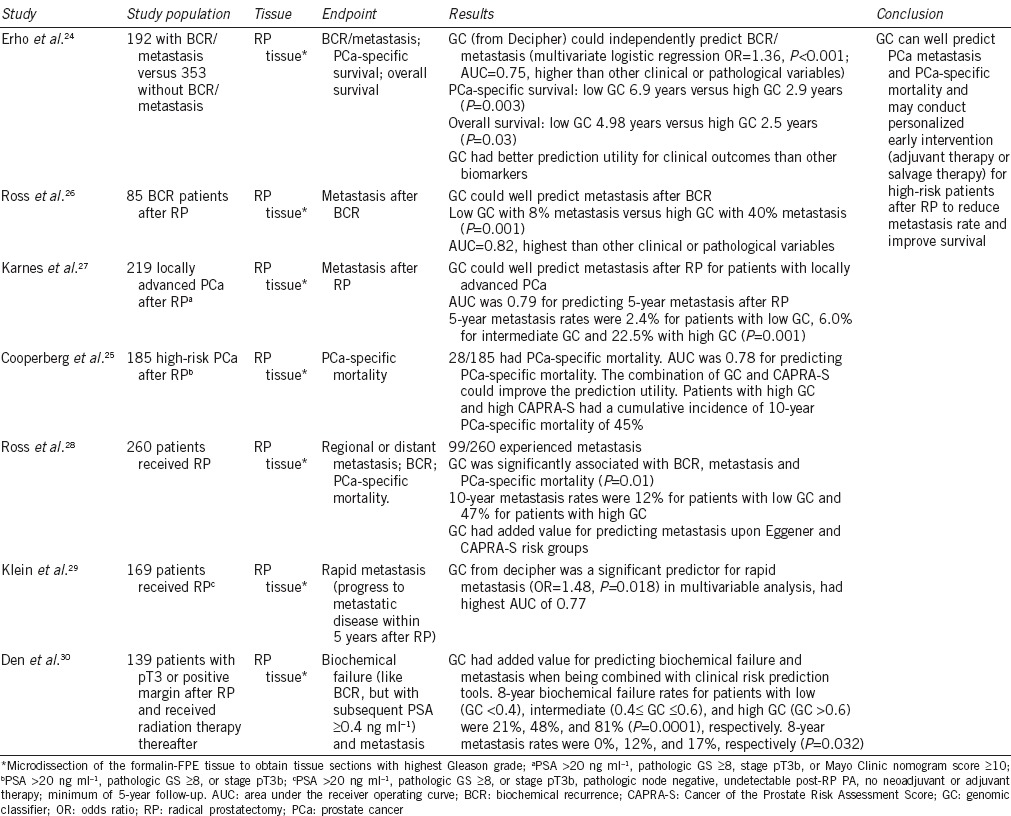
Decipher is a genetic classifier that uses an RNA profiling panel of 22 genetic markers. Before testing, microdissection of the formalin-FPE tissue from RP should be performed to obtain tissue sections with highest Gleason grade. Total RNA is extracted and tested using the Decipher panel. The panel is designed to evaluate the expression of various genetic markers associated with specific of biological processes, including cell proliferation and differentiation processes (LASP1, IQGAP3, NFIB, and S1PR4); cell structure, adhesion, and motility processes (THBS2, ANO7, PCDH7, MYBPC1, and EPPK1); the immune response process (TSBP, PBX1); cell cycle progression and mitosis processes (NUSAP1, ZWILCH, UBE2C, CAMK2N1, and RABGAP1); and other unknown functional processes (PCAT-32, GLYATL1P4/PCAT-80, and TNFRSF19); as well as three unidentified segments. The genetic markers are located in or near the gene segments (e.g., intron, exon, 3’UTR, and noncoding transcript). A genetic classifier (GC) score is then calculated, which may predict PCa metastasis and cancer-specific mortality after RP. The panel was developed by GenomeDx Biosciences Inc., Vancouver, BC, Canada.24 Previous studies have indicated that GC score may be able to predict early metastasis and cancer-specific mortality after RP, which leads to the potential to provide earlier intervention for these patients.24,25,26,27,28,29,30
A nested case–control study was performed in a subset of the population from the Mayo Clinic Radical Prostatectomy Tumor Registry from 1987 to 2001 who received RP for primary PCa as first-line treatment. A total of 639 patients were included, of which 545 patients had available samples. One hundred ninety-two patients without evidence of disease progression after at least 7 years of follow-up were identified as the control group. The remaining 353 patients had biochemical recurrences or metastases during follow-up and were identified as the case group. The investigators tested the tissue samples using an exon transcriptome chip containing approximately 1.4 million selection regions including coding and noncoding regions. Initially, 18 902 differentially expressed RNA regions were observed. Using logistic regression and random forest machine learning algorithm methods, a final set of 22 markers were selected to calculate GC scores.24
The major findings from studies of Decipher are shown in Table 3. Studies have indicated that GC scores from Decipher could predict BCR and metastasis after RP. In a retrospective case–control study based on a population of 192 BCR/metastasis patients and 353 patients without BCR/metastasis, GC had a better predictive utility for BCR/metastasis (AUC = 0.75) than other clinical or pathological variables. Investigators also found that GC had better predictive value than other existing biomarkers (e.g., PCA3, PSA, PSMA, ERG, etc.).24 In studies that investigated high-risk patients who received RP, the 5-year metastasis rates differed among patients with low GC (<0.4, 2.4%), intermediate GC (0.4–0.6, 6%), and high GC (>0.6, 22.5%); therefore, GC was found to be a significant predictor for metastasis.27,29,30 Ross et al. also observed that the 10-year metastasis rate increased for patients with higher GC (GC <0.45, 10-year metastasis rate = 12%; GC >0.5, 10-year metastasis rate = 47%).26,28 GC could also predict PCa-specific mortality and overall survival. Erho et al. found that RP patients with GC ≤0.5 had a longer median PCa-specific survival (6.9 vs 2.9 years, P = 0.003) and overall survival (4.98 vs 2.5 years, P = 0.03) than patients with GC >0.5.24 The prediction ability (assessed using AUC) of CG for PCa-specific mortality was 0.78.25 In addition, a study found that patients with high GC and high CAPRA-S have a cumulative 10-year PCa-specific mortality of up to 45%.25 Taken together, these findings indicate that Decipher GC scores have the potential to predict PCa metastasis and PCa-specific mortality, allowing for early adjuvant or salvage therapy for high GC patients after RP to improve individual prognoses.
Table 3.
Summary of decipher studies
CONCLUSIONS
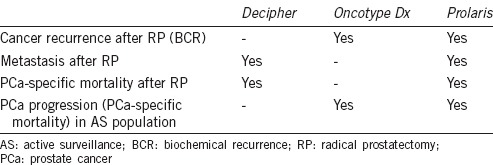
Uses of three commercially available RNA-based testing panels are summarized in Table 4. Although these RNA profiling panels have shown promising results in regards to clinical utility, several limitations are worth noting: (1) the current studies are retrospective with relatively small sample sizes, so larger-scale prospective randomized trials are necessary for validation; (2) RNA quality varies among panels (e.g., microdissection is needed for Decipher [some medical center may not have the equipment], while for Prolaris, tissue extraction relies on the instruction from pathologist, which will lead to heterogeneity of the testing results); and (3) the relatively high prices (~$1500–2000 US dollars per test if not covered by insurance) limit potential use of the panels, and it will be necessary to further evaluate their cost-effective values.
Table 4.
Uses of different commercially available RNA testing panel
Nevertheless, these commercialized RNA profiling tests provide physicians and patients with more choices for more personalized treatment rather than following one-size-fits-all clinical guidelines. Further investigations are necessary to evaluate the clinical values of these RNA-based tests (e.g., whether they can truly predict prognoses in prospective, large-scale studies; the cutoff value of the genetic classifiers) and benefits (whether they can reduce overtreatment of nonaggressive PCa and increase early treatment of aggressive PCa, as well as their medical cost-effective values).
COMPETING INTERESTS
There is no competing financial interest.
ACKNOWLEDGEMENTS
The work was supported by the National Natural Science Foundation of China (Grant No. 81402339).
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Strope SA, Andriole GL. Prostate cancer screening: current status and future perspectives. Nat Rev Urol. 2010;7:487–93. doi: 10.1038/nrurol.2010.120. [DOI] [PubMed] [Google Scholar]
- 3.Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, et al. EAU guidelines on prostate cancer. part 1: screening, diagnosis, and local treatment with curative intent-update 2013. Eur Urol. 2014;65:124–37. doi: 10.1016/j.eururo.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 4.Cooperberg MR, Carroll PR, Klotz L. Active surveillance for prostate cancer: progress and promise. J Clin Oncol. 2011;29:3669–76. doi: 10.1200/JCO.2011.34.9738. [DOI] [PubMed] [Google Scholar]
- 5.King CR, Long JP. Prostate biopsy grading errors: a sampling problem? Int J Cancer. 2000;90:326–30. doi: 10.1002/1097-0215(20001220)90:6<326::aid-ijc3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Conti SL, Dall’era M, Fradet V, Cowan JE, Simko J, et al. Pathological outcomes of candidates for active surveillance of prostate cancer. J Urol. 2009;181:1628–33. doi: 10.1016/j.juro.2008.11.107. [DOI] [PubMed] [Google Scholar]
- 7.Muntener M, Epstein JI, Hernandez DJ, Gonzalgo ML, Mangold L, et al. Prognostic significance of Gleason score discrepancies between needle biopsy and radical prostatectomy. Eur Urol. 2008;53:767–75. doi: 10.1016/j.eururo.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Eggener SE, Scardino PT, Walsh PC, Han M, Partin AW, et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol. 2011;185:869–75. doi: 10.1016/j.juro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson GP, Hussey MA, Tangen CM, Chin J, Messing E, et al. Predominant treatment failure in postprostatectomy patients is local: analysis of patterns of treatment failure in SWOG 8794. J Clin Oncol. 2007;25:2225–9. doi: 10.1200/JCO.2006.09.6495. [DOI] [PubMed] [Google Scholar]
- 10.Mullins JK, Feng Z, Trock BJ, Epstein JI, Walsh PC, et al. The impact of anatomical radical retropubic prostatectomy on cancer control: the 30-year anniversary. J Urol. 2012;188:2219–24. doi: 10.1016/j.juro.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Fraser M, Berlin A, Bristow RG, van der Kwast T. Genomic, pathological, and clinical heterogeneity as drivers of personalized medicine in prostate cancer. Urol Oncol. 2015;33:85–94. doi: 10.1016/j.urolonc.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 13.Makarov DV, Loeb S, Getzenberg RH, Partin AW. Biomarkers for prostate cancer. Annu Rev Med. 2009;60:139–51. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- 14.Klein EA, Cooperberg MR, Magi-Galluzzi C, Simko JP, Falzarano SM, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66:550–60. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Cullen J, Rosner IL, Brand TC, Zhang N, Tsiatis AC, et al. A Biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur Urol. 2015;68:123–31. doi: 10.1016/j.eururo.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 16.Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Med Genomics. 2008;1:11. doi: 10.1186/1755-8794-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 19.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooperberg MR, Simko JP, Cowan JE, Reid JE, Djalilvand A, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol. 2013;31:1428–34. doi: 10.1200/JCO.2012.46.4396. [DOI] [PubMed] [Google Scholar]
- 21.Cuzick J, Berney DM, Fisher G, Mesher D, Moller H, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106:1095–9. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishoff JT, Freedland SJ, Gerber L, Tennstedt P, Reid J, et al. Prognostic utility of the cell cycle progression score generated from biopsy in men treated with prostatectomy. J Urol. 2014;192:409–14. doi: 10.1016/j.juro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Freedland SJ, Gerber L, Reid J, Welbourn W, Tikishvili E, et al. Prognostic utility of cell cycle progression score in men with prostate cancer after primary external beam radiation therapy. Int J Radiat Oncol Biol Phys. 2013;86:848–53. doi: 10.1016/j.ijrobp.2013.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooperberg MR, Davicioni E, Crisan A, Jenkins RB, Ghadessi M, et al. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol. 2015;67:326–33. doi: 10.1016/j.eururo.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross AE, Feng FY, Ghadessi M, Erho N, Crisan A, et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2014;17:64–9. doi: 10.1038/pcan.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–53. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross AE, Johnson MH, Yousefi K, Davicioni E, Netto GJ, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2015;69:157–65. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 29.Klein EA, Yousefi K, Haddad Z, Choeurng V, Buerki C, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. 2015;67:778–86. doi: 10.1016/j.eururo.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 30.Den RB, Feng FY, Showalter TN, Mishra MV, Trabulsi EJ, et al. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys. 2014;89:1038–46. doi: 10.1016/j.ijrobp.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]


