Summary
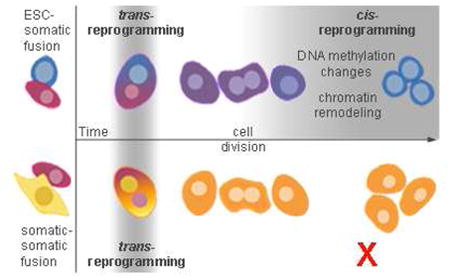
It is a long-held paradigm that cell fusion reprograms gene expression, but the extent of reprogramming and whether it is affected by the cell types employed remain unknown. We recently showed that the silencing of somatic genes is attributable to either trans-acting cellular environment or cis-acting chromatin context. Here, we examine how trans- versus cis-silenced genes in a somatic cell type behave in fusions to another somatic cell type or to embryonic stem cells (ESCs). We demonstrate that while reprogramming of trans-silenced somatic genes occurs in both types of fusions, reprogramming of cis-silenced somatic genes occurs only in somatic-ESC fusions. Importantly, ESCs reprogram somatic genome in two distinct phases: trans-reprogramming occurs rapidly independent of DNA replication, whereas cis-reprogramming occurs with slow kinetics requiring DNA replication. We also show that pluripotency genes Oct4 and Nanog are cis-silenced in somatic cells. We conclude that cis-reprogramming capacity is a fundamental feature distinguishing ESCs from somatic cells.
Graphical abstract
Introduction
Classical studies have demonstrated that genes differentially expressed between two somatic cell types tend to acquire comparable expression following cell fusion (Blau et al., 1983; Blau et al., 1985; Chiu and Blau, 1984), leading to the view that somatic cells possess a plastic transcriptome that is readily reprogrammed by trans-acting factors (Blau, 1989; Blau and Baltimore, 1991). Indeed, a recent review argued that fusions of disparate somatic cell types can reverse differentiation (Yamanaka and Blau, 2010).
In a departure from the above view, we reported recently that many genes actually fail to respond to trans-acting factors in somatic-somatic fusions, remaining silent in one fusion partner while exhibiting robust expression in the other (Lee et al., 2009a). As active copies of these genes indicate that the fused cellular milieu is supportive of expression in trans (i.e., transcriptional activators are present), it can be deduced that silent copies of these genes, in the same milieu, must be silenced by cis-acting mechanisms (Lahn, 2010; Lee et al., 2009a). We therefore concluded that two categories of silent genes exist in somatic cells. One is silenced in trans, by a cellular environment that does not support gene expression due to the lack of transcriptional activators or the presence of repressors. These genes remain competent to express if the balance of factors changes to be supportive. The other category is genes silenced in cis, by chromatin-based mechanisms that render genes silent irrespective of whether transcriptional activators are present in the milieu.
We proposed that cis-silencing of lineage-inappropriate genes serves to safeguard cellular identity, and that at a fundamental level, a given cell type is defined by the set of genes that undergo cis-silencing (Lahn, 2010). Demonstrating this empirically, we showed that when BACs corresponding to cis-silenced endogenous genes of somatic cells were transfected into these cells, the BAC transgenes were ectopically expressed in most cases while their endogenous orthologs remained silent (Gaetz et al., 2011). This indicates that the trans environment of a somatic cell is actually supportive of the expression of most cis-silenced genes, and only barrier to their expression is cis-acting chromatin mechanisms. We concluded that cis-silencing is required for maintaining cellular identity, and that its loss would lead to aberrant expression of lineage-inappropriate genes, including master regulators of alternative cell fates (Gaetz et al., 2011).
Previous studies suggested that fusion of somatic cells with ESCs can lead fused cells to adopt ESC-like features (Ambrosi et al., 2007; Bhutani et al., 2010; Cowan et al., 2005; Do et al., 2006; Do and Scholer, 2004; Han et al., 2008; Tada et al., 1997; Tada et al., 2001). However, these studies did not systematically compare reprogramming abilities between ESCs and somatic cells. Additionally, there is no genome-level data on which genes become reprogrammed in either somatic-ESC or somatic-somatic fusion, or how and when the reprogramming occurs. One somatic-ESC fusion study did examine global gene expression patterns, but because the fusion was carried out in cells of the same species, the expression of key genes, including those related to pluripotency, could not be traced to the genome of origin (Ambrosi et al., 2007). Thus, it is not known whether reprogramming in somatic-ESC fusion is fundamentally different from that in somatic-somatic fusion. Nor is there a clear understanding on the extent of reprogramming occurring in either type of fusion.
Here, we examine global gene expression changes in both somatic-somatic and somatic-ESC fusions using whole-transcriptome shotgun sequencing (RNA-Seq). We conclusively demonstrate that while the reprogramming of trans-silenced somatic genes occurs in both types of fusions, the reprogramming of cis-silenced somatic genes is a property specific to somatic-ESC fusions and absent in somatic-somatic fusions. We also present genome-wide time-course data on the kinetics of gene expression changes in somatic-ESC fusions, showing that ESCs reprogram the somatic genome in two distinct phases. The first phase is trans-reprogramming, which involves rapid activation of trans-silenced somatic genes independent of cell division. The second phase is cis-reprogramming, which involves slow activation of cis-silenced somatic genes and requires cell division. Contrary to published reports (Bhutani et al., 2010; Han et al., 2008; Tada et al., 2001), we find that the post-fusion activation of pluripotency genes Oct4 and Nanog in the somatic genome exhibits slow kinetics and requires cell division, indicating that these genes are cis-silencing in somatic cells. Finally, we address the role of Aid in cell fusions, as this gene was recently reported to be required for activation of somatic copies of Oct4 and Nanog after somatic-ESC fusion (Bhutani et al., 2010). To our surprise, Aid did not contribute to the reprogramming of somatic gene expression. Combined, our data demonstrate that in somatic-ESC fusion, ESCs reprogram the somatic genome in two phases that are kinetically and mechanistically distinct, and that the ability to reprogram cis-silenced somatic genes is a fundamental feature distinguishing ESCs from somatic cells.
Results
Terminally differentiated somatic cells cannot induce cis-reprogramming
For ease of description, we refer to trans-silenced genes as being “activatable” and cis-silenced genes as being “occluded” (Lahn, 2010; Lee et al., 2009a). We began our study by fusing two terminally differentiated somatic cells, R1A rat fibroblasts and 129TF mouse fibroblasts, and used global gene expression profiles generated through RNA-Seq to define occluded genes in R1A. To ensure multiple rounds of cell division post-fusion and to give the hybrid genome extended response time following fusion, we generated a clonal line of fused cells, called R1Ax129TF(clone), which was used for subsequent analyses.
RNA-Seq reads were mapped to a library of 16,344 genes, each of which was represented by a pair of orthologous mouse-rat open reading frames (ORFs), using sequence divergence to trace expression to either the mouse or rat genome. Quantification of expression, measured in terms of the number of transcripts per cell, was estimated for each gene. We define a gene as occluded if it is transcriptionally silent (<0.2 transcripts per cell) in one fusion partner pre- and post-fusion, while being expressed (≥2 transcripts per cell) by the other fusion partner pre- and post-fusion. Using these criteria to analyze R1Ax129TF(clone), we identified 190 occluded R1A genes (Table S1).
Because a gene could appear occluded due to chromosome loss after fusion, we used a “bioinformatic chromosome analysis” to examine total gene expression originating from each chromosome in R1Ax129TF(clone) in comparison to unfused R1A and 129TF cells. We saw no evidence of chromosome loss, as each chromosome was properly represented in the fused transcriptome (Fig. S1A).
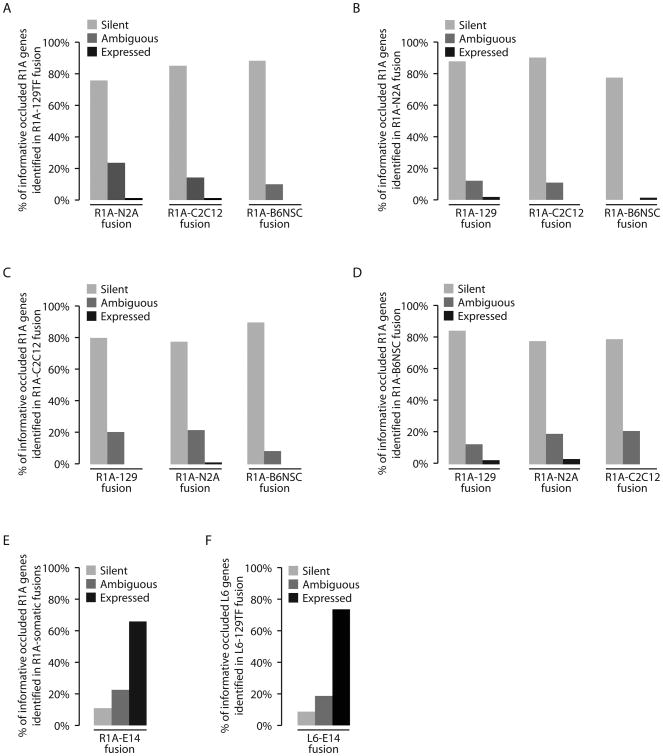
We next examined whether R1A occluded genes would become active when R1A was fused to other somatic cells: N2A neuroblastoma cells and C2C12 myoblasts. We performed RNA-Seq on clonal lines from each fusion, i.e., R1AxN2A(clone) and R1AxC2C12(clone), and on non-fused parental cells (Table S1). Again, bioinformatic analysis revealed no chromosome loss (Fig. S1B&C). Of the 190 R1A occluded genes identified in the R1A-129TF fusion, 89 were expressed from the N2A genome pre- and post-fusion. The active status of these genes in N2A provides an opportunity to assess whether the R1A copies can be activated in R1A-N2A fusions, and we therefore defined these as “informative occluded genes”. In R1AxN2A(clone), we found that only 1 of the 89 informative occluded R1A genes was activated (Fig. 1A). We performed the same analysis on R1AxC2C12(clone), again finding that only 1 occluded gene in the R1A genome became expressed in fused cells (Fig. 1A). Thus, R1A genes found to be occluded in fusion with one differentiated cell type generally remain occluded in fusions with other differentiated cells types, arguing that differentiated somatic cells are less plastic than previously assumed. Importantly, the same fusions also revealed the presence of activatable genes (Table S1), which are silenced by trans mechanisms and are competent to turn on in fused cells.
Figure 1.
Global cis-reprogramming of occluded R1A genes in somatic-ESC fusions. Very few informative occluded R1A genes identified in one fusion became expressed in another somatic-somatic fusions (A-D). The majority of informative occluded R1A genes became expressed in R1A-E14 fusion (E), and the majority of informative occluded L6 genes became expressed in L6-E14 fusion (F).
Somatic stem cells cannot induce cis-reprogramming
To test whether somatic stem cells, with their greater plasticity, possess a capacity to induce cis-reprogramming, we fused R1A with B6NSC mouse neural stem cells, and obtained a clone, R1AxB6NSC(clone), which showed no evidence of chromosome loss (Fig. S1D). We validated the stem cell nature of B6NSC by demonstrating self-renewal and confirming its potential to differentiate into all three neural lineages (Fig. S2). Of the informative occluded R1A genes (based on fusion with 129TF), none became expressed from the R1A genome in R1AxB6NSC(clone) (Fig. 1A).
We reanalyzed our data, beginning with R1A genes occluded in the fusion with N2A (or C2C12 or B6NSC) instead of 129TF, and examined their behavior in the other fusions. We saw at most a small handful of genes becoming expressed in fused cells (Fig. 1B-D), leading us to conclude that somatic cells, both differentiated and multipotent, categorically lack the ability to erase occlusion on a genome-wide scale. We cannot rule out the possibility that specific loci undergo erasure of occlusion, which could explain the few occluded genes from one fusion that turn on in other fusions. However, given the rarity of such events, they may simply be the result of experimental noise.
ESCs have the capacity to induce genome-wide cis-reprogramming
To investigate whether ESCs have the ability to erase occlusion of somatic genes, we fused R1A with E14 mouse ESCs. A clone, designated R1AxE14(clone), bearing ESC-like morphology and showing no evidence of chromosome loss (Fig. S1E), was subjected to RNA-Seq. For analysis, we pooled data from all somatic-somatic fusions (i.e., R1A fused to 129TF, N2A, C2C12 and B6NSC), identifying a total of 557 occluded R1A genes. Among them, 219 genes were expressed in ESCs and were therefore defined as informative occluded genes in this context. Of these, 146 (67%) became expressed from the R1A genome in fused cells (Fig. 1E; Table S2). The fact that occluded R1A genes are massively activated in R1AxE14(clone) indicates the erasure of their occluded status, and stands in contrast to our observations in somatic cell fusions.
To ensure that this result was representative of somatic-ESC fusions, we fused another somatic cell type, L6 rat myoblasts, to E14, derived a clonal line of fused cells, L6xE14(clone), and performed RNA-Seq. Again, there was no evidence of chromosome loss (Fig. S1F). We also fused L6 with 129TF and performed RNA-Seq to identify occluded L6 genes. We found 59 informative occluded L6 genes, and 43 (73%) of these were expressed from the L6 genome in L6xE14(clone) (Fig. 1F; Table S3). Thus, the genomes of two somatic cell types, fibroblasts (R1A) and myoblasts (L6), showed extensive activation of occluded genes when fused to ESCs. We therefore conclude that ESCs have the capacity to induce global cis-reprogramming of the somatic genome after cell fusion.
Cis-reprogramming has slower kinetics than trans-reprogramming
Occluded genes are stably maintained in the silent state by cis-acting mechanisms, whereas activatable genes are silenced by trans-acting mechanisms (Lahn, 2010; Lee et al., 2009a). Accordingly, activation of occluded genes, or cis-reprogramming, entails the erasure of the chromatin mechanisms responsible for occlusion, whereas expression of activatable genes through trans-reprogramming likely involves only the introduction of transcriptional activators. Based on this, we hypothesized that these two modes of reprogramming would have different kinetics.
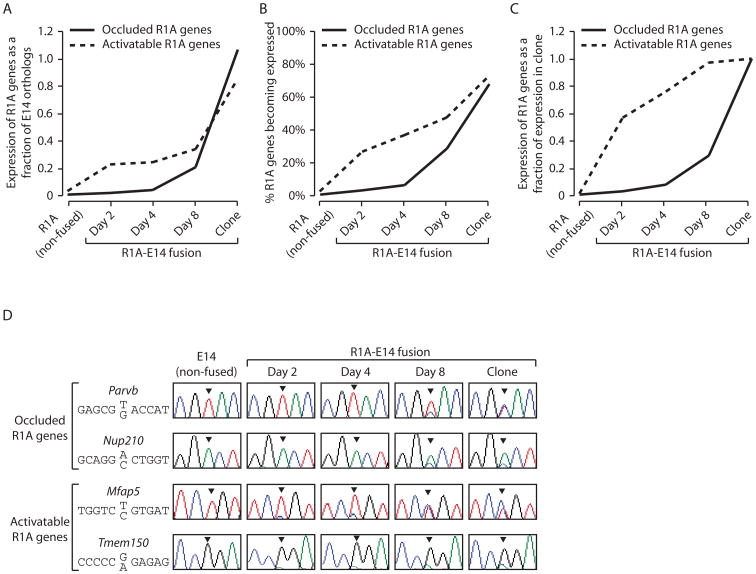
To examine the kinetics of cis-reprogramming, a time-course analysis of R1A-E14 fusions was conducted. Populations of fused cells were harvested at days 2, 4 and 8 post fusion followed by RNA-Seq. Starting with the list of occluded R1A genes identified in all somatic-somatic fusions, we measured the extent of cis-reprogramming at each time point. First, we pooled all R1A occluded genes and summed the total number of rat transcripts in fused cells. We scaled this by the total number of E14 transcripts in fused cells, allowing us to analyze rat versus mouse contribution to total gene expression at each time point (Fig. 2A; Table S2). Next, we looked at the fraction of occluded R1A genes that became expressed at each time point (Fig. 2B). Finally, we normalized expression of each reprogrammed R1A occluded gene to its expression level in R1AxE14(clone), and graphed these results over time (Fig. 2C). All three methods showed that cis-reprogramming occurred slowly. Specifically, expression of the occluded rat orthologs in fused cells was barely detectable at day 2 post fusion, became noticeable at day 4, was prominent at day 8, and only reached levels comparable to those in the mouse genome in R1AxE14(clone).
Figure 2.
Biphasic reprogramming of the somatic genome in somatic-ESC fusions. R1A-E14 fusion samples include days 2, 4 and 8 post-fusion, and R1AxE14(clone). Methods used to measure the kinetics of occluded or activatable gene expression in fusion samples include (A) pooling the R1A occluded or activatable genes, summing the total transcripts from rat orthologs, and scaling this to the total number of transcripts from mouse orthologs in fused cells, (B) calculating the percentage of informative occluded or activatable R1A genes that became expressed at each time point, and (C) analyzing the average rat ortholog expression level of each informative occluded or activatable gene scaled to its maximal rat expression level in the R1AxE14(clone). (D) RT-PCR-Seq confirms the kinetics of trans-reprogramming and cis-reprogramming. The nucleotide site that differs between R1A and E14 orthologs is indicated, with the bottom and top alleles corresponding to R1A and E14, respectively. Arrow heads indicate peaks of these sites in the chromatograms.
We next used the time-course fusion samples to investigate the kinetics by which activatable R1A genes undergo trans-reprogramming. By definition, activatable genes, also identified in the somatic-somatic fusions, are silent pre-fusion and become expressed post-fusion (Tables S1). Employing the same approaches used for occluded gene analysis, we measured the kinetics of trans-reprogramming (Fig. 2A-C; Table S4). Results showed that the rat orthologs of activatable genes began to express much more rapidly, with prominent levels of expression detected within 2 days. These data thus demonstrate that trans-reprogramming is considerably faster than cis-reprogramming.
To validate RNA-Seq data from R1A-E14 fusion samples, we carried out “RT-PCR-Seq” on occluded and activatable genes. This involved RT-PCR on fusion samples with species-common primers that flank areas of divergence between the species. Sequencing of RT-PCR products was used to determine if fused cell expression came from the mouse or rat genome, or both. This assay confirmed the RNA-Seq data (Fig. 2D).
Oct4 and Nanog are occluded in somatic cells
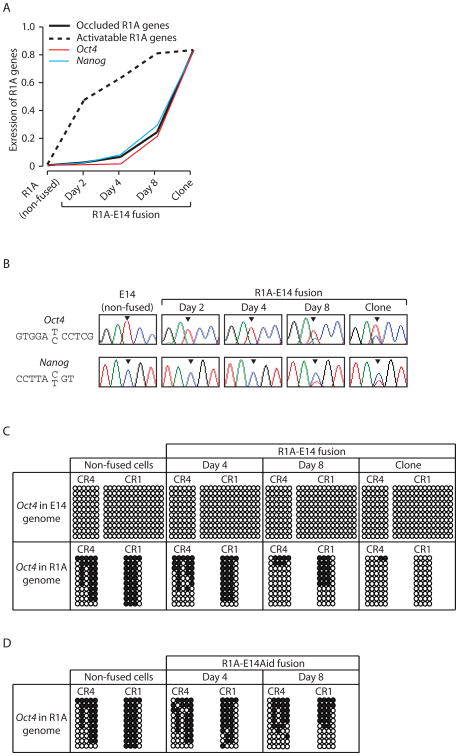
Oct4 and Nanog are expressed in ESCs but not somatic cells. Because of this, we could not determine whether R1A copies of these genes were occluded or activatable using somatic-somatic fusions. However, since somatic-ESC fusions can clearly delineate occluded and activatable genes based on their reprogramming kinetics, we sought to use these data to define the occlusion status of Oct4 and Nanog in R1A. For both genes, R1A orthologs activated slowly after fusions to ESCs, with rat transcripts representing just 0.98% (Oct4) and 2.15% (Nanog) of total gene expression in fused cells on day 2 post-fusion (mouse orthologs accounting for the rest). By day 8 post-fusion, rat Oct4 and Nanog transcripts comprised 23.69% and 20.54% of total expression in fused cells, respectively. In addition, the activation pattern of these genes, as normalized to maximal rat transcription levels in R1AxE14(clone), matched that of other occluded genes, but not activatable genes (Fig. 3A). RT-PCR-Seq on Oct4 and Nanog confirmed the RNA-Seq data (Fig. 3B). We thus conclude that Oct4 and Nanog are occluded in R1A.
Figure 3.
The behavior of Oct4 and Nanog in somatic-ESC fusion is consistent with occlusion in R1A. (A) R1A copies of Oct4 and Nanog are activated slowly after fusion. (B) Expression of R1A transcripts for Oct4 and Nanog can be detected only at day 8 (not days 2 or 4) post fusion by RT-PCR-Seq. The nucleotide site that differs between orthologs is indicated, with the bottom and top alleles corresponding to R1A and E14, respectively. Arrow heads indicate peaks of these sites in the chromatograms. (C) In R1A-E14 fusion, the R1A Oct4 promoter (CR1) and distal enhancer (CR4) in fused cells are gradually demethylated. Open and solid circles indicate unmethylated and methylated CpG sites, respectively. (D) In R1A-E14Aid fusion, there is no evidence of enhanced demethylation of the R1A Oct4 promoter (CR1) or distal enhancer (CR4).
DNA demethylation is tightly associated with Oct4 activation (Mikkelsen et al., 2008), and our previous data show DNA methylation is associated with occlusion of some genes (Lee et al., 2009b). Given this, we performed bisulfite sequencing of the promoter (CR1) and distal enhancer (CR4) of Oct4 and examined how its methylation status changed in response to somatic-ESC fusions (Fig. 3C). Pre-fusion, these regions are hypermethylated in R1A and unmethylated in E14. In R1AxE14(clone), R1A Oct4 became unmethylated like the E14 copy. In the time-course samples of R1A-E14 fusion, the E14 copy remains unmethylated, while there is gradual demethylation of the R1A copy. This pattern mirrors the gradual activation of Oct4 seen in RNA-Seq and RT-PCR-Seq. Specifically, demethylation is barely detectable at day 4 but is robust by day 8. The changes in DNA methylation are more dramatic within CR4. These data indicate that the gradual activation of R1A Oct4 post-fusion is tightly coupled to gradual demethylation of the Oct4 enhancer.
Transcriptome reprogramming is greater in somatic-ESC fusions
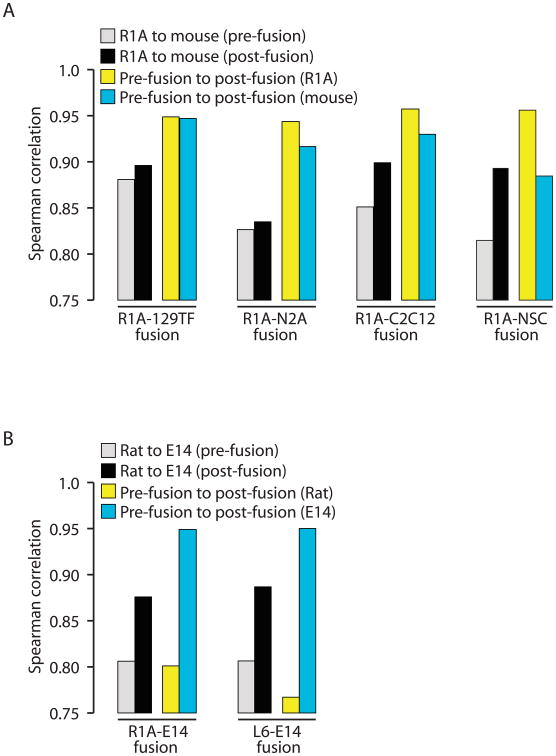
In somatic-somatic fusions, we observed that the pre- and post-fusion transcriptome profiles for a given cell are more closely correlated than their post-fusion profiles within fused cells. This is true despite the fact that the fused cell genomes share an identical trans milieu, and argues that the transcriptome profile of a somatic cell is more strongly influenced in cis by gene occlusion, than in trans (Fig. 4A). In contrast to somatic-somatic fusions, R1AxE14(clone) and L6xE14(clone) show very different patterns of correlation (Fig. 4B). For E14, the pre- and post-fusion transcriptomes are highly correlated. However, the transcriptome profiles of R1A or L6 are more similar to E14 post-fusion than to their own pre-fusion profiles. These data indicate that somatic-ESC fusions dramatically alter the somatic transcriptome to closely resemble the ESC transcriptome, highlighting the dominance of ESCs in these fusions. These analyses confirm that somatic-ESC fusions are distinct from somatic-somatic fusions due to the cis-reprogramming capability present in ESCs but not somatic cells.
Figure 4.
Cis regulation of global gene expression in somatic cells and the capacity for cis-reprogramming in ESCs. (A) In somatic-somatic fusions, the correlation of transcriptome profiles for either fusion partner, pre- and post-fusion (yellow and blue bars), is generally greater than the correlation between the two partners post-fusion (black bars), indicating the occluded portion of the genome resists reprogramming. (B) In somatic-ESC fusions (R1A-E14 and L6-E14), the post-fusion transcriptome profiles of the somatic genomes become much more closely correlated to E14 (black bar) than to their own pre-fusion profiles (yellow bar), indicating a dominance of ESC gene expression patterns. Transcriptome profiles of E14 pre- and post-fusion remain highly correlated (blue bar).
Cis-reprogramming, but not trans-reprogramming, requires DNA replication
Consistent with the literature, fused cells did not divide in the first 3-4 days following somatic-ESC fusion (Bhutani et al., 2010; Han et al., 2008). The timing of cell division appears to correlate with the slow kinetics of cis-reprogramming, suggesting that replication may be required for this process. To test this, we induced cell cycle arrest in fused R1A-E14 cells with mitomycin C (mito-C). Arrested samples were cultured under normal ESC conditions, harvested at days 4 and 8 post-fusion, and used for RNA-Seq. Remarkably, treatment with mito-C abrogated the massive activation of occluded genes seen in previous R1A-E14 fusions (Fig. 4A, Table S2). In contrast, activatable genes in R1A still underwent activation in mito-C treated samples, with kinetics similar to those in untreated samples (Fig. 4B; Table S4).
We also investigated the effect of cell cycle arrest on transcriptome correlations in somatic-ESC and somatic-somatic fusions. We found that mito-C had little effect on transcriptome correlations of treated R1A-129TF samples, as compared to untreated samples (the first panel of Fig. 5C is essentially indistinguishable from the first panel of Fig. 4A). In contrast, mito-C treatment had a profound effect on transcriptome correlations in R1A-E14 fusion samples. Here, the pre-fusion and day 8 post-fusion transcriptomes of R1A remain highly correlated, an observation that contrasts with the untreated R1A-E14 fusion (compare the second panel of Fig. 5C to the first panel of Fig. 4B). These data suggest that cis-reprogramming of the somatic genome, seen only in somatic-ESC fusions, is dependent on DNA replication, whereas trans-reprogramming, which occurs in both somatic-somatic and somatic-ESC fusions, is not.
Figure 5.
Cis-reprogramming is dependent on DNA replication. (A, B) Mito-C treatment of R1A-E14 fused cells prevents expression of occluded genes, but does not affect expression of activatable genes at days 4 and 8 post-fusion. (C) In R1A-129TF fusion, mito-C had little effect on global gene expression. Transcriptome correlations were unaltered as compared to untreated R1A-129TF fusion (Fig. 4A). Mito-C treatment had a strong effect on global gene expression in the R1A-E14 fusion, leading transcriptome correlations to resemble that of somatic-somatic fusions. This is distinct from untreated R1A-E14 fusion (Fig. 4B). (D) RT-PCR-Seq shows mito-C treatment of R1A-E14 fused cells prevents activation of R1A copies of Oct4, Nanog, and two representative occluded genes. Mito-C does not alter activation of representative R1A activatable genes. The nucleotide site that differs is indicated, with the bottom and top alleles corresponding to R1A and E14, respectively. Arrow heads indicate peaks of these sites in the chromatograms. (E, F) In both R1A-E14 and R1A-E14Aid fusion, mito-C treatment blocks demethylation of the R1A Oct4 promoter (CR1) and distal enhancer (CR4). Open and solid circles indicate unmethylated and methylated CpG sites, respectively.
We specifically examined the effect of mito-C treatment on Oct4 and Nanog using RT-PCR-Seq. We found that mito-C treatment of R1A-E14 fusion samples prevented activation of R1A copies of Oct4 and Nanog, even at day 8 post fusion (Fig. 5D). This contrasts with the untreated R1A-E14 fusion where R1A copies of Oct4 and Nanog show considerable expression at day 8 post-fusion (Fig. 3B). We also applied RT-PCR-Seq to the same occluded and activatable genes examined in Fig. 2D. As expected, mito-C treatment abolished activation of occluded R1A genes, but did not affect activatable R1A genes (Fig. 5D). Lastly, we examined the effect of mito-C treatment on the methylation status of Oct4. We found that the promoter (CR1) and distal enhancer (CR4) regions of Oct4 remained heavily methylated in mito-C treated R1A-E14 fusion samples (Fig. 5E), suggesting that mito-C blocks demethylation of the somatic copy of Oct4 in fused cells. These data further support the conclusion that cis-reprogramming, but not trans-reprogramming, requires DNA replication.
It should be noted that mito-C treatment may cause some differentiation in ESC-fused cells. However, the pluripotency genes Oct4 and Nanog remain expressed from the mouse ESC genome in our samples (Fig. 5D). Additionally, mouse copies of the Oct4 promoter and distal enhancer remain unmethylated, as in pluripotent cells, while the rat copies remain methylated (Fig. 5E). Combined, these results suggest that the failure of cis-reprogramming after treatment with mito-C is due to inhibition of DNA replication and is not a side effect of differentiation.
Behavior of chromatin modifying genes in somatic-ESC fusions
Cis-reprogramming of the somatic genome by fusion to ESCs likely involves significant remodeling of somatic chromatin, and one could reason that genes necessary for this are occluded in somatic cells. We searched for such genes in our ORF library, identifying 322 genes whose ontology descriptions include the term “chromatin modification.” Mis-annotated genes were removed, resulting in a final list of 237 chromatin modifiers. Expression of these genes was analyzed in the R1A-E14 fusion time-course data to identify genes with >10-fold increase in R1A expression between unfused and R1AxE14(clone). We identified 4 genes: Satb1, Chd7, Dnmt3b and Tet1 (Table S5). Although activation kinetics was not a factor in selecting these genes, they all displayed kinetics typical of cis-reprogramming, indicating that they are occluded in R1A. We also examined expression of these genes in the L6-E14 fusion (Table S6); despite the lack of time-course data, expression of L6 copies of these 4 genes increased dramatically in L6xE14(clone) relative to unfused L6, which is consistent with the R1A-E14 fusion.
Satb1 and Chd7 are involved in modulating chromatin structure while Dnmt3b and Tet1 are involved in regulating DNA methylation. As discussed in detail in Supplemental Text, our data and previous studies on these chromatin modifiers implicate them as players in chromatin remodeling and the resetting of DNA methylation during cis-reprogramming.
An additional chromatin modification potentially involved in cis-reprogramming is the “bivalent” domain, defined as a concurrence of the active histone mark H3K4m3 and the silent mark H3K27me3, at gene promoters (Bernstein et al., 2006). In ESCs, bivalent domains are prevalent and tend to be found in lowly expressed developmental genes, leading to speculation that they are involved in gene silencing during pluripotency and responsiveness to activating signals during differentiation. We examined how expressed, activatable, and occluded genes in R1A corresponded to the previously defined set of bivalent genes in ESCs. We found that occluded R1A genes were significantly enriched for bivalent genes as compared to activatable and expressed R1A genes (Fig. S3). This indicates that genes possessing bivalent domains in ESCs tend to be targeted for occlusion during ESC differentiation. As an extension of this idea, we suggest that reestablishment of bivalent domains on occluded genes could be an integral step in cis-reprogramming when somatic cells are experimentally dedifferentiated back toward pluripotency.
The capacity of ESCs to induce cis-reprogramming does not require Aid
Our data are at odds with a recent study showing that human copies of OCT4 and NANOG in fused cells are activated and demethylated rapidly in the absence of cell division after fusion with mouse ESCs (Bhutani et al., 2010). The study further showed that these processes require Aid, and therefore we used RNA-Seq data to examine expression of Aid in fused and unfused cells. To our surprise, we found Aid is silent in all examined cells, and this was further confirmed by RT-PCR, which failed to generate detectable product, except in activated B cells used as positive control (Muramatsu et al., 1999) (Fig. 6A). These observations indicate that Aid is unlikely to be involved in induction of global cis-reprogramming as seen in R1A-E14 and L6-E14 fusions.
Figure 6.
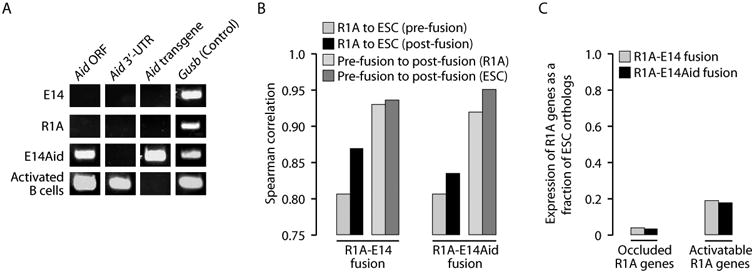
Cis-reprogramming is independent of the Aid gene. (A) RT-PCR fails to detect endogenous expression of Aid in E14 or R1A. E14Aid cells readily express Aid, as do activated B cells (positive control). Primers are specific to the Aid ORF (detect endogenous or transgene), the Aid 3′-UTR (endogenous only), or to a spacer sequence in the transgene vector (transgene only). (B) Transcriptome correlations at day 4 post-fusion are nearly identical for R1A-E14Aid fusions and R1A-E14 fusions. (C) Expression of the Aid transgene does not alter the kinetics of trans- or cis-reprogramming as the proportion of transcripts expressed from the R1A genome in R1A-E14Aid fusion at day 4 is the same as that in the R1A-E14 fusion, for either occluded or activatable genes.
Because Aid was not expressed in E14 but was expressed in the ESC line of Bhutani et al., we created a subclone of E14 expressing a mouse Aid transgene (referred to as E14Aid; Fig. 6A), and fused it with R1A. The transcriptome profile (including expression of Oct4 and Nanog) in the R1A-E14Aid fusion samples was comparable to that of R1A-E14 fusions (Fig. 6B). Additionally, Aid transgene expression did not alter the kinetics of cis-reprogramming or trans-reprogramming (Fig. 6C). At day 4 post-fusion, rat contribution to total transcript levels of occluded genes (including Oct4 and Nanog) was comparable in R1A-E14Aid and R1A-E14 samples, and the same was true for activatable genes (Fig. 6C). Finally, we examined promoter methylation of the R1A copy of Oct4 in R1A-E14Aid fusion, either with (Fig. 5F) or without mito-C treatment (Fig. 3D). There was no evidence that Aid expression promoted the demethylation of Oct4 in either case. Based on these data, it appears that within our experimental system, Aid does not play a significant role in the cis-reprogramming of occluded genes.
Discussion
Fundamental difference in reprogramming abilities between ESCs and somatic cells
Although previous studies have shown that cell fusion can reprogram gene expression, they are typically based on the assessment of a limited number of genes at a single time point post-fusion (Ambrosi et al., 2007; Bhutani et al., 2010; Blau et al., 1983; Blau et al., 1985; Chiu and Blau, 1984; Cowan et al., 2005; Do et al., 2006; Do and Scholer, 2004; Han et al., 2008; Tada et al., 1997; Tada et al., 2001). Furthermore, there has been a lack of systematic comparisons between the behavior of somatic-ESC fusions and that of somatic-somatic fusions. In contrast, our study is based on whole-transcriptome data, and it includes detailed comparisons of somatic-ESC fusions and somatic-somatic fusions. Additionally, we performed time-course analyses on the kinetics of gene expression changes post-fusion, and how it is affected by cell cycle arrest. These comprehensive approaches allowed us to uncover several insights. First, our results highlight the profound difference in reprogramming abilities between ESCs and somatic cells. Whereas the ability for trans-reprogramming is present in both ESCs and somatic cells, ESCs possess the capacity for genome-wide cis-reprogramming that is absent in somatic cells, including somatic stem cells. Indeed, this difference represents one of the most fundamental functional distinctions between ESCs and somatic cells identified thus far. It can be extrapolated that similar cis-reprogramming capacity likely exists in certain other pluripotent cells such as embryonic germ cells (EGCs) and oocytes. Second, our data show that reprogramming to pluripotency occurs in two phases: rapidly in trans, and slowly in cis. Third, we found that cis-reprogramming, but not trans-reprogramming, is dependent on cell division. Fourth, we noted that reprogramming is independent of the Aid gene, contrary to the previous report (Bhutani et al., 2010). Lastly, we demonstrated that the pluripotency genes Oct4 and Nanog undergo cis-silencing in somatic cells. Together, these insights provide an informative framework for understanding the inner workings of both differentiation and dedifferentiation in mammalian systems.
While the mechanism of cis-reprogramming remains to be elucidated, it necessarily entails the erasure of chromatin modifications that impart the occluded state to genes. This process, which can perhaps be called “deocclusion”, likely involves an active mechanism coupled to cell division. Candidate genes indentified in our study include Satb1, Chd7, Dnmt3b and Tet1, all of which appear to be occluded in somatic cells. Also implicated in the mechanism of deocclusion are the factors used to induce reprogramming of somatic cells toward iPSCs (Takahashi and Yamanaka, 2006).
Discrepancies with previous studies
Our data conflict with the report by Bhutani et al. on the rapid kinetics of somatic OCT4 and NANOG activation and the requirement of Aid in this process. A possible source for the discrepancies might lie in the fact that their study utilized the highly sensitive RT-PCR assay (including nested PCR in some cases) on both cell populations and single fused cells to examine somatic OCT4 and NANOG expression. Furthermore, they did not directly compare expression levels of somatic OCT4 and NANOG in fused cells to physiological levels in ESCs. As noted previously (Hanna et al., 2010), Bhutani et al. could have misinterpreted very low OCT4 and NANOG expression detected by RT-PCR as true reprogramming. Indeed, our RNA-Seq data showed that expression of R1A copies of Oct4 and Nanog in fused cells are only 1% and 5%, respectively, of the ESC expression levels 4 days post-fusion. Another study reported activation of a somatic Oct4-GFP transgene just two days after somatic-ESC fusion (Han et al., 2008; Tada et al., 2001), but published data have since shown that this transgene is not regulated the same way as endogenous Oct4 (Han et al., 2010). Although our observation of slow cis-reprogramming in somatic-ESC fusions conflicts with these previous studies, it is consistent with data showing that expression of OCT4 and NANOG in human B cells is at least 100-fold lower than those in human ESCs several days after fusion to mouse ESCs (Pereira et al., 2008). The slow kinetics of occluded gene expression is also consistent with our own finding that cell division is required for cis-reprogramming and with the literature on induced pluripotency. The kinetics of iPSC induction are also quite slow and there is increasing evidence for the requirement of cell division in this process (Hanna et al., 2010; Stadtfeld and Hochedlinger, 2010).
Transdifferentiation versus dedifferentiation
Our study reveals that somatic cells lack the capacity to induce cis-reprogramming of other somatic genomes upon cell fusion. This finding may seem incompatible with reports that forced expression of certain transcription factors can drive one somatic cell type, such as fibroblasts, to transdifferentiate into cells similar to another somatic cell type, such myotubes (Braun et al., 1989; Davis et al., 1987), hepatocytes (Huang et al., 2011; Sekiya and Suzuki, 2011), cardiomyocytes (Efe et al., 2011; Ieda et al., 2010), macrophages (Feng et al., 2008), and neurons (Vierbuchen et al., 2010). Indeed, one prominent view maintains that the differentiated state is dynamically and continuously regulated by a balance of transcription factors that can be tipped to result in transdifferentiation (Blau, 1989; Blau and Baltimore, 1991; Graf and Enver, 2009; Slack, 2007). We offer two explanations for the contradiction between our findings on the prevalence of occluded genes and the reported capability of somatic cells to transdifferentiate. First, claims of transdifferentiation by expression of transcription factors often examine a subset of tissue-specific markers. It is possible that despite activation of downstream activatable genes, many occluded genes remain silent. Thus, claims of transdifferentiation may actually represent a partial (and incomplete) acquisition, by the first cell type, of gene expression patterns characteristic of the second cell type. This situation, perhaps better termed “ectopic differentiation” (Lahn, 2010), is supported by our recent study which showed that although exogenous expression of Myf5 can convert fibroblasts into muscle-like cells, similar to previous report (Braun et al., 1989), the muscle-like cells do not express endogenous Myf5 (which is occluded) or other occluded genes (Gaetz et al., 2011). Thus, while these cells resemble muscle in terms of gene expression and cellular physiology, they are different from naturally derived muscle which clearly expresses endogenous Myf5. An alternative explanation for transdifferentiation is that overexpression of transcription factors beyond their physiological levels may indeed activate some occluded genes, perhaps even erasing occlusion. Given that strong promoters are often used to drive ectopic expression of transcription factor transgenes to supraphysiological levels, we feel that this artificial scenario is not directly comparable to our transcriptome-wide studies of near-physiological gene expression levels.
Our findings resolve the apparent paradox between the stability of the differentiated state under physiological conditions and the relative ease with which somatic cells can be dedifferentiated toward pluripotency by certain experimental manipulations such as SCNT into oocytes or forced expression of the iPSC factors. Specifically, we argue that the stability of the differentiated state arises from the robustness of occlusion and the lack of cis-reprogramming capability in somatic cells. We further suggest that despite this stability, somatic cells can be readily reprogrammed to pluripotency by experimental manipulations that either tap into the existing cis-reprogramming capacity of pluripotent cells (i.e., SCNT into oocytes or fusion with ESCs), or recapitulate key aspects of the cis-reprogramming machinery (i.e., the use of defined factors to create iPSCs).
Our data highlight the importance of examining occluded genes in the context of dedifferentiation and transdifferentiation, as these genes not only contribute to the phenotypic stability of somatic cells but might also play a role in “epigenetic memory” of reprogrammed cells. We suggest that the mapping of occluded genes in specific cell types and the analysis of how occlusion changes during normal differentiation or experimentally induced reprogramming will enhance our understanding of how cell fate is established, maintained, and occasionally reversed, be it in experimental settings or in disease states such as cancer.
Materials and Methods
Cell culture and fusion
Rat cells were transduced with hEF1a-dTomato lentivirus containing hygromycin resistance; mouse cells were transduced with hEF1a-EGFP containing puromycin resistance. Clonal lines were isolated in each case. Cell lines and DNA vectors used in the study are available through Cyagen Biosciences. After fusion and FACS sorting of fused cells, selection media containing puromycin (0.25 μg/ml) and hygromycin (50 μg/ml) was added to eliminate any unfused cells that might remain. For R1AxE14(clone) and L6xE14(clone), individual dual fluorescent colonies were picked manually 10-15 days post-fusion. Colonies that maintained drug resistance, dual fluorescence and continued to proliferate were expanded. R1Ax129TF(clone), R1AxC2C12(clone), R1AxN2A(clone), and R1AxB6NSC(clone) were obtained by FACS sorting of single fused cells. For cell cycle arrest, cells were treated for 2 hours with 3 μg/ml mitomycin C one day post-fusion (R1A-E14) or 2 hours post-fusion (R1A-129TF).
RNA-Seq
RNA-Seq was performed on an Illumina GA II sequencer following vendor's instructions. To align RNA-Seq reads, we generated an orthologous mouse-rat ORF library and Maq software with stringent mapping settings (-n 3 −m 1) as previously described (Li et al., 2008). Efficacy of our criteria was established by counting the number of reads mapping to the inappropriate species when aligning read sequences from a representative sample of unfused mouse or rat cells. Any genes with an error rate >2% in read mapping were removed. To quantify the number of transcripts per cell for each gene, we assumed that cells contain ∼10 pg RNA, 5% of which is poly-adenylated. For each gene, the number of transcripts per cell is proportional to the number of mapped reads and inversely proportional to the length of the ORF:
To obtain the most accurate estimate of pre-fusion transcripts, we mapped mouse reads to an ORF library containing only the mouse member of each ORF pair. An identical procedure was used for reads from each rat fusion partner. Occluded R1A genes were defined as those silent (<0.2 transcripts per cell) in unfused and fused R1 A cells, whose mouse cell counterparts were expressed (≥2 transcripts per cell) in unfused and fused samples. Activatable R1A genes were defined as changing from silent in unfused R1A cells to expressed in R1A fused to mouse partner cells. In the analysis of occluded genes for Figure 2, we began with the total number of genes occluded in all somatic-somatic fusions, but not activated in any somatic-somatic fusion (557 genes). Likewise, we used the total number of activatable genes that were never occluded in a somatic-somatic fusion (64 genes).
RT-PCR and RT-PCR-Seq
Expression of Aid in the E14Aid cells and our other cell lines was verified by RT-PCR using primers spanning the ORF (which amplify both endogenous and transgene Aid), from the ORF to the 3′UTR (endogenous only), or from the ORF into a transgene specific spacer sequence (transgene only). To assess the relative expression of mouse versus rat copies of genes in fused cells using RT-PCR-Seq, mouse-rat common primers were chosen such that all amplicons spanned at least one intron and contained at least 2 nucleotide sites that are divergent between the two species. After RT-PCR, the product was sequenced, and the sequence composition at the divergent sites was used to assess the relative expression of mouse versus rat copies of genes.
DNA methylation analysis
DNA methylation analysis was performed by bisulfite sequencing as described (Vallender and Lahn, 2006). The genomic regions of Oct4 targeted for bisulfite sequencing, CR1 and CR4, were chosen based on cross-species conservation and correlation between methylation and expression as previously defined (He et al., 2009; Imamura et al., 2006).
Additional methods
Additional details of methods are given in Supplemental Methods.
Supplementary Material
Highlights.
- Reprogramming of the somatic genome in somatic-ESC fusions is a biphasic process.
- The ability to erase cis-silencing is a unique property of ESCs.
- Oct4 and Nanog are cis-silenced in somatic cells.
- Cis-reprogramming by ESCs requires DNA replication, but not Aid expression.
Acknowledgments
For their technical assistance and/or critical discussions we thank: Liyuan (James) Cao, Ryan Duggan, David Leclerc, Katelyn Michelini, Michael Olson, Patrick Reed, Roger Sciammas, and Ann Sperling. This work was funded in part by NIH postdoctoral fellowships F32HD061205 (to KMF) and F32HL922792 (to JG), the National Natural Science Foundation of China grant 30971675,30928015 and the Key Scientific and Technological Projects of Guangdong Province grant 2007A032100003 (to APX), and the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust (to BTL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambrosi DJ, Tanasijevic B, Kaur A, Obergfell C, O'Neill RJ, Krueger W, Rasmussen TP. Genome-wide reprogramming in hybrids of somatic cells and embryonic stem cells. Stem Cells. 2007;25:1104–1113. doi: 10.1634/stemcells.2006-0532. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM. How fixed is the differentiated state? Lessons from heterokaryons. Trends Genet. 1989;5:268–272. doi: 10.1016/0168-9525(89)90100-5. [DOI] [PubMed] [Google Scholar]
- Blau HM, Baltimore D. Differentiation requires continuous regulation. The Journal of cell biology. 1991;112:781–783. doi: 10.1083/jcb.112.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, Webster C. Plasticity of the differentiated state. Science (New York, NY. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. Embo J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CP, Blau HM. Reprogramming cell differentiation in the absence of DNA synthesis. Cell. 1984;37:879–887. doi: 10.1016/0092-8674(84)90423-9. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science (New York, NY. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Do JT, Han DW, Scholer HR. Reprogramming somatic gene activity by fusion with pluripotent cells. Stem Cell Rev. 2006;2:257–264. doi: 10.1007/BF02698052. [DOI] [PubMed] [Google Scholar]
- Do JT, Scholer HR. Nuclei of embryonic stem cells reprogram somatic cells. Stem Cells. 2004;22:941–949. doi: 10.1634/stemcells.22-6-941. [DOI] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J, Clift KL, Fernandes CJ, Mao FF, Lee JH, Zhang L, Baker SW, Looney TJ, Foshay KM, Yu WH, et al. Evidence for a critical role of gene occlusion in cell fate restriction. Cell research. 2011 doi: 10.1038/cr.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Han DW, Do JT, Gentile L, Stehling M, Lee HT, Scholer HR. Pluripotential reprogramming of the somatic genome in hybrid cells occurs with the first cell cycle. Stem Cells. 2008;26:445–454. doi: 10.1634/stemcells.2007-0553. [DOI] [PubMed] [Google Scholar]
- Han DW, Tapia N, Joo JY, Greber B, Arauzo-Bravo MJ, Bernemann C, Ko K, Wu G, Stehling M, Do JT, Scholer HR. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, McHaney M, Hong J, Weiss ML. Cloning and Characterization of 3.1kb Promoter Region of the Oct4 Gene from the Fischer 344 Rat. Open Stem Cell J. 2009;1:30–39. doi: 10.2174/1876893800901010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT. The “occlusis” model of cell fate restriction. Bioessays. 2010;33:13–20. doi: 10.1002/bies.201000090. [DOI] [PubMed] [Google Scholar]
- Lee JH, Bugarija B, Millan EJ, Walton NM, Gaetz J, Fernandes CJ, Yu WH, Mekel-Bobrov N, Vallender TW, Snyder GE, et al. Systematic identification of cis-silenced genes by trans complementation. Human molecular genetics. 2009a;18:835–846. doi: 10.1093/hmg/ddn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Gaetz J, Bugarija B, Fernandes CJ, Snyder GE, Bush EC, Lahn BT. Chromatin analysis of occluded genes. Human molecular genetics. 2009b;18:2567–2574. doi: 10.1093/hmg/ddp188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome research. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. The Journal of biological chemistry. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Terranova R, Ryan NK, Santos J, Morris KJ, Cui W, Merkenschlager M, Fisher AG. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS genetics. 2008;4:e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Slack JM. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat Rev Mol Cell Biol. 2007;8:369–378. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes & development. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. Embo J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Vallender TW, Lahn BT. Localized methylation in the key regulator gene endothelin-1 is associated with cell type-specific transcriptional silencing. FEBS Lett. 2006;580:4560–4566. doi: 10.1016/j.febslet.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.