Abstract
Tetraspanins are a superfamily of small transmembrane proteins that are expressed in almost all eukaryotic cells. Through interacting with one another and with other membrane and intracellular proteins, tetraspanins regulate a wide range of proteins such as integrins, cell surface receptors, and signaling molecules, and thereby engage in diverse cellular processes ranging from cell adhesion and migration to proliferation and differentiation. In particular, tetraspanins modulate the function of proteins involved in all determining factors of cell migration including cell–cell adhesion, cell–ECM adhesion, cytoskeletal protrusion/contraction, and proteolytic ECM remodeling. We herein provide a brief overview of collective in vitro and in vivo studies of tetraspanins to illustrate their regulatory functions in the migration and trafficking of cancer cells, vascular endothelial cells, skin cells (keratinocytes and fibroblasts), and leukocytes. We also discuss the involvement of tetraspanins in various pathologic and remedial processes that rely on cell migration and their potential value as targets for therapeutic intervention.
Keywords: migration, small transmembrane proteins, therapeutics, tetraspanin
Abbreviations
- ECM
cell–extracellular matrix
- TEM
tetraspanin-enriched microdomain
- MMP
matrix metalloproteinase
- JNK
c-Jun N-terminal kinase
- RhoA
Ras homolog gene family, member A
- PI4K
phosphatidylinositol 4-kinase
- FAK
focal adhesion kinase
- lck
lymphocyte-specific protein tyrosine kinase
- p38
p38 mitogen-activated protein kinase
- HCC
hepatocellular carcinoma
- cdc42
cell division control protein 42
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- Sprouty2
Sprouty homolog 2
- ROCK
Rho-associated protein kinase
- PI3K
phosphatidylinositol-4, 5-bisphosphate 3-kinase
- ICAM-1
intercellular adhesion molecule 1
- VCAM-1
vascular cell adhesion molecule 1
- ERK
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinase
- eNOS
endothelial nitric oxide synthase
- HUVECs
human umbilical vein endothelial cells.
Introduction
Tetraspanins, also called the transmembrane 4 superfamily, are a family of small transmembrane proteins expressed in all multicellular eukaryotes. Thirty-four distinct tetraspanin family members have been found in mammals, of which 33 exist in humans. Tetraspanin proteins are structurally characterized by 4 transmembrane domains, 2 extracellular loops, and short intracellular N- and C-termini.1 One of the 2 extracellular loops is short (EC1), and the other is longer (EC2). Some tetraspanin proteins also have post-translational modifications including N-linked glycosylation on the EC2 loop and palmitoylation at a CXXC motif in their transmembrane region.2 A schematic drawing of the general structure of tetraspanins is shown in Figure 1.
Figure 1.
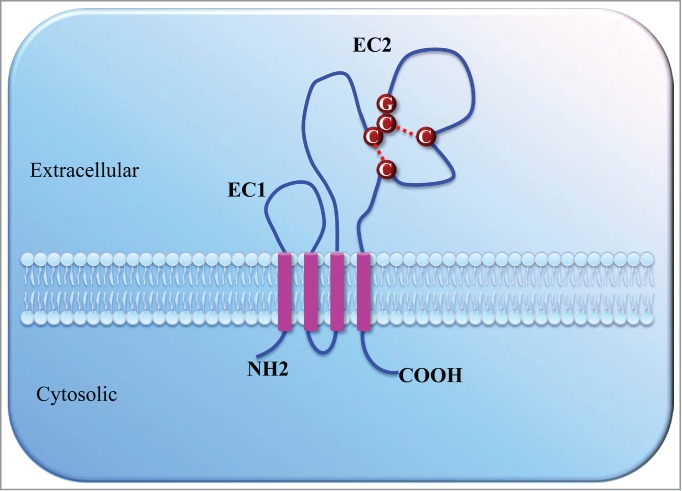
Schematic drawing of the general structure of tetraspanins. Tetraspanins are composed of 4 transmembrane domains (pink), a small (EC1) and a large extracellular domain (EC2), a very small intracellular domain, and short cytoplasmic N- and C-terminal tails. The EC2 contains a variable region presenting a conserved Cys-Cys-Gly (CCG) motif and 2–6 additional cysteine residues, which form intramolecular disulfide bonds (red dotted lines).
Although their actions and mechanisms are not fully understood, tetraspanins are known to function as scaffolding proteins in the plasma membrane of eukaryotic cells. Tetraspanins bind to one another and to numerous partner proteins, forming a "tetraspanin web" or tetraspanin-enriched microdomains (TEMs), which serve as structural and functional units in plasma membranes.3,4 Through direct protein–protein interactions and the specific organization of TEMs, tetraspanins modify the function of a wide variety of proteins including various integrins, immunoglobulin superfamily proteins, proteases, growth factor receptors, and intracellular signaling molecules.5-7 Consequently, they are engaged in a variety of cellular processes such as cell adhesion, migration, differentiation, and proliferation and are implicated in numerous pathological conditions including metastasis, inflammation, and viral infection.8-10 The four transmembrane domains of tetraspanins are involved in both intramolecular and intermolecular interactions that are crucial for the biosynthesis and assembly of TEMs. The EC2 loop is required for interactions between tetraspanins and other proteins. Despite conserved cysteine motifs, the EC2 loop is the most variable region among tetraspanin family members and likely plays a significant role in member-specific molecular recognition and function.11
Tetraspanins are found in nearly all tissues and cell types. Each member exhibits a distinct expression profile.3,12 For example, the tetraspanins CD9, CD63, CD82, and CD151 have a wide distribution among various cell types, whereas CD37 and CD53 are mainly found in leukocytes.3 The functions of a given tetraspanin are likely defined by its protein sequence, post-translational modifications, and tissue and cellular distribution. Through regulation of integrins and other adhesion- and motility-related proteins, a number of tetraspanins have emerged as key regulators of cell adhesion and migration in both normal and pathological processes. The present review focuses on research advances made in this field.
Tetraspanins in cell migration
Cell migration is a fundamental process in both normal development and pathological conditions such as cancer metastasis and inflammation. Nearly all cell migrations are driven by an extracellular signal and involve an assemblage of protein–protein interactions. The proteins that play a key role in this process include cadherins (cell–cell adhesion), integrins (cell–extracellular matrix [ECM] adhesion), Rac/Rho (cytoskeletal protrusion/contraction), and matrix metalloproteinases (MMPs) (pericellular proteolysis/proteolytic ECM remodeling).13 Numerous studies have demonstrated that tetraspanins directly interact with various integrins and modulate their membrane compartmentalization, intracellular trafficking and recycling, and subsequent downstream signaling in response to migratory signals.14,15 Tetraspanins also directly interact with MMPs and regulate their cell surface localization, trafficking, lysosomal degradation, and proteolytic activity.16-18 Interestingly, several recent studies have indicated that tetraspanin CD9 regulates the protein expression of MMP-9 via the JNK pathway.19,20 Tetraspanins also play a role in E-cadherin–based cell–cell junctions.21-23 Although the underlying mechanisms are not fully understood, direct interaction between the tetraspanin CO-029 (TSPAN8) and E-cadherin has been documented by chemical cross-linking and immunohistologic analysis in human colon carcinoma cells.24 Furthermore, augmentation or suppression of tetraspanins can alter cell motility by deregulating Rac/Rho activity. For example, CD151 silencing in epidermal carcinoma cells leads to excessive RhoA activation and loss of actin organization, resulting in destabilized cell–cell contacts and enhanced migration of tumor cell sheets.23 Likewise, the tetraspanin CD82 inhibits cancer cell retraction and motility via deregulation of the Rac1/RhoA signaling network.25,26 Interestingly, a recent study has shown evidence that CD81 directly binds to Rac in T-lymphoblast cells,27 indicating that direct protein–protein interaction may be a possible mechanism by which tetraspanins regulate the Rac/Rho signaling pathway. Taken together, these findings indicate that tetraspanins regulate the function of key proteins involved in all aspects of cell migration.
Increasing evidence also shows that tetraspanins play important roles in the migration of many different cell types, including but not limited to cancer cells, endothelial cells, keratinocytes, fibroblasts, and leukocytes, and are implicated in various normal and pathological conditions that rely on cell migration. These roles are discussed in more detail in the following sections.
Tetraspanins in cancer cell migration and metastasis
Aberrant expression of tetraspanins, especially CD151, CD9, CD82, CO-029, and CD63, is frequently detected in metastatic tumors and has been linked to cancer progression.12,28 In addition to their potential value as prognostic markers in patients with cancer, many studies have suggested that these tetraspanins also play active roles in cancer metastasis by promoting or inhibiting cancer cell migration and invasion. CD151 is the first member of the tetraspanin family to be identified as a promoter of metastasis.29 The promigratory effects of CD151 on cancer cells are mainly mediated by its association with laminin-binding integrins including α3β1, α6β4, and α6β1.30 In particular, CD151 forms a highly stoichiometric and stable association with integrin α3β1, which is linked to PI4K activation in many different cell lines.31 A considerable number of studies have shown that CD151 plays a role in metastasis of specific types of cancer; epidermoid carcinoma and breast cancer are the 2 most thoroughly investigated types of such cancers. CD151 promotes the in vitro migration and in vivo metastasis of epidermoid carcinoma cells by regulating α3β1 and α6β4 integrin-dependent cell adhesion and migration as well as the formation of Rho A-dependent cell–cell junctions.23,29,32-34 Meanwhile, the promigratory and prometastatic effects of CD151 on breast cancer cells in vitro and in vivo are associated with regulation of glycosylation of α3β1 integrin as well as growth factor-induced activation of FAK, Rac1, lck, and p38.35-39 Additionally, CD151 drives migration and metastasis of hepatocellular carcinoma (HCC) cells by enhancing β1 integrin-dependent Rac and cdc42 activation.40,41 CD151 also promotes cancer cell migration and metastasis in colon cancer, fibrosarcoma, and several other cancer types (Table 1). Interestingly, CD151-null mice exhibit reduced lung metastasis of injected cancer cells and diminished cancer cell transendothelial migration and adhesion to CD151-null lung endothelial cells, suggesting that endothelial CD151 plays a role in fostering a tumor microenvironment that facilitates cancer cell invasion.42
Table 1.
Tetrapansins in cancer cell migration and metastasis
| Tetraspanin | Cancer type | Cell line; animal model | Promoter (↑) or suppressor (↓) | References |
|---|---|---|---|---|
| CD151 (TSPAN24) | Epidermoid carcinoma | HEp-3; in vitro migration/in vivo metastasis | ↑ | 29 |
| HEp3; in vitro migration/in vivo metastasis | ↑ | 33 | ||
| A431; in vitro migration | ↑ | 32 | ||
| A431; in vitro migration | ↑ | 23 | ||
| A431; in vitro migration | ↑ | 34 | ||
| Breast cancer | MDA-MB-231; in vitro migration | ↑ | 35 | |
| MDA-MB-231; in vitro migration/in vivo progression | ↑ | 39 | ||
| MDA-MB-231; in vitro migration/in vivo metastasis | ↑ | 36 | ||
| MDA-MB-231; in vitro migration | ↑ | 37 | ||
| In vivo ErbB2+ mammary tumor metastasis | ↑ | 38 | ||
| Prostate cancer | PC3; in vitro migration | ↑ | 69 | |
| LNCap, PC3; in vitro migration | ↑ | 70 | ||
| Hepatocellular carcinoma | HCCLM3, HepG2; in vitro migration/in vivo metastasis | ↑ | 40 | |
| HepG2; in vitro migration | ↑ | 41 | ||
| Colon cancer | RPMI4788; in vitro migration/in vivo metastasis | ↑ | 71 | |
| Tongue squamous carcinoma | Tca8113; in vitro migration | ↑ | 72 | |
| Lung adenocarcinoma | A549 | ↑ | 73 | |
| Fibrosarcoma | HT1080; in vitro migration/in vivo metastasis | ↑ | 71 | |
| Glioblastoma | A172; in vitro migration | ↑ | 71 | |
| Gastric cancer | SGC7901; in vitro migration | ↑ | 74 | |
| Cervical cancer | HeLa; in vitro migration | ↑ | 29 | |
| Epithelial ovarian cancer | SKOV3, OVCAR5; in vitro migration | ↑ | 75 | |
| CD82 (KAI1, TSPAN27) | Prostate cancer | AT6.1; in vivo metastasis | ↓ | 45 |
| PC3; in vitro migration | ↓ | 46 | ||
| DU145; in vitro migration | ↓ | 47 | ||
| DU145, LNCaP; in vitro migration | ↓ | 48 | ||
| Melanoma | B16-BL6; in vitro migration/in vivo metastasis | ↓ | 49 | |
| MMRU; MMLU; in vitro migration | ↓ | 50 | ||
| UACC903M, A375M; in vitro migration/in vivo metastasis | ↓ | 22 | ||
| Non-small cell lung cancer | H1299; in vitro migration | ↓ | 25 | |
| H1299; in vitro migration | ↓ | 76 | ||
| Pancreatic cancer | PANC1, Miapaca-2; in vitro migration | ↓ | 77 | |
| Hepatocellular carcinoma | SMMC-7721; in vitro migration | ↓ | 52 | |
| Hepa1-6; in vitro migration | ↓ | 51 | ||
| HCC-LM3; in vitro migration/in vivo metastasis | ↓ | 53 | ||
| Ovarian cancer | OV-MZ-6; in vitro migration | ↓ | 78 | |
| Fibroblastoma | HT1080; in vitro migration/in vivo metastasis | ↓ | 47 | |
| CD9 (TSPAN29) | Small-cell lung cancer | OS3-R5; in vitro migration | ↓ | 59 |
| OS3-R5; in vitro migration/in vivo metastasis | ↓ | 58 | ||
| Melanoma | A375; in vitro transendothelial invasion | ↑ | 61 | |
| Early-stage VGP WM793; in vitro migration | ↓ | 60 | ||
| Breast cancer | MDA-MB-231; in vitro migration | ↑ | 62 | |
| MDA-MB-231; in vitro migration | ↓ | 63 | ||
| B02; in vivo metastasis | ↑ | 64 | ||
| Fibrosarcoma | HT1080; in vitro migration | ↓ | 56 | |
| Multiple myeloma | U266; in vitro migration | ↓ | 65 | |
| Prostate cancer | PC-3M-LN4; in vitro migration (but not in vivo metastasis) | ↑ | 66 | |
| TSPAN1 | Colon cancer | HCT-8; in vitro migration | ↑ | 79 |
| Cervical cancer | SiHa, HeLa; in vitro migration | ↑ | 80 | |
| Non-small cell lung cancer | A549, SK-MES-1; in vitro migration | ↑ | 81 | |
| Hepatocellular carcinoma | SMMC-7721; in vitro migration | ↑ | 82 | |
| Squamous cell skin carcinoma | A431; in vitro migration | ↑ | 83 | |
| TSPAN8 (CO-029) | Pancreatic adenocarcinoma | BSp73AS; in vivo metastasis | ↑ | 84 |
| Colon cancer | Isreco1; in vitro migration | ↑ | 24 | |
| HT29; in vitro migration | ↑ | 85 | ||
| Esophageal cancer | KYSE150, EC9706; in vitro migration/in vivo metastasis | ↑ | 86 | |
| CD63 (TSPAN30) | Melanoma | KM3; in vitro migration | ↓ | 87 |
| MelJuso; in vitro migration | ↓ | 88 | ||
| Colon cancer | Lovo; in vitro migration | ↓ | 89 | |
| CD81 (TSPAN28) | Hepatocellular carcinoma | HepG2, SW480, Huh7; in vitro migration/in vivo metastasis | ↓ | 90 |
| HepG2, Huh-7.5; in vitro migration | ↑ | 91 | ||
| Melanoma | MelJuSo; in vitro migration/in vivo metastasis | ↑ | 92 |
CD82, also known as KAI1, is a tetraspanin family member that functions as a metastasis suppressor.43 In addition to its association with various integrins,14,15 CD82 directly interacts with the epidermal growth factor (EGF) receptor (EGFR) and attenuates EGF-induced signaling by promoting EGFR desensitization.44 CD82 was first identified as a metastasis suppressor in prostate cancer.45 Subsequent studies have suggested that the antimetastatic effects of CD82 are mediated by inhibition of integrin-dependent activation of c-Met and Src kinases as well as suppression of fibronectin expression and β1 integrin activation.46-48 Other studies have shown that CD82 inhibits melanoma cell migration and metastasis in vitro and in vivo through suppression of Rho-associated kinase-mediated formation of stress fibers, inhibition of MMP-2, and regulation of inhibitor of growth 4.22,49,50 Further, CD82 suppresses HCC cell migration in vitro via upregulation of Sprouty2 with subsequent downregulation of sphingosine kinase 1, as well as via inhibition of EGFR and c-Met signaling.51,52 Intriguingly, CD82 is a direct target of miR-197, a metastasis promoter of HCC, and mediates the effects of miR-197 on HCC migration via regulation of Rac1 and ROCK activity.53 CD82 also suppresses migration and metastasis in several other cancer types including non-small cell lung carcinoma, pancreatic cancer, ovarian cancer, and fibroblastoma (Table 1).
CD9 is a tetraspanin family member that exhibits both promigratory and antimigratory properties. CD9 is associated with α1β1, α2β1, α3β1, α4β1, α5β1, α6β1, α7β1, αIIbβ3, and α6β4 integrins.14,15 The regulatory effects of CD9 on cell migration are mediated by integrin-dependent signaling such as phosphorylation of FAK54 and activation of PI3K, Akt, and p38 kinases.55,56 CD9 also directly interacts with EGFR in gastric cancer cells, and further expression of CD9 in EGFR/CD9-transfected HepG2 cells attenuates EGFR signaling, likely by downregulation of EGFR surface expression.57 CD9 can either promote or suppress cancer cell migration and metastasis depending on the type of cancer, the type of cells involved, and the migratory signal. CD9 inhibits both in vitro migration and in vivo metastasis of OS3-R5 cells, a small-cell lung cancer cell line.58,59 However, the effects of CD9 on melanoma migration and invasion are somewhat controversial. CD9 antagonizes osteopontin-induced migration and invasion of early-stage VGP WM793 melanoma cells,60 but supports transendothelial migration of A375 melanoma cells by strengthening interactions between tumor cells and the endothelial cell monolayer.61 Similar controversy surrounds the effects of CD9 on the migration of breast cancer cells. CD9 supports native type IV collagen-induced migration of MDA-MB-231 breast cancer cells in vitro,62 but suppresses the migration of these cells in response to fibronectin.63 An in vivo study showed that CD9 overexpression promotes bone metastasis of BO2 breast cancer cells, an osteotropic cell line derived from aggressive MDA-MB-231.64 Other studies have demonstrated that CD9 suppresses the migration of fibrosarcoma cells56 and multiple myeloma cells,65 but enhances the migration of prostate cancer cells66 (Table 1). Interestingly, the promigratory effects of CD9 on prostate cancer cells in vitro do not translate into prometastatic effects in vivo.66
Other tetraspanin family members that have roles in cancer cell migration or invasion include TSPAN1 and TSPAN8 (promigratory), CD63 (antimigratory), and CD81 (promigratory or antimigratory) (Table 1). Collectively, these data indicate that select tetraspanin family members are key regulators of cancer cell migration, invasion, and metastasis and that modulation of their activity may have promising results in the treatment of specific types of cancer. The description of specific strategies to target tetraspanins for cancer therapy is beyond the scope of the present paper and has been discussed elsewhere.67,68
Tetraspanins in endothelial cell migration and angiogenesis
Angiogenesis, the formation of new blood vessels from pre-existing ones, is an integral part of many developmental and pathological conditions including embryonic development, wound healing, tissue regeneration, and cancer progression. Migration of capillary endothelial cells is an essential component of angiogenesis and is typically driven by growth factors such as vascular endothelial growth factor or activated by integrins that bind to ECM components.93 Human endothelial cells express at least 23 tetraspanins including CD151, CD9, CD81, CD82, CD63, and TSPAN8.9 Many of these tetraspanins, especially CD151 and CD9, have been shown to regulate endothelial cell migration and angiogenesis in vitro and in vivo. In human umbilical vein endothelial cells (HUVECs), TEMs form endothelial adhesive platforms that recruit cell adhesion proteins such as ICAM-1 and VCAM-1 at cell–cell contact sites.94 Specifically, CD151 is associated with β1, β3, β4, α2, α3, α5, and α6 integrins at lateral junctions, and antibodies to CD151 inhibit HUVEC migration and in vitro angiogenesis.95,96 Subsequent studies have shown that these effects of CD151 on HUVECs are mediated by integrin-dependent activation of the PI3K/Akt and ERK signaling pathways.97-99 Additionally, CD151 promotes migration, proliferation, and tube formation of ECV304 endothelial cells by activating endothelial NO synthase (eNOS).100 CD151-null mice exhibit normal vascular development but impaired angiogenesis of pathologic conditions such as tumor growth, and CD151-null mouse lung endothelial cells display aberrant migration and tube formation in vitro, along with reduced adhesion-dependent activation of PKB/c-Akt, eNOS, Rac, and Cdc42.101 CD151 gene delivery in rats and pigs following myocardial infarction enhances both myocardial angiogenesis and cardiac function, and these effects are correlated with the activation of FAK, PI3K, and MAPK signaling.102,103 One study showed that in a rat model of hind limb ischemia, CD151 gene transfer promoted angiogenesis and improved the skin temperature of the lateral ischemic hind limb, and activated the FAK, ERK, and PI3K/Akt/eNOS pathways.104 Importantly, these effects of CD151 are abrogated by transfer of a CD151 mutant with impaired integrin association, indicating that CD151-integrin complex formation is required for CD151-induced angiogenesis.104
CD9 is another tetraspanin that plays a key role in endothelial cell migration and angiogenesis. Like CD151, tetraspanin CD9 is localized at endothelial cell–cell junctions and associates with α3β1 integrin.96 Anti-CD9 antibody inhibits the migration of human saphenous vein and mammary artery endothelial cells toward fibronectin and vitronectin via modulation of β1 or β3 integrin-dependent signaling.105 In HUVECs, CD9 forms a ternary complex with αvβ3 integrin and junctional adhesion molecule A and positively regulates basic fibroblast growth factor–induced cell migration and tube formation following release of junctional adhesion molecule A and activation of MAPK.106 GS-168AT2, a truncated form of CD9-partner 1 protein, which depletes cell surface CD151 and CD9, inhibits vascular endothelial growth factor–induced HUVEC migration and tube formation in vitro and attenuates tumor-associated angiogenesis in vivo.107 Moreover, anti-CD9 antibody was shown to inhibit tumor progression in a human gastric cancer xenograft model via inhibition of both tumor growth and tumor-associated angiogenesis.108 Interestingly, a recent study showed that CD9 deletion attenuates lymphatic endothelial cell migration and tube formation in vitro and diminishes both tumor metastasis to lymph nodes and tumor-associated lymphangiogenesis in vivo.109 These data indicate that targeting CD9 may subdue cancer progression via inhibition of both angiogenesis and lymphangiogenesis. Intriguingly, anti-CD9 antibody also inhibits the migration of microvascular endothelial cells of the bovine retina toward fibronectin,110 and intravitreous injection of siRNA-CD9 or anti-CD9 antibodies reduces laser-induced retinal and choroidal neovascularization in mice.111 These findings suggest that CD9 may be a therapeutic target for macular degeneration. Furthermore, tumor cells overexpressing rat TSPAN8 promote endothelial cell branching in vitro and induce systemic angiogenesis in vivo; these effects are driven by selective uptake of tumor cell-derived, TSPAN8-containing exosomes by endothelial cells, a process directed by exosomal TSPAN8.112,113 Other tetraspanins implicated in endothelial cell migration and possibly angiogenesis include CD81 and CD63, which have been identified as positive regulators,96,114 and CD82, which has been reported as a negative regulator.115 Therefore, accumulating evidence indicates that targeting specific tetraspanins may hold promise as a novel treatment for cancer and other conditions involving angiogenesis, such as macular degeneration and post-ischemic revascularization.
Tetraspanins in keratinocyte migration during wound healing
The wound healing process is divided into 4 sequential, yet overlapping phases: (1) hemostasis, (2) inflammation, (3) proliferation, and (4) remodeling. The entire process involves coordinated action of different cell types, including immune cells, endothelial cells, keratinocytes, and fibroblasts.116,117 Re-epithelialization of the epidermis, which involves proliferation and migration of keratinocytes from the wound edges across the wound bed to cover the injured area, is an integral part of the proliferation phase of wound healing. Several tetraspanins are expressed on the keratinocyte surface; of these, CD151, CD9, and CD81 are colocalized with α3 and β1 integrins at intercellular junctions. One study showed that antibodies to CD151, CD9, CD81, α3, and β1 inhibit the migration of human keratinocytes in an in vitro wound-healing assay.118 Consistent with these results, CD151 expression has been found to be upregulated during wound healing in C57BL/6 mice, especially within the migrating epidermal tongue at the wound edge.119 CD151-null mice show impaired wound healing that is primarily attributed to a re-epithelialization deficit,119 and CD151-null keratinocytes migrate poorly on Matrigel (a basement membrane equivalent) and laminin-332 (a key player in re-epithelialization)120 and in skin explant cultures.121 Collectively, these data indicate that CD151 positively regulates wound healing by promoting keratinocyte migration during re-epithelialization. In the proliferation phase of wound healing, fibroblasts grow, migrate, and from a new ECM by excreting collagen and fibronectin. This process is an essential prerequisite to epidermal re-epithelialization. CD151 is also expressed in normal skin fibroblasts, and CD151-null fibroblasts migrate much faster on collagen I while showing no significant changes in adhesion, proliferation, or the ability to cause contraction in response to transforming growth factor β-1 or platelet-derived growth factor.120 These results show that CD151 has a potential role in fibroblast migration during wound healing and may thus warrant further investigation.
Similar to CD151, CD9 is colocalized with α3 and β1 integrins at intercellular junctions of keratinocytes.118 Previous studies have shown that anti-CD9 antibody attenuates the migration of primary human keratinocytes;118 however, CD9 silencing enhances the migration of HaCaT cells, an immortal human keratinocyte cell line, through activation of the JNK pathway and subsequent MMP-9 expression.20 One possible explanation for these seemingly inconsistent results is that the binding of anti-CD9 antibody to CD9 does not inhibit CD9 function, but rather enhances it. The finding that CD9 is downregulated in migrating keratinocytes during wound healing both in vitro and in vivo supports the antimigratory effect of CD9 on keratinocytes under these conditions.19,20 Similar to CD151-null mice, CD9-null mice show delayed wound healing that is attributed to impaired epidermal migration. Because abnormal elevations of MMP-9 are detected in CD9-null wounds, this delayed epidermal migration may be attributed to excessive degradation of type IV collagen in the basement membrane at the wound site rather than to changes in the migrating keratinocytes themselves.19 Moreover, because CD9 promotes endothelial cell migration and angiogenesis,105,106 loss of CD9 might negatively affect angiogenesis at the wound site, additionally contributing to impaired epidermal migration and re-epithelialization. In summary, these data implicate tetraspanins CD151 and CD9 as important regulators of the wound healing process, indicating their role as potential therapeutic targets for pathological wound repair. Tetraspanins CD63 and CD81 are also found in keratinocytes,118 and their roles in wound healing may warrant future investigation.
Tetraspanins in immune cell migration
Tetraspanins were first identified as cell surface antigens in lymphocytes.2 Later studies showed that immune cells express at least 20 tetraspanins on their surface.122 In immune cells, tetraspanins interact with many key leukocyte proteins, including immunoreceptors, integrins, and signaling molecules, allowing them to regulate a range of fundamental immune cellular processes such as antigen presentation, antibody production, degranulation, proliferation, and migration/extravasation.122-124 In the present review, we focus on the role of tetraspanins in the migration and extravasation of leukocytes, a critical process in the immune response.
Dendritic cells (DCs) are antigen-presenting cells that stimulate both naive B and T cells during immune responses, and their effectiveness depends on their ability to capture, process, and present antigens and migrate to secondary lymphoid tissues.125 Tetraspanins CD63, CD9, CD81, CD82, and CD151 are expressed in immature DCs, and antibodies to CD63, CD9, CD81, and CD82 (but not CD151) enhance chemokine-induced migration of these cells.126 CD81-null DCs display drastically impaired motility because of their inability to form actin protrusions. CD81 silencing in human and mouse DCs produces a similar phenotype along with a selective loss of Rac1 activity.127 Although CD37-null DCs potently stimulate T cells in vitro,128 these cells induce poor T-cell responses when injected into wild-type mice. This is attributed to impaired migration from skin to draining lymph nodes.129 In Jurkat T lymphocytes, tetraspanin CD9 enhances cell migration, activation, and proliferation by regulating the expression and clustering of ALCAM, a member of the immunoglobulin superfamily of cell adhesion molecules.130 In mast cells, CD9 colocalizes with high-affinity IgE receptor and the transmembrane adaptor protein non-T-cell activation linker (NTAL), promoting antigen-driven chemotaxis via cross talking with these partner proteins.131 Natural killer cells show substantial expression of CD81, CD63, and CD151 on their cell surface, and stimulation of CD81 with an immobilized antibody induces phosphorylation of ezrin/radixin/moesin proteins, facilitating natural killer cell migration toward various chemokines.132 In endothelial cells, tetraspanins associate with cell adhesion proteins such as ICAM-1 and VCAM-1 at cell–cell contact sites with transmigrating leukocytes, and endothelial CD9/CD151 silencing prevents lymphocyte transendothelial migration.94,133 Additionally, CD63-null HUVECs fail to recruit leukocytes, and CD63-null mice show reduced leukocyte rolling, recruitment, and extravasation, a phenotype similar to that associated with loss of P-selectin.134 Interestingly, antibodies to CD81 and CD9 block monocyte migration across brain endothelial monolayers by acting on both leukocyte and endothelial tetraspanins.135,136 Taken together, these data indicate that both leukocyte and endothelial tetraspanins play crucial roles in leukocyte migration and extravasation during immune responses.
Leukocyte infiltration into the central nervous system is a key process in the development of demyelinating lesions in multiple sclerosis.137 In mice, administration of an anti-CD81 antibody reduces inflammation in the spinal cord and ameliorates the development of neurological symptoms of experimental autoimmune encephalomyelitis.136 These results suggest that targeting specific tetraspanins may be a novel therapeutic approach for inflammatory disorders such as multiple sclerosis.
Conclusions
The regulatory function of tetraspanin proteins in cell migration has been integrated in the present review (Fig. 2). Tetraspanins interact with a wide range of membrane proteins such as integrins, cell surface receptors, and signaling molecules. They also modulate all 4 determining factors of cell migration: cell–cell adhesion, cell–ECM adhesion, cytoskeletal protrusion/contraction, and proteolytic ECM remodeling. Numerous in vitro and in vivo studies have highlighted the important regulatory function of tetraspanins in the migration of cancer cells, vascular endothelial cells, skin cells (keratinocytes and fibroblasts), and leukocytes. Consequently, tetraspanins are implicated in many pathologic or remedial processes that rely on cell migration, such as cancer, macular degeneration, ischemic injury repair, wound healing, and inflammation. Targeting tetraspanins via small molecule agents, RNAi, or antibodies may allow the development of novel therapy for these diseases.
Figure 2.
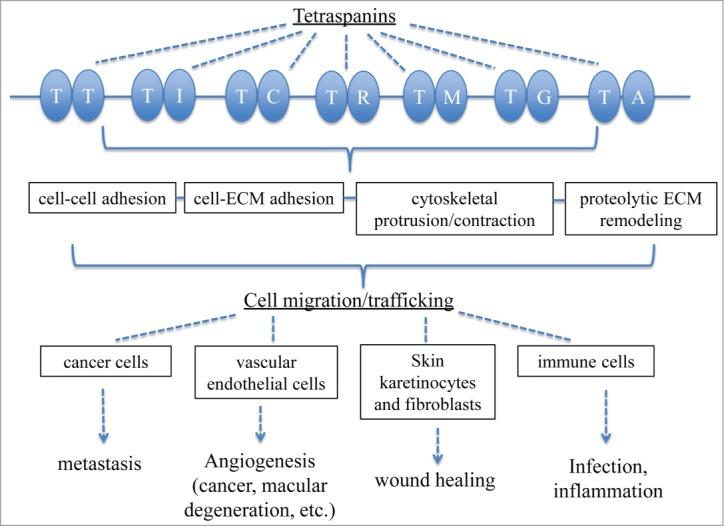
Schematic depiction of how tetraspanins regulate cell migration. T, tetraspanin; I, integrin; C, cadherin; R, Rac; M, matrix metalloproteinase; G, growth factor receptor; A, ALCAM.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported (in part) by National Natural Science Foundation of China (30973125), and the State Key Development Program for Basic Research of China (973 Program) (No. 2012CB518101).
References
- 1. Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 2005; 6:801-11; PMID:16314869; http://dx.doi.org/ 10.1038/nrm1736 [DOI] [PubMed] [Google Scholar]
- 2. Wright MD, Tomlinson MG. The ins and outs of the transmembrane 4 superfamily. Immunol Today 1994; 15:588-94; PMID:7531445; http://dx.doi.org/ 10.1016/0167-5699(94)90222-4 [DOI] [PubMed] [Google Scholar]
- 3. Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci 2001; 58:1189-205; PMID:11577978; http://dx.doi.org/ 10.1007/PL00000933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 2009; 19:434-46; PMID:19709882; http://dx.doi.org/ 10.1016/j.tcb.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 5. Yanez-Mo M, Mittelbrunn M, Sanchez-Madrid F. Tetraspanins and intercellular interactions. Microcirculation 2001; 8:153-68; PMID:11498779; http://dx.doi.org/ 10.1038/sj.mn.7800076 [DOI] [PubMed] [Google Scholar]
- 6. Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 2005; 20:218-24; PMID:16024509; http://dx.doi.org/ 10.1152/physiol.00015.2005 [DOI] [PubMed] [Google Scholar]
- 7. Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J 2009; 420:133-54; PMID:19426143; http://dx.doi.org/ 10.1042/BJ20082422 [DOI] [PubMed] [Google Scholar]
- 8. Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer 2009; 9:40-55; PMID:19078974; http://dx.doi.org/ 10.1038/nrc2543 [DOI] [PubMed] [Google Scholar]
- 9. Bailey RL, Herbert JM, Khan K, Heath VL, Bicknell R, Tomlinson MG. The emerging role of tetraspanin microdomains on endothelial cells. Biochem Soc Trans 2011; 39:1667-73; PMID:22103505; http://dx.doi.org/ 10.1042/BST20110745 [DOI] [PubMed] [Google Scholar]
- 10. van Spriel AB, Figdor CG. The role of tetraspanins in the pathogenesis of infectious diseases. Microb Infect 2010; 12:106-12; PMID:19896556; http://dx.doi.org/ 10.1016/j.micinf.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 11. Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci 2003; 28:106-12; PMID:12575999; http://dx.doi.org/ 10.1016/S0968-0004(02)00014-2 [DOI] [PubMed] [Google Scholar]
- 12. Detchokul S, Williams ED, Parker MW, Frauman AG. Tetraspanins as regulators of the tumour microenvironment: implications for metastasis and therapeutic strategies. Br J Pharmaco 2014;171:5462-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol 2010; 188:11-9; PMID:19951899; http://dx.doi.org/ 10.1083/jcb.200909003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. Journal of cell science 2001; 114:4143-51; PMID:11739647 [DOI] [PubMed] [Google Scholar]
- 15. Bassani S, Cingolani LA. Tetraspanins: Interactions and interplay with integrins. Int J Biochem Cell Biol 2012; 44:703-8; PMID:22326999; http://dx.doi.org/ 10.1016/j.biocel.2012.01.020 [DOI] [PubMed] [Google Scholar]
- 16. Takino T, Miyamori H, Kawaguchi N, Uekita T, Seiki M, Sato H. Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1 matrix metalloproteinase. Biochem Biophys Res Commun 2003; 304:160-6; PMID:12705901; http://dx.doi.org/ 10.1016/S0006-291X(03)00544-8 [DOI] [PubMed] [Google Scholar]
- 17. Lafleur MA, Xu D, Hemler ME. Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Molecular Biol Cell 2009; 20:2030-40; PMID:19211836; http://dx.doi.org/ 10.1091/mbc.E08-11-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schroder HM, Hoffmann SC, Hecker M, Korff T, Ludwig T. The tetraspanin network modulates MT1-MMP cell surface trafficking. Int J Biochem Cell Biol 2013; 45:1133-44; PMID:23500527; http://dx.doi.org/ 10.1016/j.biocel.2013.02.020 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Dong J, Gu H, Yu S, Zhang X, Gou Y, Xu W, Burd A, Huang L, Miyado K, et al. CD9 is critical for cutaneous wound healing through JNK signaling. J Invest Dermatol 2012; 132:226-36; PMID:21881583; http://dx.doi.org/ 10.1038/jid.2011.268 [DOI] [PubMed] [Google Scholar]
- 20. Jiang XP, Zhang DX, Teng M, Zhang Q, Zhang JP, Huang YS. Downregulation of CD9 in Keratinocyte Contributes to Cell Migration via Upregulation of Matrix Metalloproteinase-9. Plos One 2013; 8:e77806; PMID:24147081; http://dx.doi.org/ 10.1371/journal.pone.0077806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med 2010; 12:e3; PMID:20078909; http://dx.doi.org/ 10.1017/S1462399409001355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khanna P, Chung CY, Neves RI, Robertson GP, Dong C. CD82/KAI expression prevents IL-8-mediated endothelial gap formation in late-stage melanomas. Oncogene 2014; 33:2898-908; PMID:23873025; http://dx.doi.org/ 10.1038/onc.2013.249 [DOI] [PubMed] [Google Scholar]
- 23. Johnson JL, Winterwood N, DeMali KA, Stipp CS. Tetraspanin CD151 regulates RhoA activation and the dynamic stability of carcinoma cell-cell contacts. J Cell Sci 2009; 122:2263-73; PMID:19509057; http://dx.doi.org/ 10.1242/jcs.045997 [DOI] [PubMed] [Google Scholar]
- 24. Greco C, Bralet MP, Ailane N, Dubart-Kupperschmitt A, Rubinstein E, Le Naour F, Boucheix C. E-cadherin/p120-catenin and tetraspanin Co-029 cooperate for cell motility control in human colon carcinoma. Cancer Res 2010; 70:7674-83; PMID:20858717; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4482 [DOI] [PubMed] [Google Scholar]
- 25. Takahashi M, Sugiura T, Abe M, Ishii K, Shirasuna K. Regulation of c-Met signaling by the tetraspanin KAI-1/CD82 affects cancer cell migration. Int J Cancer 2007; 121:1919-29; PMID:17621632; http://dx.doi.org/ 10.1002/ijc.22887 [DOI] [PubMed] [Google Scholar]
- 26. Liu WM, Zhang F, Moshiach S, Zhou B, Huang C, Srinivasan K, Khurana S, Zheng Y, Lahti JM, Zhang XA. Tetraspanin CD82 inhibits protrusion and retraction in cell movement by attenuating the plasma membrane-dependent actin organization. PloS one 2012; 7:e51797; PMID:23251627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tejera E, Rocha-Perugini V, Lopez-Martin S, Perez-Hernandez D, Bachir AI, Horwitz AR, Vázquez J, Sánchez-Madrid F, Yáñez-Mo M. CD81 regulates cell migration through its association with Rac GTPase. Mol Biol Cell 2013; 24:261-73; PMID:23264468; http://dx.doi.org/ 10.1091/mbc.E12-09-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haeuw JF, Goetsch L, Bailly C, Corvaia N. Tetraspanin CD151 as a target for antibody-based cancer immunotherapy. Biochem Soc Trans 2011; 39:553-8; PMID:21428938; http://dx.doi.org/ 10.1042/BST0390553 [DOI] [PubMed] [Google Scholar]
- 29. Testa JE, Brooks PC, Lin JM, Quigley JP. Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effector of human tumor cell migration and metastasis. Cancer Res 1999; 59:3812-20; PMID:10447000 [PubMed] [Google Scholar]
- 30. Fitter S, Sincock PM, Jolliffe CN, Ashman LK. Transmembrane 4 superfamily protein CD151 (PETA-3) associates with β 1 and α IIb β 3 integrins in haemopoietic cell lines and modulates cell-cell adhesion. Biochem J 1999; 338 ( Pt 1):61-70; PMID:9931299; http://dx.doi.org/ 10.1042/0264-6021:3380061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yauch RL, Berditchevski F, Harler MB, Reichner J, Hemler ME. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol Biol Cell 1998; 9:2751-65; PMID:9763442; http://dx.doi.org/ 10.1091/mbc.9.10.2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winterwood NE, Varzavand A, Meland MN, Ashman LK, Stipp CS. A critical role for tetraspanin CD151 in alpha3beta1 and alpha6beta4 integrin-dependent tumor cell functions on laminin-5. Mol Biol Cell 2006; 17:2707-21; PMID:16571677; http://dx.doi.org/ 10.1091/mbc.E05-11-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer cell 2008; 13:221-34; PMID:18328426; http://dx.doi.org/ 10.1016/j.ccr.2008.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zevian S, Winterwood NE, Stipp CS. Structure-function analysis of tetraspanin CD151 reveals distinct requirements for tumor cell behaviors mediated by alpha3beta1 versus alpha6beta4 integrin. J Biol Chem 2011; 286:7496-506; PMID:21193415; http://dx.doi.org/ 10.1074/jbc.M110.173583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baldwin G, Novitskaya V, Sadej R, Pochec E, Litynska A, Hartmann C, Williams J, Ashman L, Eble JA, Berditchevski F. Tetraspanin CD151 regulates glycosylation of (α)3(β)1 integrin. J Biol Chem 2008; 283:35445-54; PMID:18852263; http://dx.doi.org/ 10.1074/jbc.M806394200 [DOI] [PubMed] [Google Scholar]
- 36. Sadej R, Romanska H, Kavanagh D, Baldwin G, Takahashi T, Kalia N, Berditchevski F. Tetraspanin CD151 regulates transforming growth factor β signaling: implication in tumor metastasis. Cancer Res 2010; 70:6059-70; PMID:20570898; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scales TM, Jayo A, Obara B, Holt MR, Hotchin NA, Berditchevski F, Parsons M. alpha3beta1 integrins regulate CD151 complex assembly and membrane dynamics in carcinoma cells within 3D environments. Oncogene 2013; 32:3965-79; PMID:22986527; http://dx.doi.org/ 10.1038/onc.2012.415 [DOI] [PubMed] [Google Scholar]
- 38. Deng X, Li Q, Hoff J, Novak M, Yang H, Jin H, Erfani SF, Sharma C, Zhou P, Rabinovitz I, et al. Integrin-associated CD151 drives ErbB2-evoked mammary tumor onset and metastasis. Neoplasia 2012; 14:678-89; PMID:22952421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang XH, Richardson AL, Torres-Arzayus MI, Zhou P, Sharma C, Kazarov AR, Andzelm MM, Strominger JL, Brown M, Hemler ME. CD151 accelerates breast cancer by regulating α 6 integrin function, signaling, and molecular organization. Cancer Res 2008; 68:3204-13; PMID:18451146; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Devbhandari RP, Shi GM, Ke AW, Wu FZ, Huang XY, Wang XY, Shi YH, Ding ZB, Xu Y, Dai Z, et al. Profiling of the tetraspanin CD151 web and conspiracy of CD151/integrin beta1 complex in the progression of hepatocellular carcinoma. PloS one 2011; 6:e24901; PMID:21961047; http://dx.doi.org/ 10.1371/journal.pone.0024901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fei Y, Wang J, Liu W, Zuo H, Qin J, Wang D, Zeng H, Liu Z. CD151 promotes cancer cell metastasis via integrins alpha3beta1 and alpha6beta1 in vitro. Mol Med Reports 2012; 6:1226-30; PMID:23007325 [DOI] [PubMed] [Google Scholar]
- 42. Takeda Y, Li Q, Kazarov AR, Epardaud M, Elpek K, Turley SJ, Hemler ME. Diminished metastasis in tetraspanin CD151-knockout mice. Blood 2011; 118:464-72; PMID:21536858; http://dx.doi.org/ 10.1182/blood-2010-08-302240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jackson P, Marreiros A, Russell PJ. KAI1 tetraspanin and metastasis suppressor. Int J Biochem Cell Biol 2005; 37:530-4; PMID:15618009; http://dx.doi.org/ 10.1016/j.biocel.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 44. Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Current Biol 2000; 10:1009-12; PMID:10985391; http://dx.doi.org/ 10.1016/S0960-9822(00)00652-7 [DOI] [PubMed] [Google Scholar]
- 45. Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, Barrett JC. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995; 268:884-6; PMID:7754374; http://dx.doi.org/ 10.1126/science.7754374 [DOI] [PubMed] [Google Scholar]
- 46. Sridhar SC, Miranti CK. Tetraspanin KAI1/CD82 suppresses invasion by inhibiting integrin-dependent crosstalk with c-Met receptor and Src kinases. Oncogene 2006; 25:2367-78; PMID:16331263; http://dx.doi.org/ 10.1038/sj.onc.1209269 [DOI] [PubMed] [Google Scholar]
- 47. Bari R, Zhang YH, Zhang F, Wang NX, Stipp CS, Zheng JJ, Zhang XA. Transmembrane interactions are needed for KAI1/CD82-mediated suppression of cancer invasion and metastasis. Am J Pathol 2009; 174:647-60; PMID:19116362; http://dx.doi.org/ 10.2353/ajpath.2009.080685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee HA, Park I, Byun HJ, Jeoung D, Kim YM, Lee H. Metastasis suppressor KAI1/CD82 attenuates the matrix adhesion of human prostate cancer cells by suppressing fibronectin expression and beta1 integrin activation. Cell Physiol Biochem 2011; 27:575-86; PMID:21691075; http://dx.doi.org/ 10.1159/000329979 [DOI] [PubMed] [Google Scholar]
- 49. Takaoka A, Hinoda Y, Sato S, Itoh F, Adachi M, Hareyama M, Imai K. Reduced invasive and metastatic potentials of KAI1-transfected melanoma cells. Jpn J Cancer Res 1998; 89:397-404; PMID:9617345; http://dx.doi.org/ 10.1111/j.1349-7006.1998.tb00577.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang Y, Cheng Y, Martinka M, Ong CJ, Li G. Prognostic significance of KAI1/CD82 in human melanoma and its role in cell migration and invasion through the regulation of ING4. Carcinogenesis 2014; 35:86-95; PMID:24130172; http://dx.doi.org/ 10.1093/carcin/bgt346 [DOI] [PubMed] [Google Scholar]
- 51. Li Y, Huang X, Zhang J, Li Y, Ma K. Synergistic inhibition of cell migration by tetraspanin CD82 and gangliosides occurs via the EGFR or cMet-activated Pl3K/Akt signalling pathway. Int J Biochem Cell Biol 2013; 45:2349-58; PMID:23968914; http://dx.doi.org/ 10.1016/j.biocel.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 52. Mu Z, Wang H, Zhang J, Li Q, Wang L, Guo X. KAI1/CD82 suppresses hepatocyte growth factor-induced migration of hepatoma cells via upregulation of Sprouty2. Sci China C Life Sci 2008; 51:648-54; PMID:18622748; http://dx.doi.org/ 10.1007/s11427-008-0086-1 [DOI] [PubMed] [Google Scholar]
- 53. Dai W, Wang C, Wang F, Wang Y, Shen M, Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al. Anti-miR-197 inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem Biophys Res Commun 2014; 446:541-8; PMID:24613834; http://dx.doi.org/ 10.1016/j.bbrc.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 54. Scherberich A, Giannone G, Perennou E, Takeda K, Boucheix C, Rubinstein E, Lanza F, Beretz A. FAK-mediated inhibition of vascular smooth muscle cell migration by the tetraspanin CD9. Thromb Haemost 2002; 87:1043-50; PMID:12083484 [PubMed] [Google Scholar]
- 55. Kotha J, Longhurst C, Appling W, Jennings LK. Tetraspanin CD9 regulates β 1 integrin activation and enhances cell motility to fibronectin via a PI-3 kinase-dependent pathway. Exp Cell Res 2008; 314:1811-22; PMID:18358474; http://dx.doi.org/ 10.1016/j.yexcr.2008.01.024 [DOI] [PubMed] [Google Scholar]
- 56. Chen S, Sun Y, Jin Z, Jing X. Functional and biochemical studies of CD9 in fibrosarcoma cell line. Mol Cell Biochem 2011; 350:89-99; PMID:21161334; http://dx.doi.org/ 10.1007/s11010-010-0685-1 [DOI] [PubMed] [Google Scholar]
- 57. Murayama Y, Shinomura Y, Oritani K, Miyagawa J, Yoshida H, Nishida M, Katsube F, Shiraga M, Miyazaki T, Nakamoto T, et al. The tetraspanin CD9 modulates epidermal growth factor receptor signaling in cancer cells. J Cell Physiol 2008; 216:135-43; PMID:18247373; http://dx.doi.org/ 10.1002/jcp.21384 [DOI] [PubMed] [Google Scholar]
- 58. Zheng R, Yano S, Zhang H, Nakataki E, Tachibana I, Kawase I, Hayashi S, Sone S. CD9 overexpression suppressed the liver metastasis and malignant ascites via inhibition of proliferation and motility of small-cell lung cancer cells in NK cell-depleted SCID mice. Oncol Res 2005; 15:365-72; PMID:16491954 [DOI] [PubMed] [Google Scholar]
- 59. Funakoshi T, Tachibana I, Hoshida Y, Kimura H, Takeda Y, Kijima T, Nishino K, Goto H, Yoneda T, Kumagai T, et al. Expression of tetraspanins in human lung cancer cells: frequent downregulation of CD9 and its contribution to cell motility in small cell lung cancer. Oncogene 2003; 22:674-87; PMID:12569360; http://dx.doi.org/ 10.1038/sj.onc.1206106 [DOI] [PubMed] [Google Scholar]
- 60. Yin M, Soikkeli J, Jahkola T, Virolainen S, Saksela O, Holtta E. Osteopontin promotes the invasive growth of melanoma cells by activating integrin alphavbeta3 and down-regulating tetraspanin CD9. Am J Pathol 2014; 184:842-58; PMID:24412090; http://dx.doi.org/ 10.1016/j.ajpath.2013.11.020 [DOI] [PubMed] [Google Scholar]
- 61. Longo N, Yanez-Mo M, Mittelbrunn M, de la Rosa G, Munoz ML, Sanchez-Madrid F, Sánchez-Mateos P. Regulatory role of tetraspanin CD9 in tumor-endothelial cell interaction during transendothelial invasion of melanoma cells. Blood 2001; 98:3717-26; PMID:11739177; http://dx.doi.org/ 10.1182/blood.V98.13.3717 [DOI] [PubMed] [Google Scholar]
- 62. Castro-Sanchez L, Soto-Guzman A, Navarro-Tito N, Martinez-Orozco R, Salazar EP. Native type IV collagen induces cell migration through a CD9 and DDR1-dependent pathway in MDA-MB-231 breast cancer cells. Eur J Cell Biol 2010; 89:843-52; PMID:20709424; http://dx.doi.org/ 10.1016/j.ejcb.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 63. Powner D, Kopp PM, Monkley SJ, Critchley DR, Berditchevski F. Tetraspanin CD9 in cell migration. Biochem Soc Trans 2011; 39:563-7; PMID:21428940; http://dx.doi.org/ 10.1042/BST0390563 [DOI] [PubMed] [Google Scholar]
- 64. Kischel P, Bellahcene A, Deux B, Lamour V, Dobson R, DE Pauw E, Clezardin P, Castronovo V. Overexpression of CD9 in human breast cancer cells promotes the development of bone metastases. Anticancer Res 2012; 32:5211-20; PMID:23225418 [PubMed] [Google Scholar]
- 65. Hu X, Xuan H, Du H, Jiang H, Huang J. Down-regulation of CD9 by methylation decreased bortezomib sensitivity in multiple myeloma. PloS one 2014; 9:e95765; PMID:24788635; http://dx.doi.org/ 10.1371/journal.pone.0095765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zvieriev V, Wang JC, Chevrette M. Over-expression of CD9 does not affect in vivo tumorigenic or metastatic properties of human prostate cancer cells. Biochem Biophys Res Commun 2005; 337:498-504; PMID:16198313; http://dx.doi.org/ 10.1016/j.bbrc.2005.09.073 [DOI] [PubMed] [Google Scholar]
- 67. Detchokul S, Williams ED, Parker MW, Frauman AG. Tetraspanins as regulators of the tumour microenvironment: implications for metastasis and therapeutic strategies. Br J Pharmacol 2014; 171:5462-90; PMID:23731188; http://dx.doi.org/ 10.1111/bph.12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sala-Valdes M, Ailane N, Greco C, Rubinstein E, Boucheix C. Targeting tetraspanins in cancer. Expert Opin Ther Targets 2012; 16:985-97; PMID:22880813; http://dx.doi.org/ 10.1517/14728222.2012.712688 [DOI] [PubMed] [Google Scholar]
- 69. Yang W, Li P, Lin J, Zuo H, Zuo P, Zou Y, Liu Z. CD151 promotes proliferation and migration of PC3 cells via the formation of CD151-integrin alpha3/alpha6 complex. J Huazhong Univ Sci Technolog Med Sci 2012; 32:383-8; http://dx.doi.org/ 10.1007/s11596-012-0066-y [DOI] [PubMed] [Google Scholar]
- 70. Ang J, Fang BL, Ashman LK, Frauman AG. The migration and invasion of human prostate cancer cell lines involves CD151 expression. Oncol Rep 2010; 24:1593-7; PMID:21042756 [DOI] [PubMed] [Google Scholar]
- 71. Kohno M, Hasegawa H, Miyake M, Yamamoto T, Fujita S. CD151 enhances cell motility and metastasis of cancer cells in the presence of focal adhesion kinase. Int J Cancer 2002; 97:336-43; PMID:11774285; http://dx.doi.org/ 10.1002/ijc.1605 [DOI] [PubMed] [Google Scholar]
- 72. Lan R, Liu Z, Song Y, Zhang X. Effects of rAAV-CD151 and rAAV-antiCD151 on the migration of human tongue squamous carcinoma cell line Tca8113. J Huazhong Univ Sci Technolog Med Sci 2004; 24:556-9; http://dx.doi.org/ 10.1007/BF02911353 [DOI] [PubMed] [Google Scholar]
- 73. Yamada M, Sumida Y, Fujibayashi A, Fukaguchi K, Sanzen N, Nishiuchi R, Sekiguchi K. The tetraspanin CD151 regulates cell morphology and intracellular signaling on laminin-511. FEBS J 2008; 275:3335-51; PMID:18492066; http://dx.doi.org/ 10.1111/j.1742-4658.2008.06481.x [DOI] [PubMed] [Google Scholar]
- 74. Wang X, Yu H, Lu X, Zhang P, Wang M, Hu Y. MiR-22 suppresses the proliferation and invasion of gastric cancer cells by inhibiting CD151. Biochem Biophys Res Commun 2014; 445:175-9; PMID:24495805; http://dx.doi.org/ 10.1016/j.bbrc.2014.01.160 [DOI] [PubMed] [Google Scholar]
- 75. Mosig RA, Lin L, Senturk E, Shah H, Huang F, Schlosshauer P, Cohen S, Fruscio R, Marchini S, D'Incalci M; et al. Application of RNA-Seq transcriptome analysis: CD151 is an Invasion/Migration target in all stages of epithelial ovarian cancer. J Ovarian Res 2012; 5:4; PMID:22272937; http://dx.doi.org/ 10.1186/1757-2215-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jee BK, Lee JY, Lim Y, Lee KH, Jo YH. Effect of KAI1/CD82 on the beta1 integrin maturation in highly migratory carcinoma cells. Biochem Biophys Res Commun 2007; 359:703-8; PMID:17560548; http://dx.doi.org/ 10.1016/j.bbrc.2007.05.159 [DOI] [PubMed] [Google Scholar]
- 77. Liu X, Guo XZ, Zhang WW, Lu ZZ, Zhang QW, Duan HF, Wang LS. KAI1 inhibits HGF-induced invasion of pancreatic cancer by sphingosine kinase activity. Hepatobiliary Pancreat Dis Int 2011; 10:201-8; http://dx.doi.org/ 10.1016/S1499-3872(11)60032-5 [DOI] [PubMed] [Google Scholar]
- 78. Ruseva Z, Geiger PX, Hutzler P, Kotzsch M, Luber B, Schmitt M, Gross E, Reuning U. Tumor suppressor KAI1 affects integrin alphavbeta3-mediated ovarian cancer cell adhesion, motility, and proliferation. Exp Cell Res 2009; 315:1759-71; PMID:19371633; http://dx.doi.org/ 10.1016/j.yexcr.2009.01.007 [DOI] [PubMed] [Google Scholar]
- 79. Chen L, Yuan D, Zhao R, Li H, Zhu J. Suppression of TSPAN1 by RNA interference inhibits proliferation and invasion of colon cancer cells in vitro. Tumori 2010; 96:744-50; PMID:21302622 [DOI] [PubMed] [Google Scholar]
- 80. Holters S, Anacker J, Jansen L, Beer-Grondke K, Durst M, Rubio I. Tetraspanin 1 promotes invasiveness of cervical cancer cells. Int J Oncol 2013; 43:503-12; PMID:23754316 [DOI] [PubMed] [Google Scholar]
- 81. Chen Y, Peng W, Lu Y, Chen J, Zhu YY, Xi T. MiR-200a enhances the migrations of A549 and SK-MES-1 cells by regulating the expression of TSPAN1. J Biosci 2013; 38:523-32; PMID:23938385; http://dx.doi.org/ 10.1007/s12038-013-9351-6 [DOI] [PubMed] [Google Scholar]
- 82. Wang GL, Chen L, Wei YZ, Zhou JM, Wu YY, Zhang YX, Qin J, Zhu YY. The effect of NET-1 on the proliferation, migration and endocytosis of the SMMC-7721 HCC cell line. Oncol Rep 2012; 27:1944-52; PMID:22378020 [DOI] [PubMed] [Google Scholar]
- 83. Chen L, Zhu Y, Li H, Wang GL, Wu YY, Lu YX, Qin J, Tuo J, Wang JL, Zhu J. Knockdown of TSPAN1 by RNA silencing and antisense technique inhibits proliferation and infiltration of human skin squamous carcinoma cells. Tumori 2010; 96:289-95; PMID:20572588 [DOI] [PubMed] [Google Scholar]
- 84. Herlevsen M, Schmidt DS, Miyazaki K, Zoller M. The association of the tetraspanin D6.1A with the alpha6beta4 integrin supports cell motility and liver metastasis formation. J Cell Sci 2003; 116:4373-90; PMID:13130099; http://dx.doi.org/ 10.1242/jcs.00760 [DOI] [PubMed] [Google Scholar]
- 85. Guo Q, Xia B, Zhang F, Richardson MM, Li M, Zhang JS, Chen F, Zhang XA. Tetraspanin CO-029 inhibits colorectal cancer cell movement by deregulating cell-matrix and cell-cell adhesions. Plos One 2012; 7:e38464; PMID:22679508; http://dx.doi.org/ 10.1371/journal.pone.0038464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhou Z, Ran YL, Hu H, Pan J, Li ZF, Chen LZ, Sun LC, Peng L, Zhao XL, Yu L, et al. TM4SF3 promotes esophageal carcinoma metastasis via upregulating ADAM12m expression. Clin Exp Metastasis 2008; 25:537-48; PMID:18365756; http://dx.doi.org/ 10.1007/s10585-008-9168-0 [DOI] [PubMed] [Google Scholar]
- 87. Radford KJ, Thorne RF, Hersey P. Regulation of tumor cell motility and migration by CD63 in a human melanoma cell line. J Immunol 1997; 158:3353-8 [PubMed] [Google Scholar]
- 88. Jang HI, Lee H. A decrease in the expression of CD63 tetraspanin protein elevates invasive potential of human melanoma cells. Exp Mol Med 2003; 35:317-23; PMID:14508073; http://dx.doi.org/ 10.1038/emm.2003.43 [DOI] [PubMed] [Google Scholar]
- 89. Sordat I, Decraene C, Silvestre T, Petermann O, Auffray C, Pietu G, Sordat B. Complementary DNA arrays identify CD63 tetraspanin and alpha3 integrin chain as differentially expressed in low and high metastatic human colon carcinoma cells. Lab Invest 2002; 82:1715-24; PMID:12480921; http://dx.doi.org/ 10.1097/01.LAB.0000044350.18215.0D [DOI] [PubMed] [Google Scholar]
- 90. Mazzocca A, Liotta F, Carloni V. Tetraspanin CD81-regulated cell motility plays a critical role in intrahepatic metastasis of hepatocellular carcinoma. Gastroenterology 2008; 135:244-56 e1; PMID:18466772; http://dx.doi.org/ 10.1053/j.gastro.2008.03.024 [DOI] [PubMed] [Google Scholar]
- 91. Brimacombe CL, Wilson GK, Hubscher SG, McKeating JA, Farquhar MJ. A role for CD81 and hepatitis C virus in hepatoma mobility. Viruses 2014; 6:1454-72; PMID:24662676; http://dx.doi.org/ 10.3390/v6031454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hong IK, Byun HJ, Lee J, Jin YJ, Wang SJ, Jeoung DI, Kim YM, Lee H. The Tetraspanin CD81 Protein Increases Melanoma Cell Motility by Up-regulating Metalloproteinase MT1-MMP Expression through the Pro-oncogenic Akt-dependent Sp1 Activation Signaling Pathways. J Biol Chem 2014; 289:15691-704; PMID:24733393; http://dx.doi.org/ 10.1074/jbc.M113.534206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lamalice L, Le Boeuf F, Huot J. Endothelial cell migration during angiogenesis. Circ Res 2007; 100:782-94; PMID:17395884; http://dx.doi.org/ 10.1161/01.RES.0000259593.07661.1e [DOI] [PubMed] [Google Scholar]
- 94. Barreiro O, Zamai M, Yanez-Mo M, Tejera E, Lopez-Romero P, Monk PN, Gratton E, Caiolfa VR, Sánchez-Madrid F. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol 2008; 183:527-42; PMID:18955551; http://dx.doi.org/ 10.1083/jcb.200805076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK. PETA-3/CD151, a member of the transmembrane 4 superfamily, is localised to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci 1999; 112 ( Pt 6):833-44; PMID:10036233 [DOI] [PubMed] [Google Scholar]
- 96. Yanez-Mo M, Alfranca A, Cabanas C, Marazuela M, Tejedor R, Ursa MA, Ashman LK, de Landázuri MO, Sánchez-Madrid F. Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA-1 and CD151/PETA-3 with alpha3 beta1 integrin localized at endothelial lateral junctions. J Cell Biol 1998; 141:791-804; PMID:9566977; http://dx.doi.org/ 10.1083/jcb.141.3.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Peng D, Zuo H, Liu Z, Qin J, Zhou Y, Li P, Wang D, Zeng H, Zhang XA. The tetraspanin CD151-ARSA mutant inhibits angiogenesis via the YRSL sequence. Mol Med Rep 2013; 7:836-42; PMID:23292489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zuo HJ, Lin JY, Liu ZY, Liu WF, Liu T, Yang J, Liu Y, Wang DW, Liu ZX. Activation of the ERK signaling pathway is involved in CD151-induced angiogenic effects on the formation of CD151-integrin complexes. Acta Pharmacol Sin 2010; 31:805-12; PMID:20581856; http://dx.doi.org/ 10.1038/aps.2010.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zheng ZZ, Liu ZX. Activation of the phosphatidylinositol 3-kinase/protein kinase Akt pathway mediates CD151-induced endothelial cell proliferation and cell migration. Int J Biochem Cell Biol 2007; 39:340-8; PMID:17045834; http://dx.doi.org/ 10.1016/j.biocel.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 100. Zheng ZZ, Liu ZX. CD151 gene delivery increases eNOS activity and induces ECV304 migration, proliferation and tube formation. Acta Pharmacol Sin 2007; 28:66-72; PMID:17184584; http://dx.doi.org/ 10.1111/j.1745-7254.2007.00490.x [DOI] [PubMed] [Google Scholar]
- 101. Takeda Y, Kazarov AR, Butterfield CE, Hopkins BD, Benjamin LE, Kaipainen A, Hemler ME. Deletion of tetraspanin Cd151 results in decreased pathologic angiogenesis in vivo and in vitro. Blood 2007; 109:1524-32; PMID:17023588; http://dx.doi.org/ 10.1182/blood-2006-08-041970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang L, Yang J, Liu ZX, Chen B, Liu J, Lan RF, Qin J. [Gene transfer of CD151 enhanced myocardial angiogenesis and improved cardiac function in rats with experimental myocardial infarction]. Zhonghua Xin Xue Guan Bing Za Zhi 2006; 34:159-63; PMID:16626587 [PubMed] [Google Scholar]
- 103. Zuo H, Liu Z, Liu X, Yang J, Liu T, Wen S, Zhang XA, Cianflone K, Wang D. CD151 gene delivery after myocardial infarction promotes functional neovascularization and activates FAK signaling. Mol Med 2009; 15:307-15; PMID:19603100; http://dx.doi.org/ 10.2119/molmed.2009.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Liu WF, Zuo HJ, Chai BL, Peng D, Fei YJ, Lin JY, Yu XH, Wang DW, Liu ZX. Role of tetraspanin CD151-alpha3/alpha6 integrin complex: Implication in angiogenesis CD151-integrin complex in angiogenesis. Int J Biochem Cell Biol 2011; 43:642-50; PMID:21237282; http://dx.doi.org/ 10.1016/j.biocel.2011.01.004 [DOI] [PubMed] [Google Scholar]
- 105. Klein-Soyer C, Azorsa DO, Cazenave JP, Lanza F. CD9 participates in endothelial cell migration during in vitro wound repair. Arterioscler Thromb Vasc Biol 2000; 20:360-9; PMID:10669631; http://dx.doi.org/ 10.1161/01.ATV.20.2.360 [DOI] [PubMed] [Google Scholar]
- 106. Peddibhotla SS, Brinkmann BF, Kummer D, Tuncay H, Nakayama M, Adams RH, Gerke V, Ebnet K. Tetraspanin CD9 links junctional adhesion molecule-A to alphavbeta3 integrin to mediate basic fibroblast growth factor-specific angiogenic signaling. Mol Biol Cell 2013; 24:933-44; PMID:23389628; http://dx.doi.org/ 10.1091/mbc.E12-06-0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Colin S, Guilmain W, Creoff E, Schneider C, Steverlynck C, Bongaerts M, Legrand E, Vannier JP, Muraine M, Vasse M, et al. A truncated form of CD9-partner 1 (CD9P-1), GS-168AT2, potently inhibits in vivo tumour-induced angiogenesis and tumour growth. Br J Cancer 2011; 105:1002-11; PMID:21863033; http://dx.doi.org/ 10.1038/bjc.2011.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Nakamoto T, Murayama Y, Oritani K, Boucheix C, Rubinstein E, Nishida M, Katsube F, Watabe K, Kiso S, Tsutsui S, et al. A novel therapeutic strategy with anti-CD9 antibody in gastric cancers. J Gastroenterol 2009; 44:889-96; PMID:19468669; http://dx.doi.org/ 10.1007/s00535-009-0081-3 [DOI] [PubMed] [Google Scholar]
- 109. Iwasaki T, Takeda Y, Maruyama K, Yokosaki Y, Tsujino K, Tetsumoto S, Kuhara H, Nakanishi K, Otani Y, Jin Y, et al. Deletion of tetraspanin CD9 diminishes lymphangiogenesis in vivo and in vitro. J Biol Chem 2013; 288:2118-31; PMID:23223239; http://dx.doi.org/ 10.1074/jbc.M112.424291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Deissler H, Kuhn EM, Lang GE, Deissler H. Tetraspanin CD9 is involved in the migration of retinal microvascular endothelial cells. Int J Mol Med 2007; 20:643-52; PMID:17912457 [PubMed] [Google Scholar]
- 111. Kamisasanuki T, Tokushige S, Terasaki H, Khai NC, Wang Y, Sakamoto T, Kosai K. Targeting CD9 produces stimulus-independent antiangiogenic effects predominantly in activated endothelial cells during angiogenesis: a novel antiangiogenic therapy. Biochem Biophys Res Commun 2011; 413:128-35; PMID:21875571; http://dx.doi.org/ 10.1016/j.bbrc.2011.08.068 [DOI] [PubMed] [Google Scholar]
- 112. Gesierich S, Berezovskiy I, Ryschich E, Zoller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res 2006; 66:7083-94; PMID:16849554; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-0391 [DOI] [PubMed] [Google Scholar]
- 113. Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, Lochnit G, Preissner KT, Zöller M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res 2010; 70:1668-78; PMID:20124479; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2470 [DOI] [PubMed] [Google Scholar]
- 114. Tugues S, Honjo S, Konig C, Padhan N, Kroon J, Gualandi L, Li X, Barkefors I, Thijssen VL, Griffioen AW, et al. Tetraspanin CD63 promotes vascular endothelial growth factor receptor 2-beta1 integrin complex formation, thereby regulating activation and downstream signaling in endothelial cells in vitro and in vivo. J Biol Chem 2013; 288:19060-71; PMID:23632027; http://dx.doi.org/ 10.1074/jbc.M113.468199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nagao K, Oka K. HIF-2 directly activates CD82 gene expression in endothelial cells. Biochem Biophys Res Commun 2011; 407:260-5; PMID:21382346; http://dx.doi.org/ 10.1016/j.bbrc.2011.03.017 [DOI] [PubMed] [Google Scholar]
- 116. Broughton G, 2nd, Janis JE, Attinger CE. The basic science of wound healing. Plast Reconstr Surg 2006; 117:12S-34S; PMID:16799372; http://dx.doi.org/ 10.1097/01.prs.0000225430.42531.c2 [DOI] [PubMed] [Google Scholar]
- 117. Stadelmann WK, Digenis AG, Tobin GR. Physiology and healing dynamics of chronic cutaneous wounds. Am J Surg 1998; 176:26S-38S; PMID:9777970; http://dx.doi.org/ 10.1016/S0002-9610(98)00183-4 [DOI] [PubMed] [Google Scholar]
- 118. Penas PF, Garcia-Diez A, Sanchez-Madrid F, Yanez-Mo M. Tetraspanins are localized at motility-related structures and involved in normal human keratinocyte wound healing migration. J Invest Dermatol 2000; 114:1126-35; PMID:10844555; http://dx.doi.org/ 10.1046/j.1523-1747.2000.00998.x [DOI] [PubMed] [Google Scholar]
- 119. Cowin AJ, Adams D, Geary SM, Wright MD, Jones JC, Ashman LK. Wound healing is defective in mice lacking tetraspanin CD151. J Invest Dermatol 2006; 126:680-9; PMID:16410781; http://dx.doi.org/ 10.1038/sj.jid.5700142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Geary SM, Cowin AJ, Copeland B, Baleato RM, Miyazaki K, Ashman LK. The role of the tetraspanin CD151 in primary keratinocyte and fibroblast functions: implications for wound healing. Exp Cell Res 2008; 314:2165-75; PMID:18534576; http://dx.doi.org/ 10.1016/j.yexcr.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 121. Wright MD, Geary SM, Fitter S, Moseley GW, Lau LM, Sheng KC, Apostolopoulos V, Stanley EG, Jackson DE, Ashman LK. Characterization of mice lacking the tetraspanin superfamily member CD151. Mol Cell Biol 2004; 24:5978-88; PMID:15199151; http://dx.doi.org/ 10.1128/MCB.24.13.5978-5988.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Tarrant JM, Robb L, van Spriel AB, Wright MD. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol 2003; 24:610-7; PMID:14596886; http://dx.doi.org/ 10.1016/j.it.2003.09.011 [DOI] [PubMed] [Google Scholar]
- 123. van Spriel AB. Tetraspanins in the humoral immune response. Biochem Soc Trans 2011; 39:512-7; PMID:21428930; http://dx.doi.org/ 10.1042/BST0390512 [DOI] [PubMed] [Google Scholar]
- 124. Veenbergen S, van Spriel AB. Tetraspanins in the immune response against cancer. Immunol lett 2011; 138:129-36; PMID:21497620; http://dx.doi.org/ 10.1016/j.imlet.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 125. Yoneyama H, Matsuno K, Matsushimaa K. Migration of dendritic cells. Int J Hematol 2005; 81:204-7; PMID:15814331; http://dx.doi.org/ 10.1532/IJH97.04164 [DOI] [PubMed] [Google Scholar]
- 126. Mantegazza AR, Barrio MM, Moutel S, Bover L, Weck M, Brossart P, Teillaud JL, Mordoh J. CD63 tetraspanin slows down cell migration and translocates to the endosomal-lysosomal-MIICs route after extracellular stimuli in human immature dendritic cells. Blood 2004; 104:1183-90; PMID:15130945; http://dx.doi.org/ 10.1182/blood-2004-01-0104 [DOI] [PubMed] [Google Scholar]
- 127. Quast T, Eppler F, Semmling V, Schild C, Homsi Y, Levy S, Lang T, Kurts C, Kolanus W. CD81 is essential for the formation of membrane protrusions and regulates Rac1-activation in adhesion-dependent immune cell migration. Blood 2011; 118:1818-27; PMID:21677313; http://dx.doi.org/ 10.1182/blood-2010-12-326595 [DOI] [PubMed] [Google Scholar]
- 128. Sheng KC, van Spriel AB, Gartlan KH, Sofi M, Apostolopoulos V, Ashman L, Wright MD. Tetraspanins CD37 and CD151 differentially regulate Ag presentation and T-cell co-stimulation by DC. Eur J Immunol 2009; 39:50-5; PMID:19089816; http://dx.doi.org/ 10.1002/eji.200838798 [DOI] [PubMed] [Google Scholar]
- 129. Gartlan KH, Wee JL, Demaria MC, Nastovska R, Chang TM, Jones EL, Apostolopoulos V, Pietersz GA, Hickey MJ, van Spriel AB, et al. Tetraspanin CD37 contributes to the initiation of cellular immunity by promoting dendritic cell migration. Eur J Immunol 2013; 43:1208-19; PMID:23420539; http://dx.doi.org/ 10.1002/eji.201242730 [DOI] [PubMed] [Google Scholar]
- 130. Gilsanz A, Sanchez-Martin L, Gutierrez-Lopez MD, Ovalle S, Machado-Pineda Y, Reyes R, Swart GW, Figdor CG, Lafuente EM, Cabañas C. ALCAM/CD166 adhesive function is regulated by the tetraspanin CD9. Cell Mol Life Sci 2013; 70:475-93; PMID:23052204; http://dx.doi.org/ 10.1007/s00018-012-1132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Halova I, Draberova L, Bambouskova M, Machyna M, Stegurova L, Smrz D, Dráber P. Cross-talk between tetraspanin CD9 and transmembrane adaptor protein non-T cell activation linker (NTAL) in mast cell activation and chemotaxis. J Biol Chem 2013; 288:9801-14; PMID:23443658; http://dx.doi.org/ 10.1074/jbc.M112.449231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kramer B, Schulte D, Korner C, Zwank C, Hartmann A, Michalk M, Söhne J, Langhans B, Nischalke HD, Coenen M, et al. Regulation of NK cell trafficking by CD81. Eur J Immunol 2009; 39:3447-58; PMID:19830727; http://dx.doi.org/ 10.1002/eji.200939234 [DOI] [PubMed] [Google Scholar]
- 133. Barreiro O, Yanez-Mo M, Sala-Valdes M, Gutierrez-Lopez MD, Ovalle S, Higginbottom A, Monk PN, Cabañas C, Sánchez-Madrid F. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood 2005; 105:2852-61; PMID:15591117; http://dx.doi.org/ 10.1182/blood-2004-09-3606 [DOI] [PubMed] [Google Scholar]
- 134. Doyle EL, Ridger V, Ferraro F, Turmaine M, Saftig P, Cutler DF. CD63 is an essential cofactor to leukocyte recruitment by endothelial P-selectin. Blood 2011; 118:4265-73; PMID:21803846; http://dx.doi.org/ 10.1182/blood-2010-11-321489 [DOI] [PubMed] [Google Scholar]
- 135. Schenk GJ, Dijkstra S, van het Hof AJ, van der Pol SM, Drexhage JA, van der Valk P, Reijerkerk A, van Horssen J, de Vries HE. Roles for HB-EGF and CD9 in multiple sclerosis. Glia 2013; 61:1890-905; PMID:24038577 [DOI] [PubMed] [Google Scholar]
- 136. Dijkstra S, Kooij G, Verbeek R, van der Pol SM, Amor S, Geisert EE, Jr., Dijkstra CD, van Noort JM, Vries HE. Targeting the tetraspanin CD81 blocks monocyte transmigration and ameliorates EAE. Neurobiol Dis 2008; 31:413-21; PMID:18586096; http://dx.doi.org/ 10.1016/j.nbd.2008.05.018 [DOI] [PubMed] [Google Scholar]
- 137. Kipp M, van der Valk P, Amor S. Pathology of multiple sclerosis. CNS Neurol Disord Drug Targets 2012; 11:506-17; PMID:22583433; http://dx.doi.org/ 10.2174/187152712801661248 [DOI] [PubMed] [Google Scholar]


