Summary
To demonstrate effects of aging visually requires a robust technique that can reproducibly detect small differences in efficiency or kinetics between groups. Investigators of aging will greatly appreciate the benefits of Amnis ImageStream technology (www.amnis.com/), which combines quantitative flow cytometry with simultaneous high-resolution digital imaging. Imagestream is quantitative, reproducible, feasible with limited samples, and it facilitates in-depth examination of cellular mechanisms between cohorts of samples.
Keywords: immune response, signal transduction, aging, monocyte, dendritic cell, neutrophil
1. Introduction
Tremendous advances in imaging techniques and analysis modalities present an unprecedented ability to view cellular events and trace intracellular signaling pathways and mechanisms (1). Using transfected fluorescent proteins, it is now possible to resolve kinetics of cellular processes, organelle localization, and dynamic interactions in living cells by fluorescence recovery after photobleaching (FRAP). Fluorescence resonance energy transfer (FRET) can localize molecules interacting within 2–6 nm or resolve temporal interactions in living cells (1). Super-Resolution microscopy combines increased sensitivity of imaging hardware with novel image analysis to achieve nanometer resolution at the light level (2), and multiphoton imaging allows deep penetration (500 nm) into living tissue with minimal damage to surrounding tissue and may be especially valuable for investigation of differences in murine models of aging (3). These dramatic advances in imaging are now feasible for many general uses in cell biology, but remain for the most part impractical for the study of aging, although multiphoton imaging may be valuable for investigation of differences in murine models of aging (3). The use of imaging to demonstrate effects of aging requires a robust visualization technique that can detect small differences (10–30%) in efficiency or kinetics between groups and can be repeated routinely so that samples from older or younger subjects may be directly compared. The technique should be able to be conducted with minimal manipulation of cells ex vivo and should not introduce new sources of variability. Both FRAP and FRET entail introducing fluorescently labeled components into cells – by transfection or injection -- to detect their localization or interaction in the cells. The variability associated with adding these manipulations -- potentially different in cells of different ages -- into the experiments may mask the ability to detect effects of aging.
For these reasons the benefits of Amnis ImageStream technology (www.amnis.com/) will be greatly appreciated among investigators of aging. Imagestream combines quantitative flow cytometry, which can be employed without manipulation of cells and has sufficient sensitivity to assess differences from only a few thousand cells, and combines it with simultaneous high-resolution digital imaging. ImageStream allows gating of cell subsets directly from peripheral blood cells and imaging of cell responses from gated, but unsorted cell populations. Imagestream has been employed to elucidate many cellular functions, e.g. to quantify nuclear translocation of transcription factors (4), bacterial phagocytosis and oxidative burst (5), as well as differences in signaling in studies of aging subjects and disease cohorts (6–8). Imagestream’s advantages include quantitation, reproducibility, feasibility with limited samples, and ability to promote in depth examination of cellular mechanisms between cohorts of samples.
2. Materials
All samples must be collected in accordance with appropriate IACUC, IRB, and biosafety regulations governing laboratory investigation. Comparison groups must be carefully selected to reduce any variations other than age such as manner of sample collection and handling. Sufficient samples must be collected to support robust statistical analysis such as mixed effects modeling (9) of factors such as gender, race, and co-morbid conditions.
AMNIS ImageStream X imaging cytometer (Amnis.com) equipped with ≥ 4 lasers (Excitation lasers: 488 nm blue laser, 561 nm green laser, 642 nm red laser, 405 nm violet laser and a Darkfield (SSC) laser 785 nm laser).
Computer: The Amnis IDEAS software currently requires a 32-or 64-bit Windows computer with recommended minimum specifications as follows: a quad core processor with 4GB of RAM and a PC bus speed of at least 1066Mhz. We have not found the Imagestream analysis program files to be compatible with MAC computers.
Fresh blood samples from younger and older human subjects collected in anticoagulated vacutainer tubes or BD Vacutainer® CPT™ Cell Preparation Tubes; or equivalent source of murine cells.
Cell culture media and sera: RPMI, MEM, pooled human serum or FBS for murine cells. We pre-screen aliquots of multiple test lots of serum from commercial sources (Lonza Group Ltd, Gemini Bio-Products, Valley Biomedical Products & Services, Inc.) to identify sera that optimize cell conditions. We culture monocytes from 2–5 donors in each sera and assess viability, morphology, and cytokine production in response to ligand stimulation.
Buffers for flow cytometry: We use phosphate-buffered saline (PBS) to wash cells, BD Staining Buffer and Perm/Wash buffer (BD Biosciences, NJ) for antibody labeling, we fix cells in 4% paraformaldehyde (Electron Microscopy Sciences) in PBS (PFA/PBS), freezing buffer is 90% FBS containing 10% DMSO.
Fluorescently conjugated monoclonal antibodies: lineage markers Lin-1 (lineage-1 marker includes CD3, CD14, CD19, CD20, CD56), HLA-DR, CD45, CD11c, CD123. Functional markers: TLRs, signaling components (e.g., IRF1, IRF3, IRF5, IRF7, NF-κB). Commercial sources for these antibodies include BD Biosciences, BD Pharmingen, eBioscience, Invitrogen, CA, SantaCruz Biotechnology, CA (Table 1).
Table 1.
Antibody panel for cell lineage staining of signaling pathways
| Cell type | FITC | PE | Alexa 647 | PacBlue | Catalogue number |
|---|---|---|---|---|---|
| Monocyte | CD14 | NF-κB (p65) |
eBioscience 12-0149-41; SantaCruz SC- 372 |
||
| Neutrophil | CD15 | BD Biosciences 551376 | |||
| mDC | Lin1 | CD11c | NF-κB (p65) |
CD4 | BD Biosciences 340546; BD Biosciences 347637; SantaCruz SC-372; BD Pharmingen 68436 |
| pDC | Lin1 | CD123 | NF-κB (p65) |
CD4 | BD Biosciences 340546; BD Biosciences 340545; SantaCruz SC-372; BD Pharmingen 68436 |
Monocyte: CD14+
Neutrophil CD15+
mDC: Lin1-, CD4dim, CD11c+
pDC: Lin1-, CD4dim, CD123+
Lin1 (lineage) marker includes CD3, CD14, CD19, CD20, CD56
3. Methods
Procedures with human blood should be conducted in a laminar flow hood for Biosafety level 2 containment. Carry out procedures at room temperature unless otherwise specified. To minimize technical variability, samples can be collected and treated as available and frozen to be assessed together at one time.
Design your experimental plan so that you can efficiently handle the number of samples sufficiently quickly. Examining samples from multiple study subjects at once reduces variability but only when the protocol can be managed smoothly. If many conditions per subject are required you may need to cap the maximum number of subjects you can assess per study day.
A panel of lineage markers and activation markers is shown in Table 1, many from BD Biosciences, BD Pharmingen, eBioScience. To design a labeling panel, markers should be tested singly and in combination to build a panel with specific signal after subtraction of isotype staining, consistent detection across the channels, and faithful quantitation of each marker (6). For transcription factors that translocate into the nucleus, it is better to use a small molecule dye such as Alexa 647 or FITC. Larger dyes such as PE and PE-Texas Red may reduce efficiency of translocation. Choices for additional antibodies to include in the panel will be driven by the availability of the antibody of choice in a given fluorochrome. Custom conjugations may be necessary for less common or lab-specific markers. It may be necessary to design separate panels to detect markers that form a complex in the cells of interest. For example, we have found that we cannot detect authentic levels of TLR4 in cells labeled with CD14, and we have substituted CD4 dim or CD11c staining to define cell lineage for samples in which we measure TLR4.
Obtain 10–50 ml heparinized blood from study volunteers after written informed consent under the guidelines of the local Institutional Review Board.
Dilute blood 1:1 with PBS.
Prepare 50 ml tubes with 12.5 ml filtered Ficoll-Hypaque Plus gradient separation gradient gel (GE Healthcare, NJ, density 1.077 g/ml). Dispense approximately 35 ml diluted blood very slowly and gently down the side of tube so that it forms a layer of diluted blood over the ficoll. Take care to minimize any blood entering into the ficoll material.
Centrifuge at 800 × g for 20 min at RT with brake off; for CPT tubes, centrifuge at 1500–1800 × g for 20 minutes at RT.
Peripheral blood mononuclear cells (PBMCs) will form a white band in the middle of the tube. Using a pipette, remove and discard the upper supernatant to within 0.5 cm of the cell layer. Collect cells from the white layer into a new 15 ml tube containing 5–7 ml RPMI.
Pellet cells at 500 × g for 10 min at 4°C, discard supernatant, and resuspend PBMC pellet in culture medium. For human cells we use RPMI with 20% human serum. With murine cells, we use MEM with 10% FBS.
Count cells and divide for treatment groups, ~1×106 cells/condition. You may use wells of a 96 well plate or autoclaved 1.5 ml eppendorf tubes.
To study kinetics of cell responses, we begin stimulation by adding the stimulating agent to the series of samples at staggered intervals and then harvest all the samples together at one end point. We have found this is more efficient for labeling all samples together at the final time point. In the example shown in Figures 1 and 2, we show mock treated cells and cells stimulated with flagellin (2.5 µg/ml), a TLR5 agonist. We added Flg at time 0, 30, and 45 min for harvest at 60 min. In this way we determine changes evident after 60, 30, and 15 min of stimulation.
Incubate cells at 37 °C in 5 % CO2 incubator for the desired time(s). If cells are to be incubated in PBS rather than tissue culture medium, incubations should not be done in a 5% CO2 atmosphere as PBS will not have sufficient buffering capacity to maintain cultures at physiological pH. Instead when using PBS choose a warm room or incubator with room air at 37 °C.
We label surface markers on the day of isolation and then cells are fixed and stored in freezing buffer at −80°C until all the samples are collected. Intracellular markers are labeled in batches of cells at one time to minimize assay variability.
To label cell surface lineage markers, we first remove the stimulation medium by centrifuging samples at 300 × g for 10 min at 4°C, then remove the supernatant taking care to avoid disturbing the pellet.
Wash cells once by resuspending samples in 200 µl cold Staining Buffer, centrifuge at 300 × g for 10 min at 4°C, and discard the supernatant.
Label cells by resuspending samples in 50 µl Staining Buffer containing specific antibodies or isotype controls and incubate for 20 min at 4°C protected from light.
At the end of the staining period, wash cells by adding 200 µl cold Staining Buffer, and centrifuge at 300 × g for 10 min at 4°C. Remove the supernatant.
Fix cells by adding 0.1 ml 4% paraformaldehyde in PBS for 10 min at RT protected from light.
At the end of the fixation period, centrifuge samples at 500 × g for 10min at 4°C. Remove the supernatant and resuspend cells in 0.1 ml 90% fetal calf serum/10% DMSO for storing at −80°C.
To minimize technical variability, batches of untreated and stimulated cells from a group of donors should be processed together. On the day of analysis, thaw cells quickly in 37 °C water bath, centrifuge at 500 × g to remove freezing buffer.
Wash cells by adding 0.3 ml BD Staining Buffer, and centrifuge at 500 × g for 10 min at 4°C. Remove the supernatant.
Before intracellular staining, resuspend cells in 0.2 ml PermWash Buffer. Centrifuge at 500 × g for 10 min at 4°C. Remove the supernatant.
Resuspend cells in 0.1 ml PermWash Buffer and incubate for 15 min at 4°C. Centrifuge at 500 × g for 10 min at 4°C. Remove the supernatant.
Label intracellular signaling components or organelle structures with specific antibodies and isotype controls in PermWash Buffer for 20 min at RT, e.g. rabbit anti- NF-κB (p65) antibody (10 µg/mL, SantaCruz Biotechnology, CA), detected using F(ab’)2 goat anti-rabbit IgG-Alexa647 (Invitrogen, CA). These incubations are conducted at RT according to the instrument manufacturer.
Immediately prior to imaging, counterstain nuclei with DAPI (final concentration 0.2 µg/ml, Invitrogen, CA) or propidium iodide (PI; final concentration 20 ng/ml, Invitrogen, CA). These concentrations should be optimized for new stocks of reagents and for different cell types.
Power up ImageStream, launch INSPIRE software and load default template as the setting. The calibration steps are automated by the instrument software. Initialize Fluidics and load SpeedBeads until the Flow Speed CV is consistently less than 0.2%. To calibrate and test the instrument, using ‘Start All’ command in the ASSIST tab.
After the ImageStream system is calibrated, define instrument settings using a sample that is bright in all the fluorescent channels employed for the study samples. Load the experiment’s brightest sample and set the laser power for each fluorochrome to show maximum pixel values between 100 and 4000 counts, as measured in the dot plots. To normalize settings of batches of samples, primary cells may be used when the supply is plentiful, or stored aliquots of frozen cells may be used. Compensation beads are not recommended by Amnis because speed control beads are used during the sample runs.
Set the ‘Cell Classification’ criteria to eliminate collection of unwanted events, and set a number of events to acquire (a minimum of 5000 events for monocytes).
Click ‘FLL: Flush, Lock, Load’. The sample pathway will be flushed first so wait until prompted before placing the sample on the left sample uptake line. To initiate readings, load labeled cells into sample chambers, either place a 96 well plate of labeled cells in the sample plate dock for autoplate reader instruments, or add tubes to the loading rack for instruments that use tubes.
Begin collecting sample data using the ‘Run Acquire’ command to collect the data and save the file. Once acquisition finishes, click ‘FLL’ to load the next sample. When finished collecting data on the last sample, click ‘FLL’ and leave the system on. When the last user of the day is finished, choose Sterilize System from the instrument menu to prepare for shutting down.
Design a gating strategy template for the study and apply the gates equivalently across all study samples. While automated gating strategies are becoming more accurate and gaining popularity, we find at present that individual variation between subjects is best appreciated by manual gating.
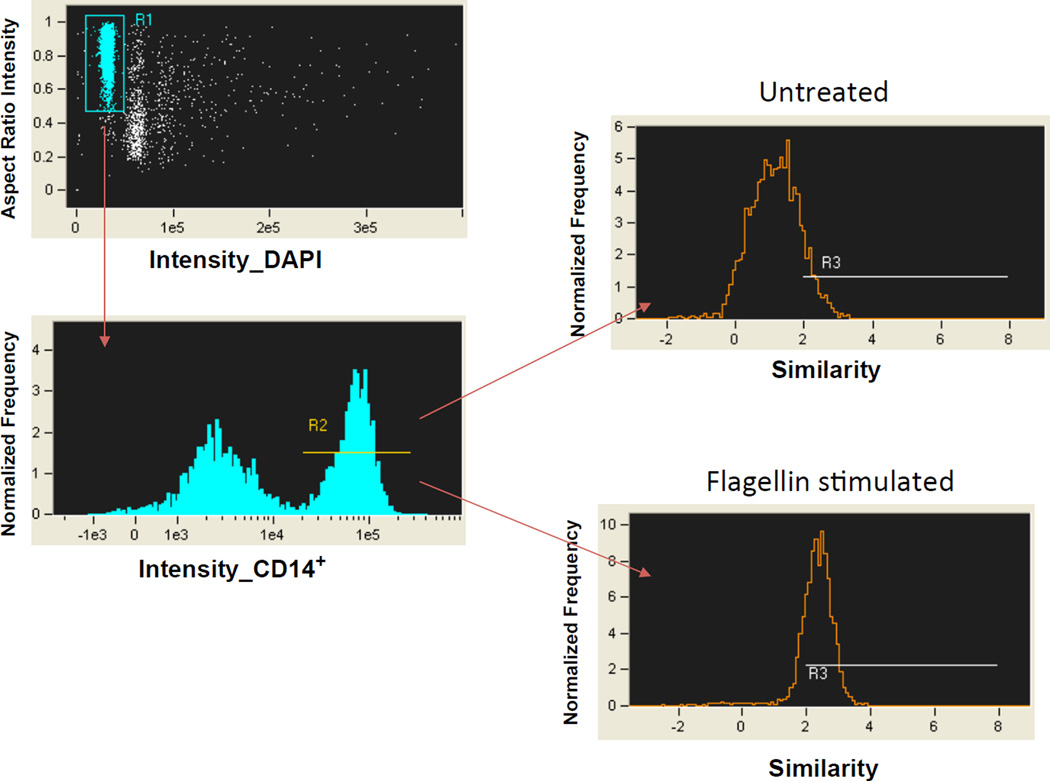
Data processing. Identify monocytes using Amnis IDEAS software and follow gates as shown in Figure 1. To distinguish in-focus single cells from debris, gate on events with normal nuclear dye DAPI intensity and high aspect ratio (defined as the ratio of the width of a shape to its height) for DAPI (R1).
To identify monocytes, choose cells gated with high intensity labeling of the CD14 marker (R2).
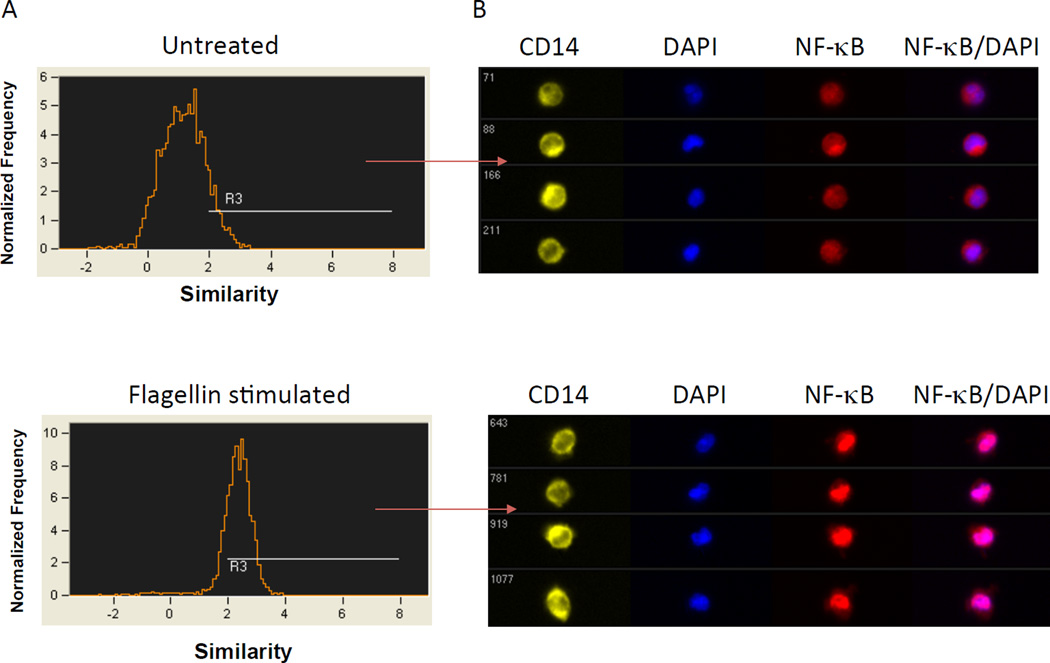
To identify activated cells after stimulation, the similarity score of NF-κB and nuclear dye DAPI will be calculated in CD14+ cells. Similarity measures the degree to which two images (NF-κB/DAPI) are correlated on a pixel-by-pixel basis. A high correlation of NF-κB with DAPI, indicating co-localization of the two markers, is reflected in a high ‘Similarity’ score (R3). After stimulation, the treated cells were determined to have a significantly higher similarity score of NF-kB p65 with nuclear stain DAPI, indicating that NF-κB p65 translocated into the nucleus after stimulation. Representative images of cells are shown in Figure 2.
The labeling and gating strategy described here works well for both monocytes and dendritic cells and should be effective for other cell types that show nuclear translocation of NF-κB after stimulation.
Differences between subject groups will be readily apparent through dot plots, histograms and cell images. Data representative of the study cohort may be presented as populations in histogram format, and/or representative images (where they are particularly compelling). Data from within a defined gate -- such as the ‘Similarity’ score -- can be tabulated for each study subject and/or group and is suitable for statistical determinations.
Figure 1. Gating Strategy for human monocytes from PBMCs.
Monocytes are distinguished in gates with normal nuclear dye DAPI intensity and high DAPI aspect ratio (R1) which are labeled with lineage marker CD14 (R2). A high correlation of NF-κB with the nuclear dye DAPI localization is reflected in a high similarity score and indicates the degree of activation in CD14+ monocytes (R3).
Figure 2. Translocation of NF-κB in human monocytes after stimulation.
The translocation of NF-κB (p65) into the nucleus after stimulation (R3) is depicted in populations of untreated and stimulated cells. The median value of similarity scores is 1.16 in untreated sample and 2.34 in stimulated sample (A). Digital images collected simultaneously of the untreated or flagellin - stimulated cell populations show representative cells and the intensity of NF-κB translocated into the cell nucleus (B).
Acknowledgments
Funding: This work was supported in part by the National Institutes of Health (HHS N272201100019C, U19AI089992).
The authors are grateful to the Yale Human Immunophenotyping Consortium and IMAGIN teams for insightful discussions.
Footnotes
The authors declare no commercial or other association that might pose a conflict of interest for this work.
References
- 1.Higashi T, Watanabe W, Matsunaga S. Application of visualization techniques for cell and tissue engineering. J Biosci Bioeng. 2012 doi: 10.1016/j.jbiosc.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Herbert S, Soares H, Zimmer C, Henriques R. Single-Molecule Localization Super-Resolution Microscopy: Deeper and Faster. Microscopy and microanalysis : the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 2012:1–11. doi: 10.1017/S1431927612013347. [DOI] [PubMed] [Google Scholar]
- 3.Masedunskas A, et al. Intravital microscopy: A practical guide on imaging intracellular structures in live animals. Bioarchitecture. 2012;2(5) doi: 10.4161/bioa.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George TC, et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Meth. 2006;311(1–2):117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Ploppa A, George TC, Unertl KE, Nohe B, Durieux ME. ImageStream cytometry extends the analysis of phagocytosis and oxidative burst. Scan J Clin Lab Invest. 2011;71(5):362–369. doi: 10.3109/00365513.2011.572182. [DOI] [PubMed] [Google Scholar]
- 6.Qian F, Montgomery RR. Quantitative imaging of lineage specific Toll-like receptor mediated signaling in monocytes and dendritic cells from small samples of human blood. JoVE. 2012;62:e3741. doi: 10.3791/3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian F, et al. Age-associated elevation in TLR5 leads to increased inflammatory responses in the elderly. Aging Cell. 2012;11:104–110. doi: 10.1111/j.1474-9726.2011.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone RC, et al. Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arth Rheum. 2012;64(3):788–798. doi: 10.1002/art.33395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schluchter MD, Elashoff JD. Small-sample adjustments to tests with unbalanced repeated measures assuming several covariance structures. J Stat Comp Simul. 1990;37:69–87. [Google Scholar]