Abstract
Pantocin A (PA) is a member of the growing family of ribosomally encoded and post-translationally modified peptide natural products (RiPPs). PA is much smaller than most known RiPPs, a tripeptide with a tight bicyclic core that appears to be cleaved from the middle of a larger 30-residue precursor peptide. We show here that the enzyme PaaA catalyzes the double dehydration and decarboxylation of two glutamic acid residues in the 30-residue precursor PaaP. Further truncates of PaaP leader and follower peptide sequences demonstrate the different impacts of these two regions on PaaA-mediated tailoring and delineate an essential role for the follower sequence in the decarboxylation step. The crystal structure of apo PaaA is reported, allowing identification of structural features that set PaaA apart from other homologous enzymes that typically do not catalyze such extended post-translational chemistry. Together, these data reveal how additional chemistry can be extracted from a ubiquitous enzyme family toward ribosomally derived peptide natural product biosynthesis and suggest that more examples of such enzymes likely exist in untapped genomic space.
RiPPs, ribosomally synthesized and post-translationally modified peptides, are a growing family of natural products.1 RiPPs have gained substantial attention due to the relative promiscuity of their biosynthetic enzymes.2–7 RiPP enzymes modify short, typically 30–50 amino acid, peptides comprised of combinations of recognition (RS) and core (CS) sequences; the CS undergoes modification while the RS directs these modifications. The RS may be a leader and/or a follower peptide depending on its relative location with respect to the CS. The catalog of RiPP chemistries is still somewhat limited, but advances in genome mining suggest that there may be many more such leader peptide-directed post-translational modifying enzymes involved in bacterial biosynthetic pathways.8–10 Due to their substrate promiscuity, RiPP enzymes have promise as biocatalysts and there is great interest in expanding knowledge to new members of this class that may catalyze novel chemistry.
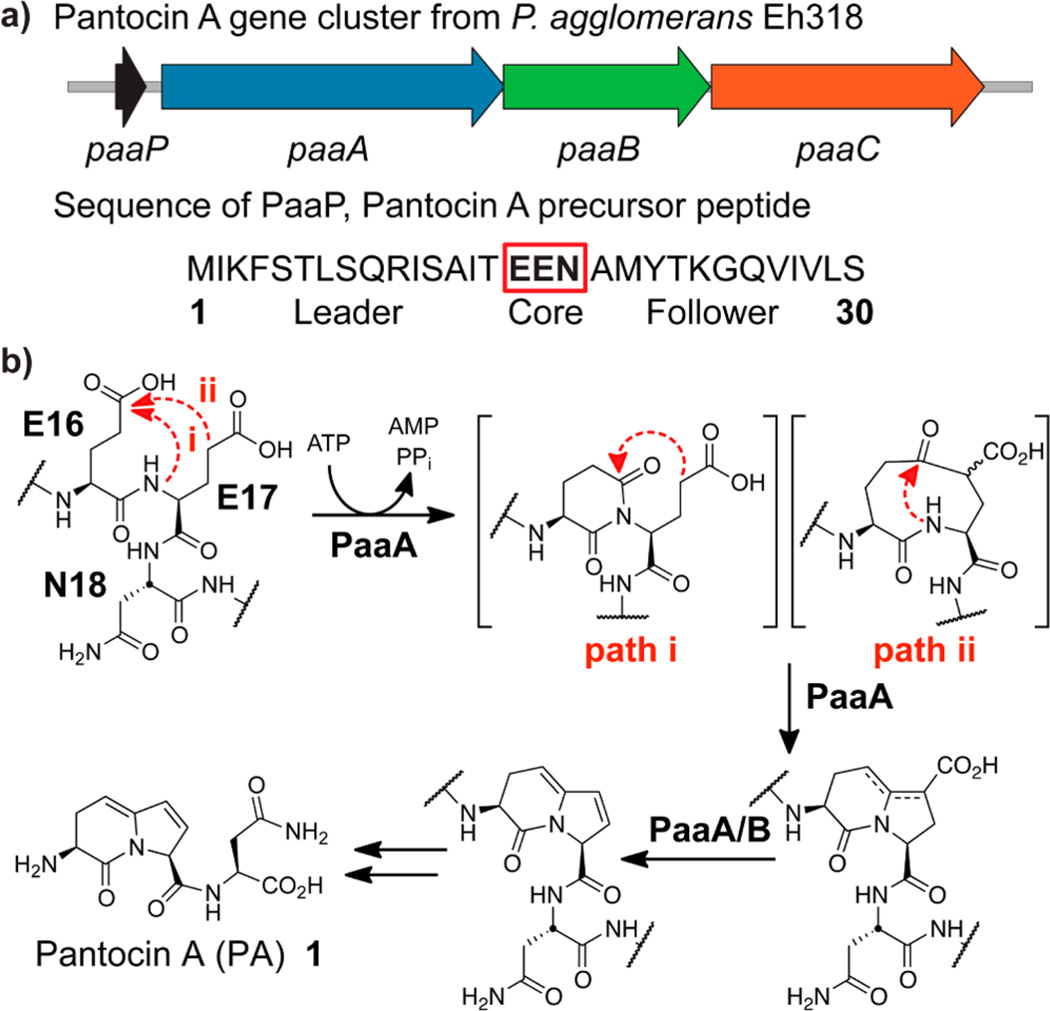
Pantocin A (PA) is a small tripeptide inhibitor of l-histidinol phosphate aminotransferase and was first isolated from Pantoea agglomerans by Clardy and co-workers in 2003.11,12 In tandem with the structural elucidation, the researchers reported the biosynthetic gene cluster from a cosmid clone, indicating that PA is a RiPP natural product (Figure 1a). PA appears to be carved from a three residue Glu-Glu-Asn motif at the center of a 30-residue precursor peptide, PaaP. Three ORFs are involved in the post-translational modification and host immunity: a predicted ATP-dependent enzyme of the ThiF/E1-like super-family, paaA, an α-ketoglutarate-dependent iron oxidase, paaB, and an efflux pump, paaC. The ability of PaaC to confer host immunity was confirmed by genetic experiments. However, the exact biosynthetic scheme and molecular functions of PaaA and PaaB in post-translational fashioning of PA remained undetermined.
Figure 1.
(a) Pantocin (PA) biosynthetic cluster from P. agglomerans and (b) proposed biosynthesis of Pantocin A. Biosynthesis requires condensation onto the peptide backbone and Claisen-like condensation between the two glutamic acids. Logically, either condensation (path i or ii) could occur first and either glutamate residue could provide the putative enolate nucleophile. A subsequent oxidative decarboxylation will yield the same bicyclic core. A detailed mechanistic proposal is given in Scheme S1 in the Supporting Information.
Spurred by the potential for new leader peptide-directed chemistry, we sought to interrogate the pantocin pathway by in vitro reconstitution of the activity of one or both of the PaaA and PaaB enzymes. Members of the ThiF/E1-like enzymes superfamily are known to activate carboxyl functional groups by adenylation followed by elimination.13 In particular, PaaA shows homology to the E1-like enzyme MccB, another RiPP enzyme, which catalyzes two consecutive adenylation steps at a C-terminal asparagine in MccA to generate a unique phosphoramide linkage.14,15 We therefore hypothesized that PaaA could act first in the pathway to effect backbone condensation of one glutamic acid and subsequent Claisen-like condensation with an intermediate imidate (Figure 1b, path i). It is equally possible that these two steps proceed in the reverse order, with backbone cyclization of an intermediate ketone (Figure 1b, path ii); both mechanisms would ultimately provide the same bicyclic product; similarly, either Glu16 or Glu17 could logically be activated as the Claisen electrophile (see SI Scheme S1). Subsequent oxidative decarboxylation of the intermediate enamine tautomers, by either PaaA or PaaB or some combination thereof, would yield the same PA-precursor regardless. Finally, this intermediate would need only proteolytic cleavage to provide the mature natural product. Based on this biosynthetic hypothesis, we began our efforts to understand the pathway with PaaA.
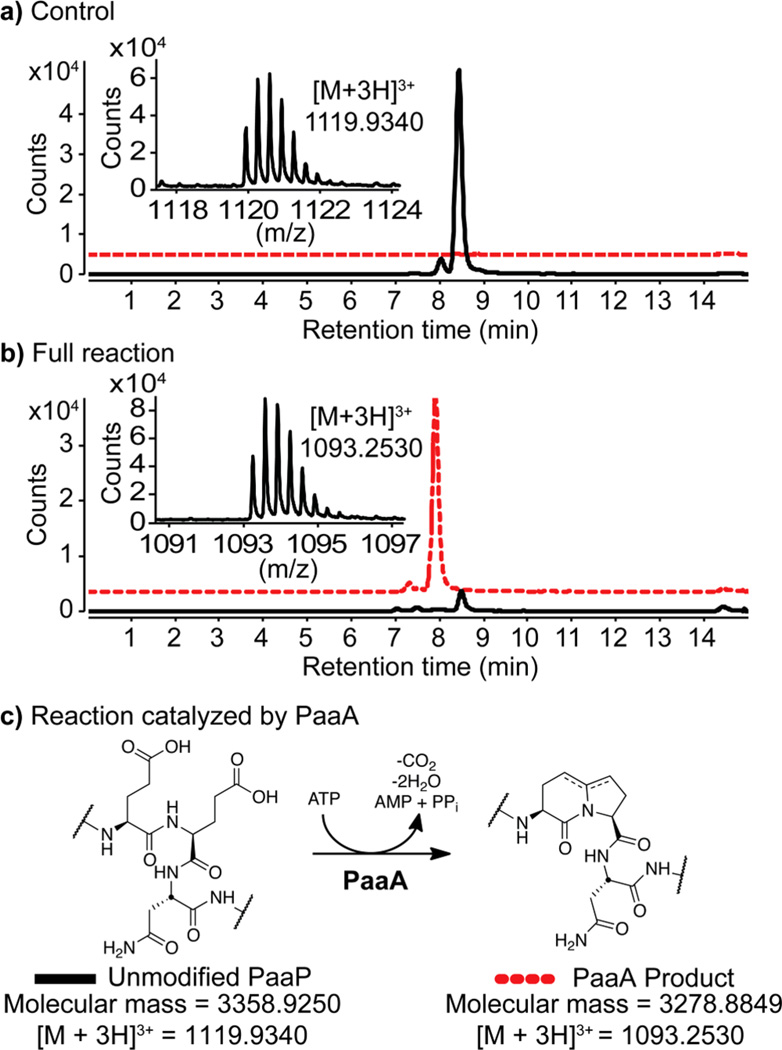
The 1.2kb paaA gene was cloned from the original cosmid pCPP702.12 Recombinant PaaA was expressed with a C-terminal 6xHis-tag; the 30-residue full length PaaP was synthesized in parallel via solid-phase peptide synthesis. Upon incubation of purified PaaA (2.5 µM) with substrate (25 µM), 4 mM MgCl2, 1 mM ATP, and 0.5 mM TCEP, we were surprised to see neither of the anticipated singly or doubly dehydrated products. In comparison to the control (Figure 2a) we observed a new mass corresponding to loss of two waters and a carboxylate (Figure 2b). This suggests that PaaA catalyzes both dehydrations plus the decarboxylation as depicted in Figure 2c and that PaaB may perform a two-electron dehydrogenation to give the conjugated olefin in Pantocin A (1). We could further confirm the unusual combination of modifications by MS/MS (see SI Figure S1), as well as the formation of new signals in the 1H NMR (see SI Figure S2). Assays run without ATP and/or MgCl2 demonstrate that PaaA needs both to perform this chemistry (see SI Figure S3).
Figure 2.
Extracted ion chromatograms for synthetic PaaP (black line, m/z 1119.9340) and PaaA product (red line, m/z 1093.2530). (a) Control PaaP (25 µM) without enzyme. (b) PaaP (25 µM) after incubation with PaaA (2.5 µM), 4 mM MgCl2, 1 mM ATP, and 0.5 mM TCEP for 15 h. (c) Reaction catalyzed by PaaA.
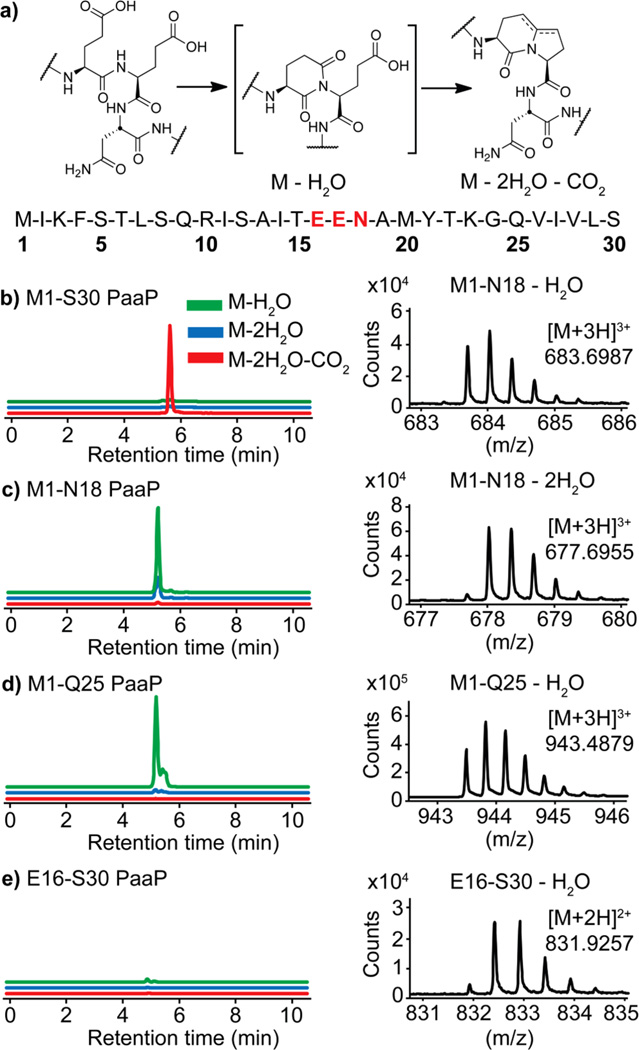
We next sought to probe the importance of the N-terminal leader and C-terminal follower portions of PaaP in PaaA modification (Figure 3). A series of truncates were prepared by solid phase peptide synthesis. When the last five residues from the PaaP primary sequence are deleted, the corresponding substrate M1-Q25 is consumed, but there was no detectable formation of the (PaaP – 2H2O – CO2) species. The same observation applies to the M1-N18 PaaP truncate in which the entire follower peptide sequence is missing. In both cases, we instead observed the formation of a dehydrated species (Figure 3c and 3d, respectively). This suggests that the second dehydration is not spontaneous. Incubation of the enzyme with the M1-N18 PaaP truncate in trans with the follower peptide sequence A19-S30 gave the same product as M1-N18 alone. Incubation of E16-S30 PaaP, a truncate with the full-length follower peptide but no leader peptide, with PaaA, displayed no detectable transformation of the substrate (Figure 3e). Shorter truncates containing the leader peptide, R10-N18, N-acyl-I11-N18, N-acyl-S12-N18, and N-acyl-A13-N18, were not processed by the enzyme.
Figure 3.
Effects of PaaP truncations on enzymatic modification by PaaA. Full length PaaP (b) is dehydrated twice and decarboxylated. The complete (c) and partial (d) C-terminal truncates only undergo dehydration. The N-terminal, leader peptide truncate (e) does not appear to undergo enzymatic processing. Insets: MS of select ions of respective products.
The results of the substrate truncates highlight the importance of both leader and follower sequences in post-translational processing by PaaA.16 While the leader sequence (M1-T15) is necessary for even partial processing, the absence of the follower allows at least the first condensation in the putative mechanism. There are other examples of follower peptides in RiPP natural product biosynthetic pathways, perhaps most notably in the cyanobactins, where the 4-residue follower peptide acts primarily as a recognition motif for protease/cyclase enzymes.1,17,18 Bottromycin precursor peptides also exhibit a long follower peptide, similar to PaaP, but its role in processing has not yet been elucidated.6,7,19–21 The mechanistic role of the PaaP follower peptide requires further investigation, but could stand as a model for enzymes capable of performing multiple chemistries using one active site.
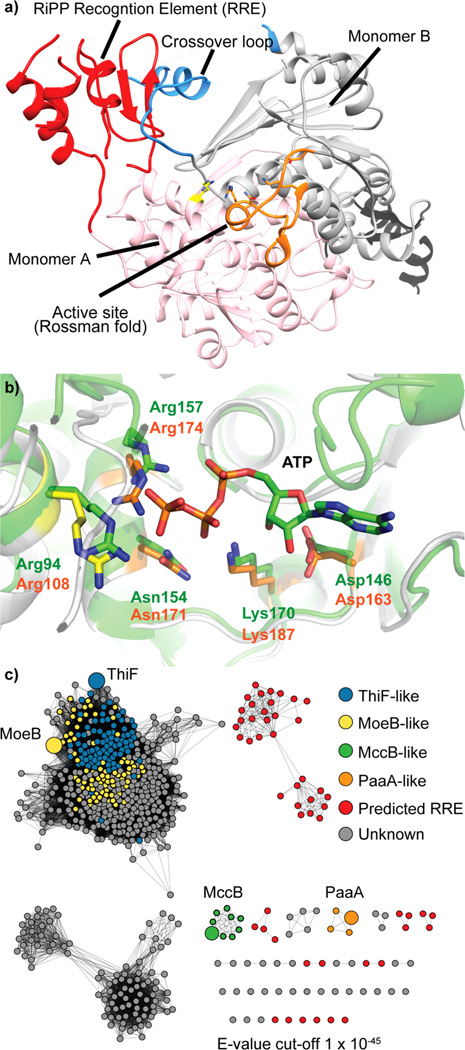
The existence of additional decarboxylative activity of PaaA, as well as the key role of the follower peptide, prompted us to examine PaaA by means of X-ray crystallography. The apo-structure of PaaA was determined at 2.9 Å resolution. Similar to MccB and the cyanobactin heterocyclase, TruD, the asymmetric unit of PaaA is comprised of a homodimer.14,22 Each homodimer further has two parallel, two-domain active units: an adenylation domain (residues 100–281, 321–371) and a smaller, globular domain (1–98), with similarity to recently identified RiPP precursor peptide recognition elements or RREs.8 Overall, the adenylation domain exhibits close structural similarity to the MccB structure, with an RMSD of 1.8 Å over 232 equivalent Cα positions. As in MccB, each half includes a five stranded β-sheet that forms a concave peptide-binding surface, leading from the RRE into a Rossman-fold ATP-binding cleft. Overlay with the ATP-bound structure of MccB allows identification of key ATP-binding residues (Figure 4b), and accordingly, an R174A mutation substantially diminished PaaA activity while K187A completely abrogated substrate turnover (see SI Figure S5). PaaA is also characterized by a wider and more accessible active site, relative to MccB: estimation of the cavity volume by the method of Voss et al. shows the PaaA active site to be almost twice as large as MccB (1908 and 1100 Å3, respectively).13,23 Presumably, the larger active site allows for threading of the follower peptide, as well as greater movement of the substrate during modification. Indeed, the active site further opens up on the backside of PaaA into a relatively shallow groove, which could accommodate the follower peptide.
Figure 4.
Structure of PaaA and sequence similarity network (SSN) of PaaA homologues (a). The two subunits are shown in cartoon representation in pink (monomer A) and gray (monomer B). Dashed lines represent connections between sequences not visible in the electron density. RiPP recognition domains are indicated in red (A) and black (B) and ATP binding residues in orange (B) and yellow (A). (b) Overlay of PaaA apo (orange and yellow, PDB: 5FF5) and MccB-ATP complex (green, PDB: 3H5N) ATP-binding residues, showing conserved residues and orientation. (c) SSN based on PaaA homology.
The presence of an RRE domain distinguishes both MccB and PaaA from the broader family of E1-like activating enzymes such as MoeB and ThiF. In the MccB structure, the N-terminus RRE domain of one subunit interacts with a “crossover loop” from the other, to form the peptide clamp domain. We could resolve a similar cooperative architecture in the structure of PaaA, but observed substantial disorder relative to the same motif in MccB (Figure 4a and SI Figure S7). We anticipate that binding of PaaP organizes and stabilizes the RRE and feeds the prepropeptide into the active site. This stabilizing effect could be confirmed by limited proteolysis of PaaA in both the presence and absence of PaaP (see SI Figure S8). PaaP significantly protects PaaA from both tryptic and chymotryptic digestion, suggesting a dynamic interaction between enzyme and substrate.
The E1 superfamily is large, even within prokaryotes alone; a sequence similarity network (SSN)14,15,24 based on PaaA homology at an e-value cutoff of 10−45 shows several significant subfamilies (Figure 4c). ThiF, MoeB, and MccB are all present in the SSN, with ThiF and MoeB, mutually defining a large cluster. There is a clear bimodal distribution in sequence length of the predicted homologues; a large group is comprised of proteins with approximately 250 aa, like ThiF (253 aa) and MoeB (249 aa) and a second group of proteins, roughly 370 aa in size, of which PaaA and MccB are members. Further inspection reveals that the additional 120 aa of the latter group are, in all cases, predicted to contain an RRE, suggesting the broader adaptation of E1-like chemistry in the post-translational modification of short peptides. Given the high degree of structure level homology between PaaA, MccB, ThiF, and MoeB catalytic domains and the broad divergence in their chemistry (SI Figure S9), these uncharacterized E1-RiPP fusions could harbor untapped leader-peptide directed chemistry.
In conclusion, we have characterized a new RiPP enzyme, PaaA, which catalyzes new chemistry in the double cyclization–condensation and decarboxylation of its precursor peptide. This single enzyme requires both leader and follower sequences for complete transformation. Additionally, structural characterization demonstrates the presence of an RRE, which is stabilized by interaction with the substrate peptide. This RRE combines with a canonical E1-like catalytic domain to effect the new chemistry of PaaA. Bioinformatics suggests that this formula is broadly reproduced in other biosynthetic pathways. Future efforts will focus on dissecting the mechanism of this unique enzymatic transformation and uncovering the chemistry of other members of this family.
Supplementary Material
Acknowledgments
We thank S. Allen, B. Li, and C. Neumann, for informative discussions and S. V. Beer for the gift of pCPP702. We also thank G. Young, P. Sapienza, and K. Koshlap for their help in the acquisition and processing of NMR data. Part of this work was supported by NIH Grant CA098468 (M.R.R.). A.A.B. is a Beckman Young Investigator and acknowledges support by the Arnold & Mabel Beckman Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Coordinates and structure factors are deposited in the Research Collaboratory for Structural Biology (http://www.rcsb.org); PDB ID code is 5FF5; Experimental details, synthetic schemes, and figures (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Ortega MA, van der Donk WA. Cell Chem. Biol. 2016;23:31. doi: 10.1016/j.chembiol.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowers AA, Acker MG, Koglin A, Walsh CT. J. Am. Chem. Soc. 2010;132:7519. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young TS, Dorrestein PC, Walsh CT. Chem. Biol. 2012;19:1600. doi: 10.1016/j.chembiol.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruffner DE, Schmidt EW, Heemstra JR. ACS Synth. Biol. 2015;4:482. doi: 10.1021/sb500267d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deane CD, Melby JO, Molohon KJ, Susarrey AR, Mitchell DA. ACS Chem. Biol. 2013;8:1998. doi: 10.1021/cb4003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piscotta FJ, Tharp JM, Liu WR, Link AJ. Chem. Commun. 2015;51:409. doi: 10.1039/c4cc07778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oman TJ, van der Donk WA. Nat. Chem. Biol. 2010;6:9. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA. Nat. Chem. Biol. 2015;11:564. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. Cell. 2014;158:412. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. Cell. 2014;158:1402. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin M, Liu L, Wright SAI, Beer SV, Clardy J. Angew. Chem., Int. Ed. 2003;42:2898. doi: 10.1002/anie.200351053. [DOI] [PubMed] [Google Scholar]
- 12.Jin M, Wright SAI, Beer SV, Clardy J. Angew. Chem. Int. Ed. 2003;42:2902. doi: 10.1002/anie.200351054. [DOI] [PubMed] [Google Scholar]
- 13.Hochstrasser M. Nature. 2009;458:422. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regni CA, Roush RF, Miller DJ, Nourse A, Walsh CT, Schulman BA. EMBO J. 2009;28:1953. doi: 10.1038/emboj.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roush RF, Nolan EM, Löhr F, Walsh CT. J. Am. Chem. Soc. 2008;130:3603. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 16.Lautenschlager CL. Ph.D. Thesis. Ithaca, NY: Cornell University; 2005. [Google Scholar]
- 17.Sardar D, Pierce E, McIntosh JA, Schmidt EW. ACS Synth. Biol. 2015;4:167. doi: 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto Y, Ito Y, Kato Y, Tsunoda S, Suga H. Chem. Biol. 2014;21:766. doi: 10.1016/j.chembiol.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Crone WJ, Leeper FJ, Truman AW. Chem. Sci. 2012;3:3516. [Google Scholar]
- 20.Huo L, Rachid S, Stadler M, Wenzel SC, Muller R. Chem. Biol. 2012;19:1278. doi: 10.1016/j.chembiol.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Escribano JP, Song L, Bibb MJ, Challis GL. Chem. Sci. 2012;3:3522. [Google Scholar]
- 22.Koehnke J, Mann G, Bent AF, Ludewig H, Shirran S, Botting C, Lebl T, Houssen WE, Jaspars M, Naismith JH. Nat. Chem. Biol. 2015;11:558. doi: 10.1038/nchembio.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voss NR, Gerstein M. Nucleic Acids Res. 2010;38:W555. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL. Biochim. Biophys. Acta, Proteins Proteomics. 2015;1854:1019. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.