Abstract
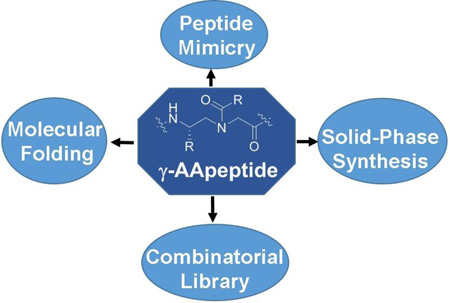
Sequence-specific peptidomimetics are molecules that mimic structure and function of peptides and proteins. With new backbones and molecular frameworks, peptidomimetics are of considerable interest to address challenges encountered in chemical biology and biomedical sciences. Based on the γ-PNA backbone, a new class of peptidomimetics - "γ-AApeptides" were recently developed. Both linear and cyclic γ-AApeptides can be synthesized with high efficiency. Compared with α-peptides, γ-AApeptides are resistant to enzymatic degradation, and amendable to diversification with a variety of chemical groups. Moreover, they could mimic primary and secondary structure, as well as function of peptides, and show promise in biological applications, such as development of new agents combating bacteria, cancer, and Alzheimer’s disease (AD). A few research outcomes of γ-AApeptides are highlighted in this concept article, and a future perspective is also proposed.
Keywords: Peptidomimetics, γ-AApeptides, folding, antimicrobial, combinatorial chemistry, anti-Aβ aggregation
Graphical Abstract
Introduction
Peptidomimetics are designed to capture structure and function of bioactive peptides and proteins, and have found important biological applications in chemical biology and biomedical sciences, such as protein surface mimicry and recognition, modulation of protein-protein interactions, catalysis, etc.[1] Peptidomimetic oligomers developed in the last decade, including β-peptides,[2] peptoids,[3] azapeptides,[4] oligoureas,[5] are excellent examples depicting the features of peptidomimetics. For example, they are built on different unnatural backbones, resistant to enzymatic hydrolysis, and have enormous chemodiversity.[1] They have been designed to mimic α-helix and β-sheet so as to disrupt critical protein-protein interactions. However, as proteins display an endless set of structure and function, there is constant desire for the development of peptidomimetics with new backbones and molecular frameworks. [1] To this end, we recently developed "γ-AApeptides", a new class of peptidomimetics based on the chiral PNA backbone.[6] The terminology of γ-AApeptides is given because they are oligomers of γ-substituted-N-acylated-N-aminoethyl amino acids (Figure 1). In addition to common features of peptidomimetics such as enzymatic resistance and chemodiversity,[6–7] γ-AApeptides have also exhibited some unique properties. In this concept article we introduce strategies for the synthesis of various types of γ-AApeptides, along with a few examples of their structural studies and biological applications. We also provide perspectives on the potential directions of γ-AApeptides in the future research.
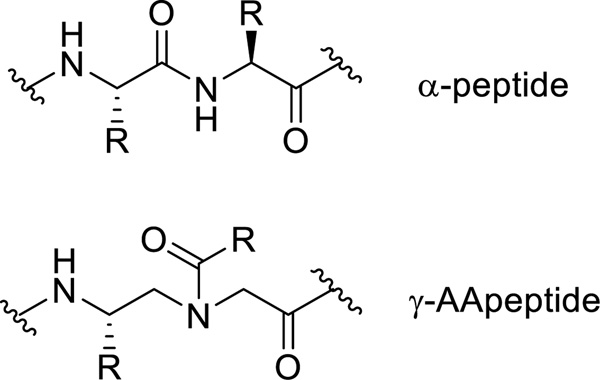
Figure 1.
The chemical structure of α-peptide and γ-AApeptide.
Synthesis of γ-AApeptides
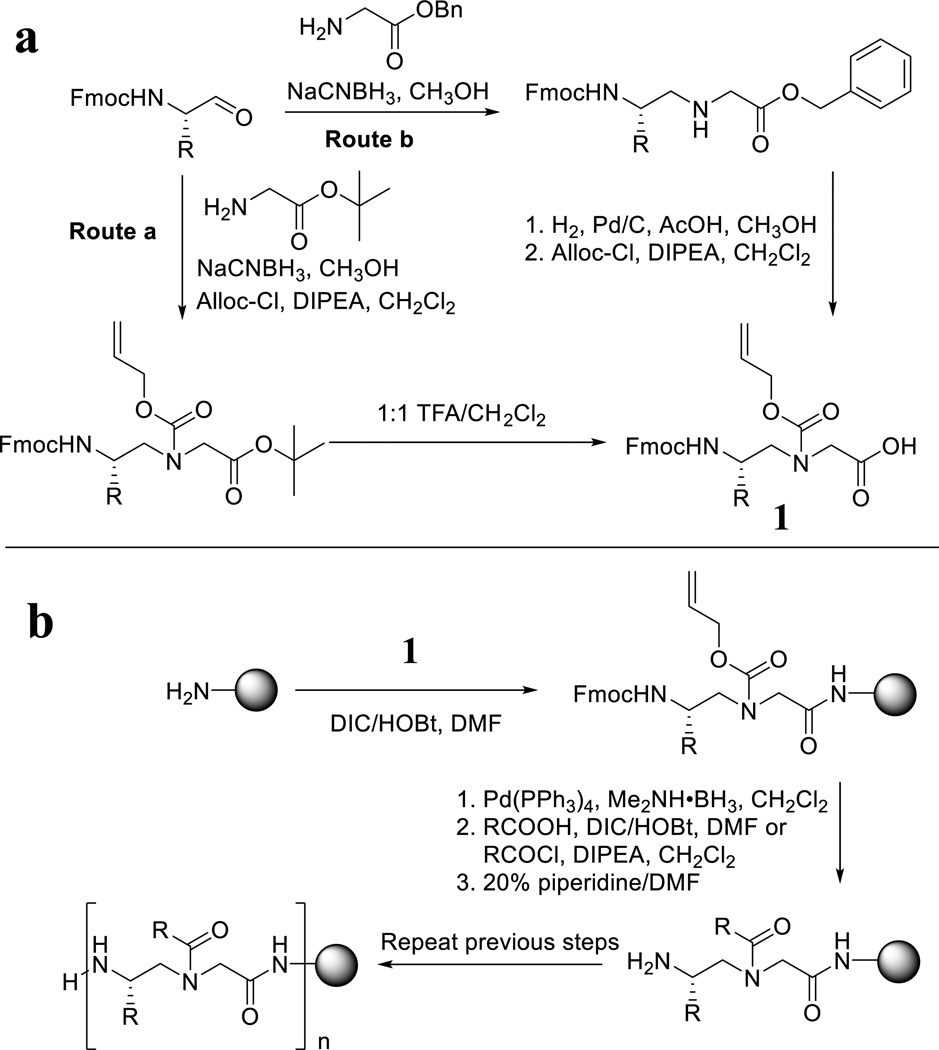
Linear γ-AApeptides are synthesized on the solid phase by adapting the standard protocol of Fmoc-chemistry for peptide synthesis. [8] The Fmoc protected N-alloc γ-AApeptide building block 1 is prepared via the route a or the route b (Figure 2a). In the route a, Fmoc amino aldehyde reacts with t-butyl glycinate through reductive amination, followed by alloc protection and TFA hydrolysis to provide 1. In the route b, Fmoc amino aldehyde reacts with benzyl glycinate to form secondary amine, followed by removal of the benzyl group through hydrogenation, and subsequent alloc protection. Next, the building block 1 is used for the synthesis of linear γ-AApeptides as shown in Figure 2b. After a building block is coupled to the resin (Rink-amide resin is most frequently used), the alloc protecting group is removed. Then the secondary amine on the backbone is acylated with a variety of acids, acyl chlorides, as well as other acylating agents such as sulfonyl chlorides, isocyanates, etc. The cycle is repeated until the desired sequence is assembled on the solid phase, which is subsequently cleaved by 95:2.5:2.5 TFA/TIS/H2O, and purified by HPLC.
Figure 2.
a, synthesis of γ-AApeptide building blocks; b, synthesis of linear γ-AApeptides.
Cyclic peptides and peptidomimetics are known to be more resistant to enzymatic degradation than their linear counterparts. They are also more cell permeable and could be more active towards targets because functional groups in their structures are constrained. Although stability is not a concern for γ-AApeptides, cyclization of γ-AApeptides could enhance the rigidity of the sequences, and thus significantly improve their activity. To date, the synthesis of cyclic γ-AApeptides with both head-to-tail cyclization and head-to-side-chain cyclization have been achieved.
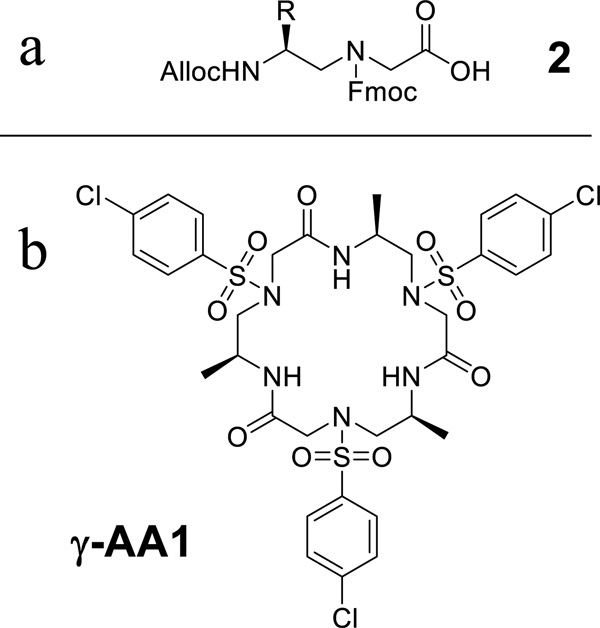
The head-to-tail cyclization is carried on the CTC (2-chlorotrityl chloride) resin. [9] The use of the building block 1 as the first building block attached to the resin was not successful due to the formation of ketopiperazine side products which self-cleave from the solid phase during the Fmoc deprotection step. Alternatively, an N’-Fmoc-N-Alloc γ-AApeptide building block 2 (Figure 3a) is now used as the first building block. Since the removal of the alloc protecting group is under neutral condition, the side reaction of ketopiperazine formation is minimized. After the desired sequence is prepared, it is cleaved from the resin by weak acid cocktail (acetic acid /trifluoroethanol/ dichloromethane 1:1:8) which keeps side chains still protected. Next, the cyclization is achieved with high efficiency in the presence of TBTU, HOBt and DMAP in dichloromethane. Figure 3b shows an example of the head-to-tail γ-AApeptide γ-AA1 which contains three γ-AApeptide building blocks.
Figure 3.
a, the structure of Alloc-N’-Fmoc-N-AApeptide building block 2; b, the structure of the γ-AA1.
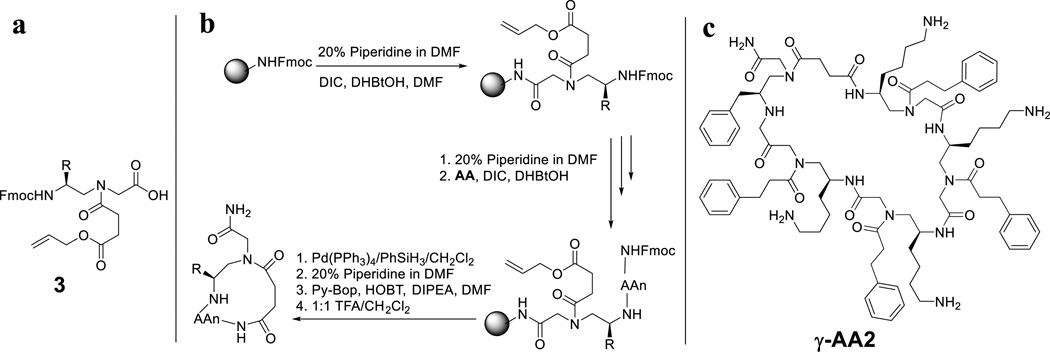
The synthesis of head-to-side-chain cyclic γ-AApeptides can be accomplished on the solid phase without solution reaction as observed in the head-to-tail cyclization (Figure 4). [10] To facilitate on-resin cyclization, a γ-AApeptide building block with the mono-allyl succinate group 3 (Figure 4a) is prepared and used as the first building block attached to the solid phase. After the desired sequence is assembled, the allyl group of 3 is removed by Pd(PPh3)4/PhSiH3, followed by the removal of the Fmoc group on the N-terminal building block (Figure 4b). Finally, the carboxyl group of 3 reacts with the N-terminal amino group to form the cyclic sequence, which is subsequently cleaved by TFA. An example of head-to-side-chain cyclic γ-AApeptide γ-AA2 is shown in Figure 4c.
Figure 4.
a, the structure of γ-AApeptide building block 3 which contains an allyl succinate ester group; b, the synthetic scheme of head-to-side-chain cyclic γ-AApeptides; c, the structure of the γ-AA2.
The structure of γ-AApeptides
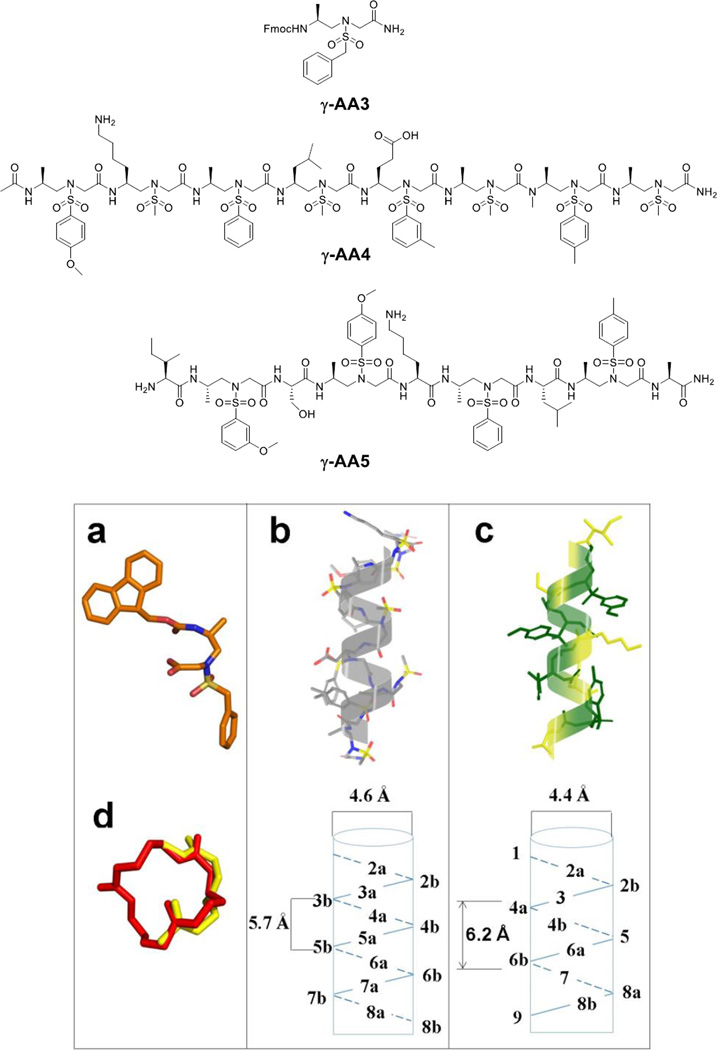
It is known that natural biomacromolecules including nucleic acids and proteins possess a myriad of functions, because these molecules fold into diverse three dimensional structures. Unnatural peptidomimetics have been studied and developed for their folding propensities, in order to understand protein structure-function relationship, and even to evolve new functions.[1] Similarly, the study of the folding conformation of γ-AApeptides has also been carried out. Canonical γ-AApeptides contain tertiary amide bonds which lead to cis/trans isomerization, resulting in significant ambiguity in the elucidation of the 2D-NMR solution structure. However, the obstacle is diminished in a subclass of γ-AApeptides - sulfono-γ-AApeptides, in which tertiary amide moieties are replaced with tertiary sulfonamide moieties (Figure 5). To date, both sulfono-γ-AApeptides[11] and 1:1 α/sulfono-γ-AA hybrid peptides[12] have been investigated for their folding conformations.
Figure 5.
a, the crystal structure of the sulfono-γ-AApeptide γ-AA3; b, NMR structure of the sulfono-γ-AApeptide γ-AA4 in MeOH; c, NMR structure of the 1:1 α/sulfono-γ-AA hybrid peptide γ-AA5 in MeOH; d, the crystal structure of γ-AA1 (red), and its superimposition with a type II β turn (yellow, PDB: 1YCC). Reproduced with permission from ref 9 and 11, Copyright 2015 Royal Society of Chemistry and Wiley.
γ-AA3, a sulfono-γ-AApeptide building block, forms a turn structure, possibly due to the bulkiness of the sulfonamide group (Figure 5a). [11] Both sulfono-γ-AApeptide (γ-AA4, Figure 5b) [11] and 1:1 α/sulfono-γ-AA hybrid peptide (γ-AA5, Figure 5c) [12] adopt right-handed helical conformations in solution according to 2D-NMR analysis, albeit their helical pitch and diameter differ slightly. These sequences contain secondary amide moieties identical to those of α-peptides, and therefore intramolecular hydrogen bonds are speculated to form, which facilitates the helix formation. It is also likely that the sulfonyl groups may directly participate in hydrogen bonding to further stabilize the helical propensity. The folding conformations are also supported by CD and Small Angle X-ray Scattering (SAXS).[11–12] Although more sequences needed to be investigated to prove the helical formation is general, the folding propensity of sulfono-γ-AApeptides are likely to be coherent in this class of peptidomimetics because sequences used in the study bear random side chains.
Cyclic γ-AApeptides are also found to form defined secondary structures. For instance, the head-to-tail cyclic sulfono-γ-AApeptide γ-AA1 forms a turn structure analogous to the type II β turn (Figure 5d). [9]
The biological applications of γ-AApeptides
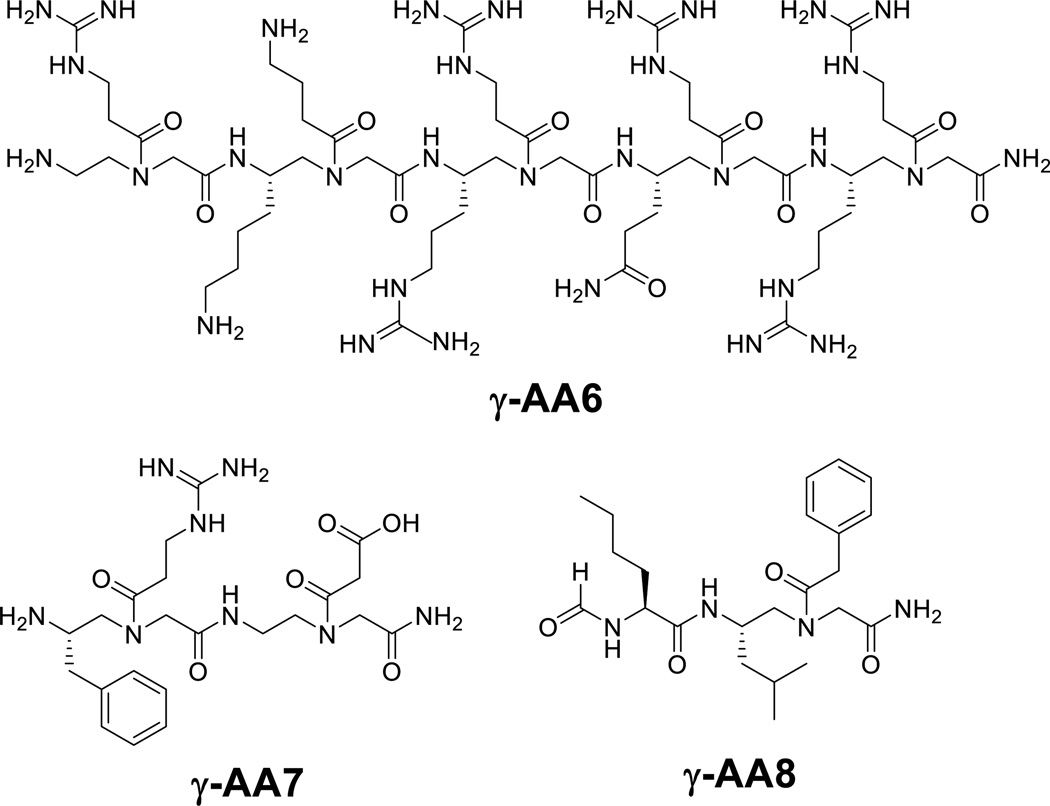
To assess the potential of γ-AApeptides for their application in chemical biology and biomedical sciences, the function of γ-AApeptides was explored recently. To date, γ-AApeptides have been developed to mimic peptides and modulate important biological processes. Our early efforts were focused on the identification of γ-AApeptides that mimic the primary structure of bioactive peptides, in order to demonstrate the proof of principle for the design of γ-AApeptides for biological applications. In those attempts, γ-AApeptides were designed to project functional side chains similar to those of bioactive peptides, and their biological activity was assessed afterwards. For instance, γ-AA6, a sequence bearing functional groups resembling the Tat 48–57 peptide, show capability of HIV binding and membrane translocation akin to the Tat peptide counterpart.[13] γ-AA7 is a peptide designed to mimic the RGD motif, and indeed displays a similar efficacy to c(RGDyK) for glioblastoma tumor-targeted imaging after conjugation with the Cu64-DOTA moiety.[14] It is noted that the γ-AApeptide based RGD mimetics is much more resistant to proteolytic degradation than c(RGDyK).[14] Another example is the γ-AApeptide γ-AA8, which was designed to mimic the short peptide fMLF, showing ability to effectively activate calcium mobilization and trigger the production of ROS and chemotaxis.[15]
Our recent effort has also been extended to studying the capability of γ-AApeptides to mimic three dimensional structures of bioactive peptides. One of the ongoing endeavors is to design γ-AApeptides that can mimic the globally amphipathic structure of host-defense peptides (HDPs) and kill bacteria via membrane disruption.[16] Antimicrobial resistance is recognized as one of the three greatest threats to humankind in the 21st century. Conventional antibiotics generally function on specific targets including bacterial cell wall, ribosome, and nucleic acids, leading to development of resistance in bacteria which evade from the antimicrobial mechanism.[17] It is imperative to search antibiotics with new mechanisms so as to combat antibiotic resistance. HDPs are the first line of defense in the innate immune system of virtually all living organisms, and recently are revisited for their potential as alternative therapeutic strategies. It is interesting that HDPs have a variety of three dimensional structures such as extended structures, helices, and β-sheets, however, they share a common characteristic of adopting globally amphipathic conformations when interacting with bacterial membrane.[18] The amphipathic structures contain both hydrophobic and cationic patches, although the actual secondary structures can vary. It is generally believed that HDPs selectively reach the surface of bacterial membranes (which is always negatively charged) due to electrostatic attraction of their cationic regions, and then disrupt the membranes by their hydrophobic domains. Although no therapeutic agents are devoid of resistance eventually, HDPs may be less prone to the development, because the interactions between bacteria and HDPs are rather biophysical and lack specific membrane or intracellular targets. [19] Nonetheless, HDPs may not be ideal for antibiotic development, as they are susceptible to proteolytic degradation, and also do not have strong activity. Alternatively, peptidomimetics may be viable because they can overcome these drawbacks. As such, a few subclasses of γ-AApeptides have been designed and evaluated for their ability to mimic HDPs and kill drug-resistant bacteria. [20]
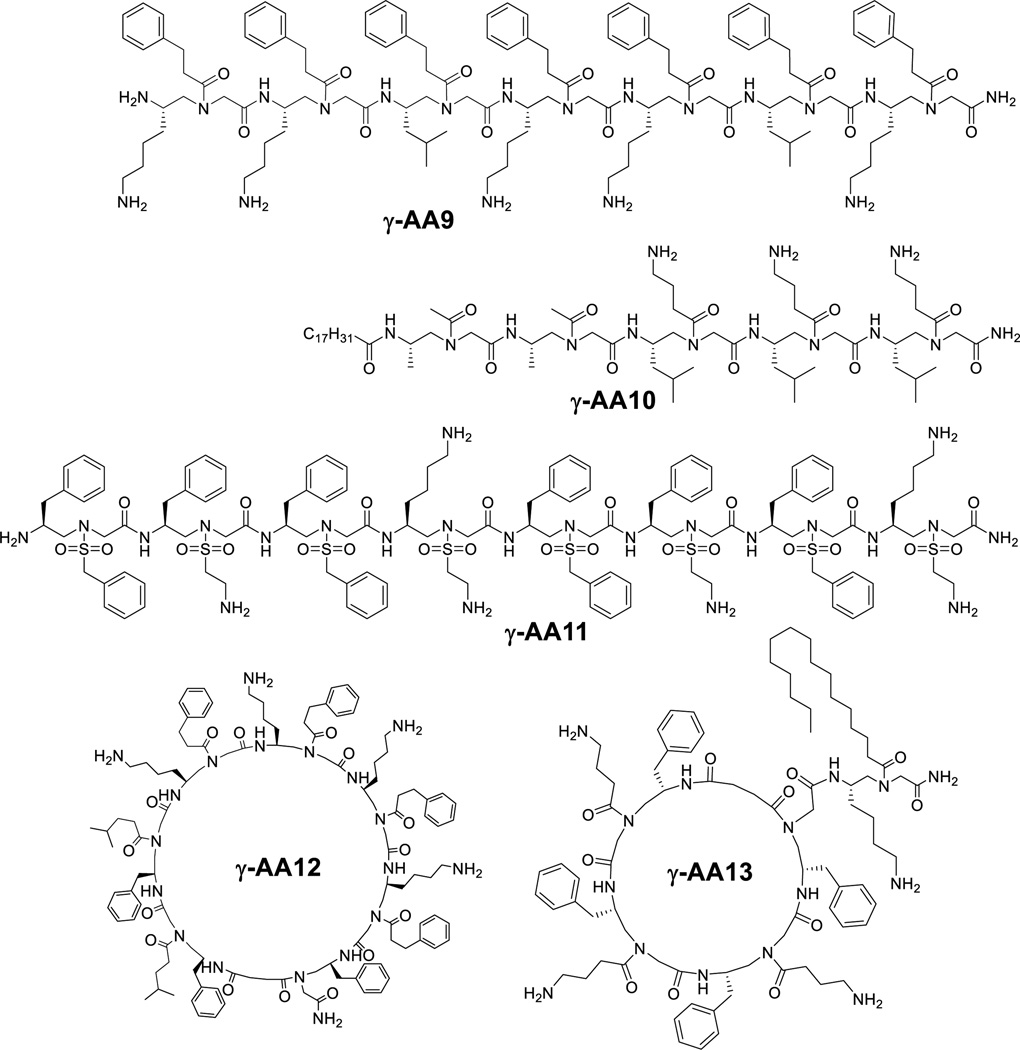
Our initial effort was focused on the design of linear antimicrobial γ-AApeptides (Figure 7). γ-AA9 is such a sequence containing mostly amphiphilic building blocks (containing one hydrophobic and one cationic group in each building block), as well as two hydrophobic building blocks (containing two hydrophobic side chains in each building block).[21] Amphiphilic building blocks were postulated to direct the formation of globally amphipathic structure in the sequence. The hydrophobic building blocks were included to adjust the ratio of hydrophobicity and hydrophilicity. Indeed, γ-AA9, in which two amphiphilic building blocks were replaced with two hydrophobic building blocks (containing two hydrophobic side chains in each building block), show broad-spectrum activities against both Gram-positive and Gram-negative bacteria.[21]
Figure 7.
Structures of antimicrobial γ-AApeptides.
Linear peptides with lipid tails are found widely in nature as antimicrobial peptides, although they may not kill bacteria via the mechanism of membrane disruption observed for HDPs. The lipid tails could enhance the lipophilicity of the peptides, and thus intensify interactions with bacterial membranes. As shown in Table 1, the lipo-linear γ-AApeptide γ-AA10 is even more broadly active than γ-AA9.[22] Interestingly, the unsaturated alkyl tail is found to be more selective towards bacteria than mammalian cells, which may indicate that bacterial membranes are more susceptible to disruption by unsaturated alkyl tails.
Table 1.
Antimicrobial activity of γ-AApeptides.
| Organism | MIC (µg/mL) | ||||
|---|---|---|---|---|---|
| γ-AA9 | γ-AA10 | γ-AA11 | γ-AA12 | γ-AA13 | |
| Gram-negative | |||||
| E. Coli | 5 | 3 | 4 | - | 2 |
| P. aeruginosa | >50 | 3 | 4 | 8 | 5 |
| Gram-Positive | |||||
| MRSE | 5 | 3 | 2 | 2 | 1 |
| E. faecalis | 5 | 4 | 2 | 5 | 3 |
| MRSA | 5 | 3 | 2 | 1 | 2 |
| Hemolysis (HC50) | 300 | >500 | >500 | 150 | 250 |
As some HDPs such as magainin 2 adopt helical amphipathic structures on the bacterial surface, it is conceived that helical sulfono-γ-AApeptides could also be designed to mimic helical HDPs. Indeed, γ-AA11 displays excellent antimicrobial activity (Figure 7, Table 1) against both Gram-positive and –negative bacteria.[7] It is interesting that when the N-terminus of γ-AA11 is capped with the acetyl group, it forms a more defined helical structure, however, its antimicrobial activity is worse. Our result is consistent to previous findings that more defined secondary structure could lead to less selective antimicrobial agents. It should be noted that this sulfono-γ-AApeptide is completely resistant against proteolytic degradation.
In addition to linear γ-AApeptides, we also extend our effort to the development of cyclic antimicrobial γ-AApeptides. Similar to the design of linear sequences, cyclic γ-AApeptides are mainly composed of amphiphilic building blocks, and hydrophobic building blocks are included to fine tune the activity and selectivity. γ-AA12 (Figure 7, Table 1) is such a cyclic γ-AApeptide which show broad-spectrum activity,[10] especially towards Gram-positive pathogens. Akin to lipidated linear γ-AApeptides, lipo-cyclic γ-AApeptides were also prepared to mimic the structure motif of lipo-cyclic peptide antibiotics such as daptomycin and polymyxin. As seen in Figure 7 and Table 1, the lipo-cyclic γ-AApeptide γ-AA13 is even more potent towards Gram-negative pathogens than non-alkylated cyclic γ-AApeptides. It is intriguing that lipo-cyclic γ-AApeptides could modulate immune response and antagonize lipopolysaccharide (LPS) stimulated Toll-like receptor 4 (TLR4) cell signaling, indicating that lipo-cyclic γ-AApeptides could potentially be used as both anti-inflammatory and antimicrobial agents.[23]
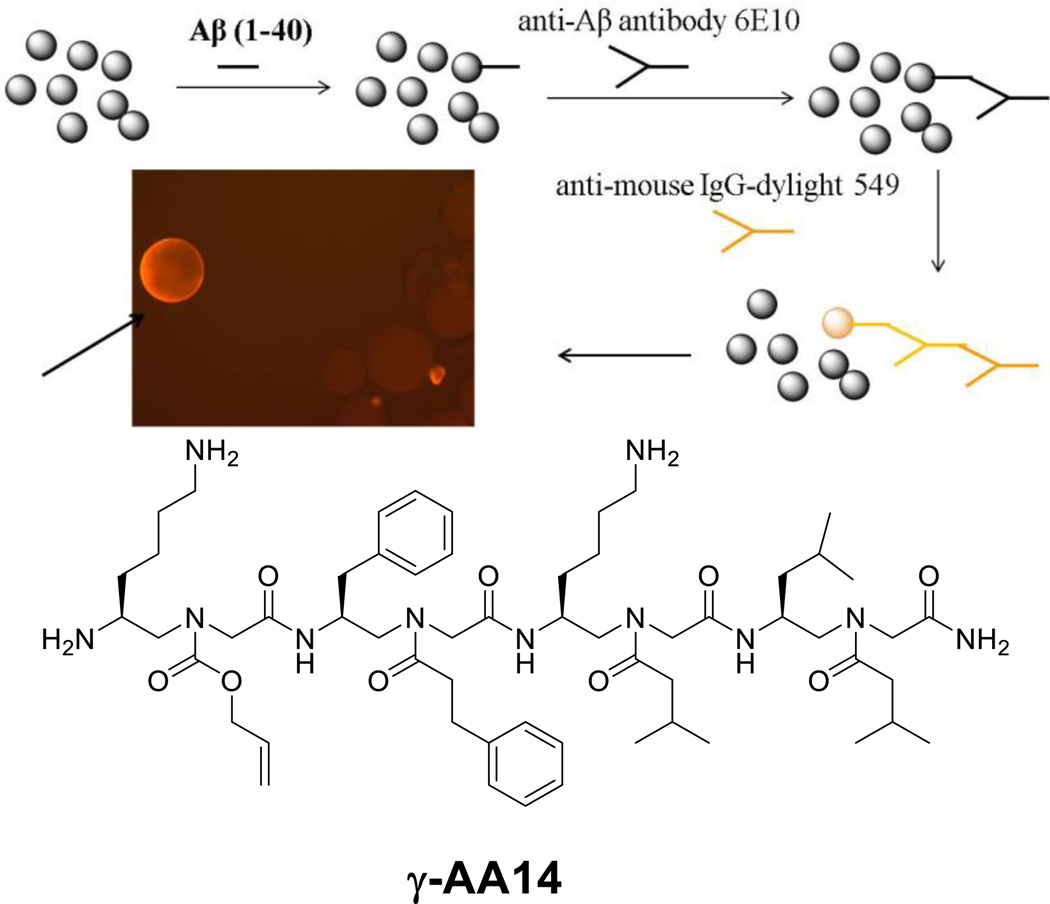
In addition to rational design of γ-AApeptides for the mimicry of peptides, γ-AApeptides are also applicable to the development of combinatorial chemistry, in order to identify molecular ligands that specifically target proteins of interest. In one example, a one-bead-one-compound (OBOC) γ-AApeptide combinatorial library containing 192000 compounds was prepared to select for ligands targeting the Aβ40 peptide, a biomarker that plays a critical role in AD pathogenesis.[24] Beads were first incubated with the Aβ40 peptide, and then subject to anti-Aβ antibody, which was subsequently recognized by the secondary antibody with a fluorescent tag. The putative hits (fluorescent beads) were separated and the sequences were analyzed by MS/MS (Figure 8).[24] One hit γ-AA14 was successfully identified and shown to be at least 100-fold potent as a regular peptide KLVFF, which is a pentamer derived from the hydrophobic core of the Aβ peptide. Indeed, γ-AA14 could prevent and even reverse the aggregation of the Aβ40 peptide. The sequence also exhibits inhibitory activity in cellular assays. The Aβ42 aggregates show considerable toxicity towards N2a neuroblastoma cells. When one equivalent of γ-AA26 was incubated with the Aβ42 peptide, cells almost completely regained their viability, indicating the toxicity of the Aβ42 peptide was removed. It indirectly supports that the aggregation of the Aβ42 peptide was prevented.[24]
Figure 8.
Screening of the OBOC γ-AApeptide library against the Aβ40 peptide. Reproduced with permission from ref 24. Copyright 2014 Royal Society of Chemistry.
In addition to interrogation of Alzheimer’s disease, the OBOC γ-AApeptide library was also used to identify potential anti-cancer agents antagonizing STAT3-mediated cell signaling. The signal transducer and activator of transcription 3 (STAT3) is a transcription factor that is constantly activated in many solid tumors and hematological cancers. Although theoretically STAT3 signaling can be inhibited by blocking either STAT3 dimerization or STAT3-DNA interaction, the latter strategy is much rarer because rational design of agents targeting protein/DNA interface is challenging. [25]
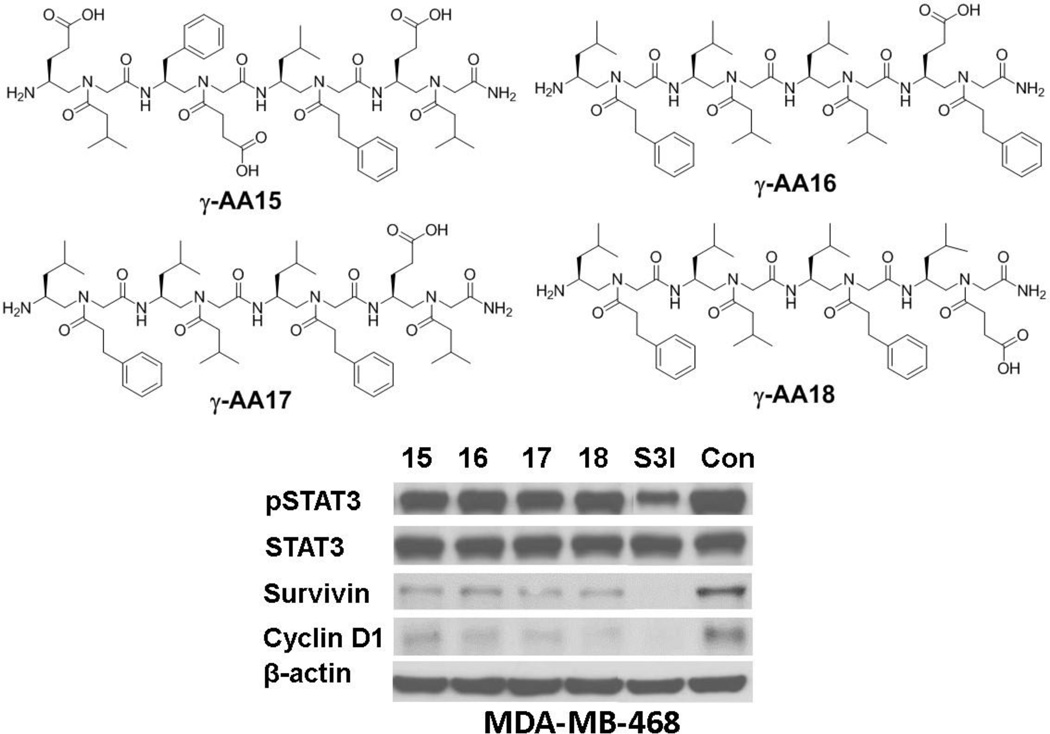
The OBOC library was thus prepared and screened against STAT3 protein using the approach similar to the strategy show in Figure 8. Four sequences γ-AA15-18 were identified (Figure 9). None of these sequences show any activity to inhibit STAT3 dimerization in a fluorescence polarization assay. However, they exhibited considerable activity to disrupt STAT3-DNA interaction in a STAT3-DNA binding filter assay. Furthermore, they were shown to suppress the expression of STAT3 regulated genes including survivin and cyclin D1 in whole cells, suggesting molecules modulating STAT3-DNA interaction could be an alternative approach for the development of potential anti-cancer agents.[25]
Figure 9.
The chemical structure of γ-AA15-18 and their activity on STAT3-mediated cell signaling. Reproduced with permission from ref 25. Copyright 2014 Royal Society of Chemistry.
Conclusion and Outlook
Our studies on γ-AApeptides suggest that this class of peptidomimetics can be easily synthesized. They can fold into defined secondary structures such as helices and turns, and therefore they can be rationally designed to mimic bioactive peptides in the near future. In the meantime, their modular synthesis allows the development of combinatorial library, from which molecular probes and potential drug leads can be identified. More applications of γ-AApeptides are expected to emerge if the following aspects are further addressed. First, more structural studies should be conducted for γ-AApeptides with diverse functional groups, in order to assess if the folding is consistent in the same class of backbones. It is envisioned that proper distribution of hydrophobic, polar and charged groups on the folding scaffold could further stabilize the secondary structure. Thus, the folding principle could be elucidated. In addition, computer modeling maybe employed to guide the rational design. Although some solution structures have been investigated, it is imperative to obtain crystal structures to gain insight into folding propensities. To move forward, in vivo studies may have to be carried out so as to evaluate the biological potential of γ-AApeptides. Combinatorial library of γ-AApeptides can be improved by developing cyclic γ-AApeptide libraries, which is expected to improve the binding affinity and specificity of ligands towards targets of interest.
Figure 6.
γ-AApeptides that mimic primary structures of bioactive peptides.
Acknowledgments
We thank financial support from NSF 1351265 and NIH 1R01GM112652-01A1.
REFERENCES
- 1.Goodman CM, Choi S, Shandler S, DeGrado WF. Nat. Chem. Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 3.Zuckermann RN, Kodadek T. Curr. Opin. Mol. Ther. 2009;11:299–307. [PubMed] [Google Scholar]
- 4.Broadrup RL, Malachowski WP. Abstr. Pap. Am. Chem. S. 2004;228:U92–U92. [Google Scholar]
- 5.Fischer L, Semetey V, Lozano JM, Schaffner AP, Briand JP, Didierjean C, Guichard G. Eur. J. Org. Chem. 2007:3944–3944. [Google Scholar]
- 6.Niu Y, Hu Y, Li X, Chen J, Cai J. New J. Chem. 2011;35:542–545. [Google Scholar]
- 7.Li Y, Wu H, Teng P, Bai G, Lin X, Zuo X, Cao C, Cai J. J. Med. Chem. 2015;58:4802–4811. doi: 10.1021/acs.jmedchem.5b00537. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Teng P, Cai J. Eur. J. Org. Chem. 2014:1760–1765. [Google Scholar]
- 9.Wu H, She F, Gao W, Prince A, Li Y, Wei L, Mercer A, Wojtas L, Ma S, Cai J. Org. Biomol. Chem. 2015;13:672–676. doi: 10.1039/c4ob02232g. [DOI] [PubMed] [Google Scholar]
- 10.Wu H, Niu Y, Padhee S, Wang RE, Li Y, Qiao Q, Ge B, Cao C, Cai J. Chem. Sci. 2012;3:2570–2575. [Google Scholar]
- 11.Wu H, Qiao Q, Hu Y, Teng P, Gao W, Zuo X, Wojtas L, Larsen RW, Ma S, Cai J. Chem. Eur. J. 2015;21:2501–2507. doi: 10.1002/chem.201406112. [DOI] [PubMed] [Google Scholar]
- 12.Wu H, Qiao Q, Teng P, Hu Y, Antoniadis D, Zuo X, Cai J. Org. Lett. 2015;17:3524–3527. doi: 10.1021/acs.orglett.5b01608. [DOI] [PubMed] [Google Scholar]
- 13.a) Niu Y, Bai G, Wu H, Wang RE, Qiao Q, Padhee S, Buzzeo R, Cao C, Cai J. Molecular pharmaceutics. 2012;9:1529–1534. doi: 10.1021/mp300070w. [DOI] [PubMed] [Google Scholar]; b) Niu Y, Jones AJ, Wu H, Varani G, Cai J. Org. Biomol. Chem. 2011;9:6604–6609. doi: 10.1039/c1ob05738c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Niu Y, Hong H, Wu H, Zhang Y, Engle J, Barnhart T, Cai J, Cai W. Chem. Commun. 2012;48:7850–7852. doi: 10.1039/c2cc33620k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, Cheng N, Wu H, Kang S, Ye RD, Cai J. Chem. Bio. Chem. 2014 doi: 10.1002/cbic.201402396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock RE, Sahl HG. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 17.Marr AK, Gooderham WJ, Hancock RE. Curr. Opin. Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Brown KL, Hancock REW. Curr. Opin. Immunol. 2006;18:24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Hancock REW, Brown KL, Mookherjee N. Immunobiology. 2006;211:315–322. doi: 10.1016/j.imbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 20.a) Niu Y, Wang RE, Wu H, Cai J. Future Med Chem. 2012;4:1853–1862. doi: 10.4155/fmc.12.111. [DOI] [PubMed] [Google Scholar]; b) Niu Y, Wu H, Li Y, Hu Y, Padhee S, Li Q, Cao C, Cai J. Org. Biomol. Chem. 2013;11:4283–4290. doi: 10.1039/c3ob40444g. [DOI] [PubMed] [Google Scholar]
- 21.Niu Y, Padhee S, Wu H, Bai G, Harrington L, Burda WN, Shaw LN, Cao C, Cai J. Chem. Commun. 2011;47:12197–12199. doi: 10.1039/c1cc14476f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu Y, Padhee S, Wu H, Bai G, Qiao Q, Hu Y, Harrington L, Burda WN, Shaw LN, Cao C, Cai J. J. Med. Chem. 2012;55:4003–4009. doi: 10.1021/jm300274p. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Smith C, Wu H, Padhee S, Manoj N, Cardiello J, Qiao Q, Cao C, Yin H, Cai J. ACS Chem. Biol. 2014;9:211–217. doi: 10.1021/cb4006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Li Y, Ge B, Niu Y, Qiao Q, Tipton J, Cao C, Cai J. Chem. Commun. 2014;50:5206–5208. doi: 10.1039/c3cc46685j. [DOI] [PubMed] [Google Scholar]
- 25.Teng P, Zhang XL, Wu HF, Qiao Q, Sebti SM, Cai JF. Chem. Commun. 2014;50:8739–8742. doi: 10.1039/c4cc03909b. [DOI] [PMC free article] [PubMed] [Google Scholar]