Abstract
The rational design of inhibitors of the bHLH-ZIP oncoprotein c-Myc is hampered by a lack of structure in its monomeric state. We describe herein the design of novel, low-molecular-weight, synthetic α-helix mimetics that recognize helical c-Myc in its transcriptionally active coiled-coil structure in association with its obligate bHLH-ZIP partner Max. These compounds perturb the heterodimer’s binding to its canonical E-box DNA sequence without causing protein–protein dissociation, heralding a new mechanistic class of “direct” c-Myc inhibitors. This model was corroborated by additional biophysical methods including NMR spectroscopy and surface plasmon resonance. Several compounds demonstrated a 2-fold or greater selectivity for c-Myc–Max heterodimers over Max–Max homodimers with IC50 values as low as 5.6 µM. Finally, these compounds inhibited the proliferation of c-Myc-over-expressing cell lines in a concentration-dependent manner that correlated with the loss of expression of a c-Myc-dependent reporter plasmid despite the fact that c-Myc–Max heterodimers remained intact.
Keywords: c-Myc, Max, Protein–Protein Interaction, α-Helix Mimetic, 10074-G5, JY-3-094, JKY-2-169, JQ-1
Graphical abstract
Introduction
c-Myc is a basic helix-loop-helix leucine zipper (bHLH-ZIP) transcription factor that, in addition to being oncogenic when over-expressed, affects many transformation-related processes such as proliferation, apoptosis, differentiation and metabolism.1–10 As an intrinsically disordered protein, c-Myc becomes transcriptionally functional only after heterodimerizing with its obligate bHLH-ZIP partner Max to assume a coiled-coil structure that recognizes the E-box sequence 5’-CACGTG-3’.11 Indeed, this event is required for all known biological functions of c-Myc, including its oncogenic activity.1–5,12 c-Myc dysregulation is associated with most human cancers, including lung, pancreatic and colorectal cancers, as well as leukaemias and lymphomas,13–19 and numerous murine cancer models are c-Myc-dependent, even when c-Myc is not the primary oncogenic driver.20–22 Together, these studies suggest that c-Myc inhibition, either direct or indirect (for example, via inhibition of the BET bromodomains BRD2–423–27), is a viable therapeutic strategy towards the development of new and targeted antineoplastics. Indeed, both genetic and pharmacologic inhibition of c-Myc, or its closely-related cousin N-Myc, in vivo have resulted in tumour regression and prolonged survival.22,28,29 Moreover, despite the widespread expression of c-Myc by normal proliferating cells, recent studies have demonstrated that long-term, whole-body genetic silencing of c-Myc in vivo leads to remarkably mild and reversible side effects,30–33 providing further rationale that c-Myc inhibition is a clinically feasible strategy to expand the anti-cancer drug arsenal.
In contrast to the bromodomain proteins BRD2–4 that exhibit well-defined, acetyl-lysine binding sites that are tractable targets for small-molecule drug design and provide an indirect approach to the inhibition of oncogenic c-Myc function,23–27 the rational design of direct c-Myc inhibitors is complicated by the intrinsic disorder of the bHLH-ZIP domain, although some progress has been made.34–49 Several of the more potent compounds that have been identified bind to distinct segments of this region and cause highly localized distortions that prevent productive interaction with Max’s bHLH-ZIP domain.45,46 For example, 10058-F4 (1) binds c-Myc402–410, whilst 10074-G5 (2) binds c-Myc363–381. However, these compounds generally exhibit only low affinities to c-Myc, double-digit micromolar IC50 values for the prevention of c-Myc–Max heterodimerization in vitro and similar activities in cellular proliferation assays. Attempted optimizations of 2 (for example, JY-3-094 (3) and its ester prodrugs) and 10058-F4 have met with limited success, and new leads are urgently required. In contrast to monomeric c-Myc, the c-Myc–Max heterodimer exists as a structured coiled coil with ~70% α-helical content that increases to 84% upon DNA binding.50 We, therefore, reasoned that molecules appropriately crafted to recognize this α-helical content might perturb the protein–protein interaction (PPI). Precedent for this approach has been described by Hamilton wherein synthetic α-helix mimetics have successfully disrupted the assembly of multiple helices involved in HIV-1 infection51 and abrogated the aggregation of transient helical forms of amyloid52 that would otherwise lead to amyloid fibrillogenesis. More generally, we53–55 and others56,57 have demonstrated that synthetic α-helix mimetics are effective inhibitors of a range of helix-mediated PPIs in which only one of the protein partners uses an α-helical recognition domain, including Bcl-xL–Bak, Mcl-1–Bim and HDM2–p53.58 In light of these observations, we hypothesized that rationally engineered α-helix mimetics might disrupt the coiled coil of the c-Myc–Max heterodimer, thus providing a novel therapeutic strategy to inhibit the oncogenic function of c-Myc.
Results
Design
We have previously shown there to be at least three independent small-molecule binding sites on the c-Myc bHLH-ZIP domain.45,46 We have also recently completed a structure–activity relationship (SAR) analysis of the direct c-Myc inhibitor 2,47 which binds one of these sites centered around residues 363–381 corresponding to the junction between the basic domain and helix 1. NMR analysis of the interaction of 2 with a c-Myc363–381 synthetic peptide allowed the delineation of a more refined binding site, and our SAR work was largely in agreement with this model. Interestingly, NOESY experiments indicated that 2 induced helicity in the c-Myc363–381 peptide. The successful targeting of c-Myc363–381 with congeners of 2 coupled with c-Myc’s acquisition of considerable helicity upon heterodimerization with Max suggests that synthetic α-helix mimetics designed to recognize this region of c-Myc in its helical form might perturb the c-Myc–Max heterodimer and impair DNA binding. This may be envisaged to occur either through directly interfering with the construction of the c-Myc–Max interface when c-Myc is mostly helical and complex formation is almost complete or through disruption of c-Myc’s HLH motif within the c-Myc–Max heterodimer. Accordingly, we designed the α-helix mimetic 4 depicted in Figure 1A. Analogous to our oligoamide-based α-helix mimetics of the Bak-BH3 α-helix in which the side chains of each of the three subunits mimic the i, i + 3/4 and i + 7 side chains on one face of the native helix,53 the hydrophobic R1 and R2 groups were designed to recognize Phe374 (i) and Leu377 (i + 3), respectively, of helical c-Myc in late stage complex assembly. Alternatively, these hydrophobic groups may recognize the tetramethylene side-chain of Lys371 (i) and Phe375 (i + 4) of the assembled complex. Since this hydrophobic domain is flanked by arginines Arg366/Arg367/Arg372 and Arg378 at the N- and C-terminal ends, respectively, we incorporated groups into the periphery of 4 that would complement these basic side chains. For inspiration, we looked to previous small-molecules that also bind this region: the electron rich nitro group of 2 is believed to interact with Arg366, Arg367 and Arg372,45,46 whereas the carboxylic acid in the second-generation 2 congener JY-3-09447 possibly interacts with Arg378. Since the nitro group of 2 and that of JY-3-094 along with its carboxylic acid were instrumental in furnishing the small-molecules with c-Myc inhibitory activities, we, therefore, selected a nitro group to recognize Arg366, Arg367 and/or Arg372 and a carboxylic acid to recognize Arg378. This would allow the bis-benzamides to be prepared from the readily available starting material 3-fluoro-4-nitrobenzoic acid (2). In addition, it was anticipated that the carboxylic acid would serve to promote the solubilities of the helix mimetics. A putative binding mode of target molecule 4da (“JKY-2-169”) is given in Figure 1B.
Figure 1.
A. A generic α-helix mimetic designed to recognize the Arg366–Arg378 region of c-Myc in the helical c-Myc–Max heterodimer. R1 and R2 are hydrophobic groups, such as isobutyl and benzyl (see Scheme 1). Bubbles with two sets of residues indicate side chains targeted (outside parentheses) at the c-Myc–Max interface in late stage complex assembly and (inside parentheses) at the HLH region of c-Myc in the c-Myc–Max complex. B. Chemical structure of 4da (“JKY-2-169”). C. Putative binding mode of compound 4da indicating the region of the c-Myc–Max PPI to be targeted by synthetic α-helix mimetics (PDB ID: 1NKP). The binding site was delineated by Arg372, Phe374, Leu377 and Arg378. D. Zoom-in of binding mode. Computational modeling was achieved with Molegro Virtual Docker. Figure 1B was generated with PyMOL: red = c-Myc; blue = Max; green = DNA; cyan, coloured by atom type = c-Myc residues interacting significantly with ligand; yellow, coloured by atom type = 4da (“JKY-2-169”).
Chemistry
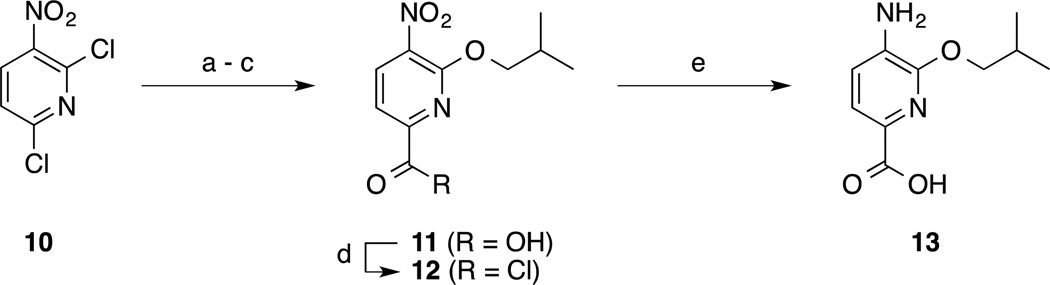
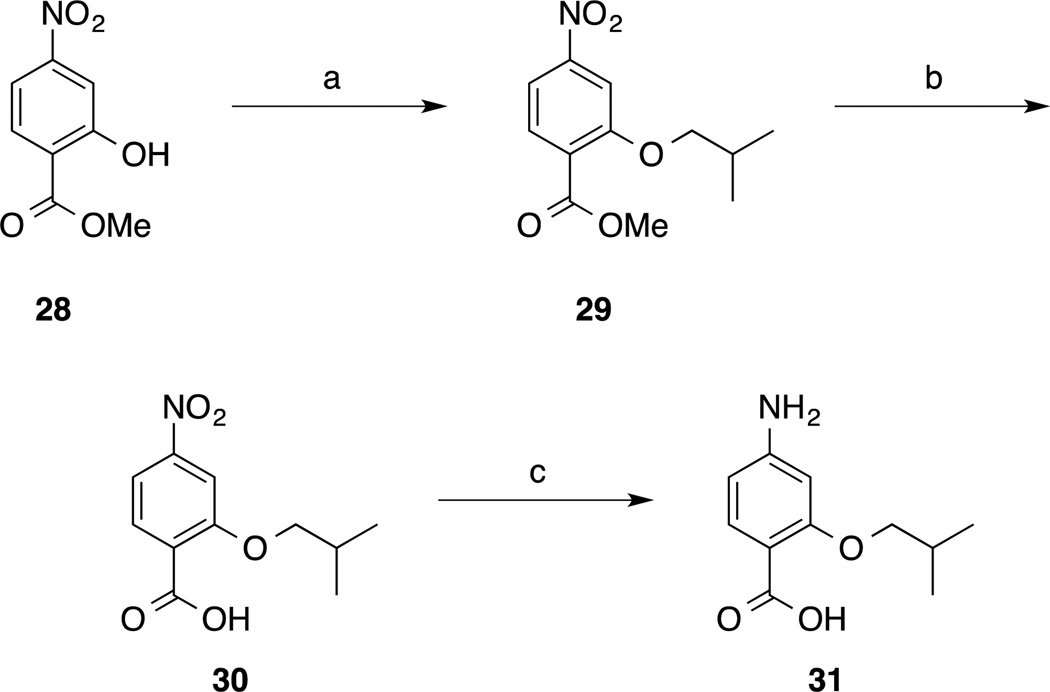
Target molecules 4 were readily generated in a short, convergent synthesis. Subunits were synthesized as described in Schemes 1 and 2. The benzene-based subunits were synthesized starting from 3-fluoro-4-nitrobenzoic acid (5). Nucleophilic aromatic substitution (SNAr) reactions of 5 with either alcohols R1OH or R2OH (R1 and R2 group identities given in Scheme 3) with an excess of NaH furnished the 3-alkoxy derivatives 6. Treatment of acids 6 with oxalyl chloride delivered acid chlorides 7, whilst reduction by catalytic hydrogenation or tin (II) chloride yielded anilines 8. Esterification of acids 6 followed by nitro group reduction delivered anilines 9. Two pyridine-based subunits were also prepared (Scheme 2). Briefly, 2,6-dichloro-3-nitropyridine (10) was regioselectively 2-isobutoxylated, and then the 6-chloro group was converted to a carboxylic acid (compound 11) via a two-step Stille reaction with tributyl(vinyl)tin followed by oxidative cleavage of the newly installed vinyl group. Acid chloride generation or nitro group reduction as before then furnished subunits 12 and 13, respectively. The bis-benzamides 4, 14–16 were then prepared by condensations of acid chlorides 7 and 12 with anilines 8, 9 and 13 in the presence of N,N-dimethylaniline as base (Scheme 3). In order to determine the importance of the R1 and R2 side-chains, bis-benzamides 4aj, 4ja and 4jj were also prepared according to Scheme 3 but in which at least one of the subunits carried no side-chain (i.e. 4-aminobenzoic acid and/or 4-nitrobenzoyl chloride). To investigate the significance of the nitro group, 3-isobutoxybenzoyl chloride was coupled to 4-amino-3-isobutoxybenzoic acid (8a) to afford compound 17, again essentially as described in Scheme 3. Additional analogues to test the importance of the nitro group were prepared as shown in Scheme 4. Specifically, the nitro group of compounds 16, 4aa and 4ea was reduced with tin (II) chloride to deliver anilines 18, 19 and 20, respectively. Acylation of 19 with ethyl chlorooxoacetate or diglycolic anhydride generated compounds 21 and 22, respectively, and then the former underwent saponification to deliver compound 23 as shown in Scheme 5. Modifications to the C-terminus are depicted in Scheme 6 where the proximity of the acidic group, as well as the number of acidic groups, would allow further investigation of the structure-activity relationships (SAR) of these compounds, as in compounds 26 and 27. Finally, relocation of the side chains to the 2-position of the benzene ring was effected starting from commercially available compound 28 (Scheme 7). Appropriate subunits (4aa, 30 and 31) were then coupled together using similar chemistry to that described in Scheme 3 to afford target molecules 32–34.
Scheme 1.
(a) R1OH or R2OH, NaH, THF, RT, 0 °C to RT, 2 h; (b) (COCl)2, cat. DMF, CH2Cl2, RT, 2 h; (c) H2, 10% Pd/C, EtOH, RT, 1–5 h; or SnCl2.2H2O, EtOAc, 50 °C, 3 h; (d) SOCl2, MeOH, 60 °C, 6 h; or MeI, K2CO3, DMF, RT, 4 h.
Scheme 2.
(a) Isobutanol, NaH, THF, RT, 0 °C to RT, 4 h; (b) Bu3SnCH=CH2, Pd(PPh3)4, toluene, 110 °C, 5 h; (c) KMnO4, H2O/acetone, RT, 6 h; (d) (COCl)2, cat. DMF, CH2Cl2, RT, 2 h; (e) SnCl2.2H2O, EtOAc, 50 °C, 3 h.
Scheme 3.
(a) N,N-dimethylaniline, acetone, RT, 0 °C to RT, 16 h.
Scheme 4.
(a) H2, 10% Pd/C, EtOH, RT, 1–5 h.
Scheme 5.
(a) ClCOOEt, pyridine, CH2Cl2, 0 °C to RT, 16 h; or diglycolic anhydride, Et3N, DIPEA, 50 °C, 16 h; (b) LiOH.H2O, THF/MeOH/H2O, 3:1:1, 24 h, RT.
Scheme 6.
(a) Glycine tert-butyl ester•HCl or di-tert-butyliminodiacetate, HBTU, DIPEA, DMF, RT, 16 h; (b) 20% TFA/CH2Cl2, RT, 3 h.
Scheme 7.
(a) Isobutyl iodide, K2CO3, DMF, 50 °C, 16 h; (b) NaOH, THF/MeOH/H2O, 3:1:1, RT, 1 h; (c) H2, 10% Pd/C, EtOH, RT, 16 h.
Biology
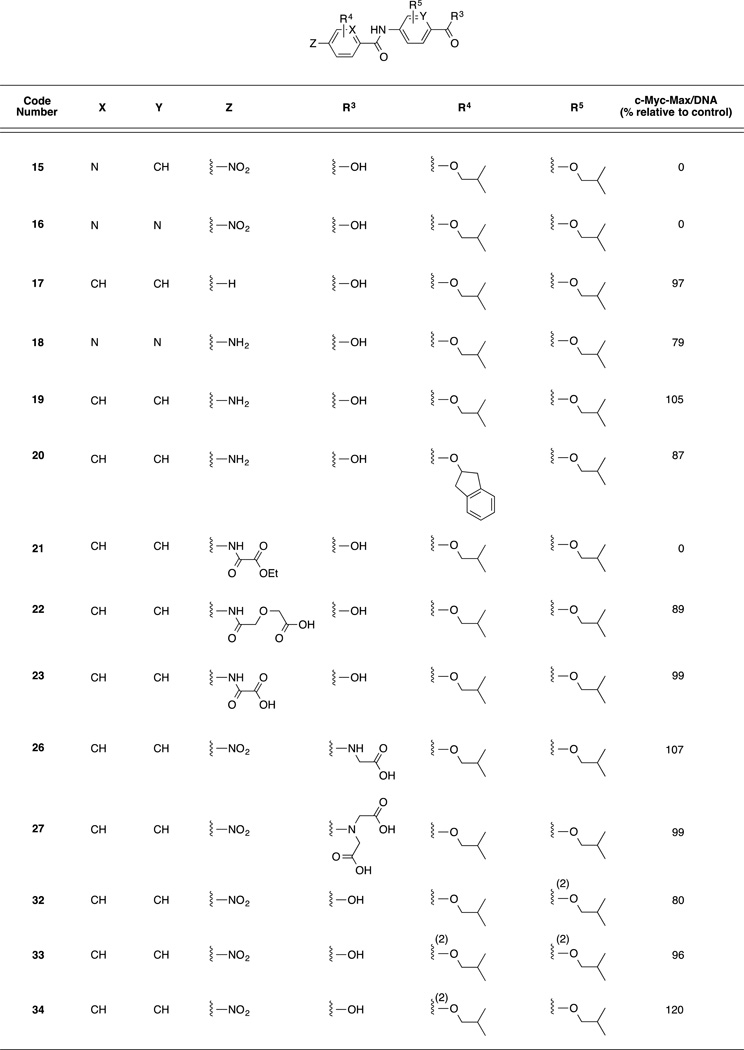
Initially, a focused library of derivatives of 4 was prepared, and the compounds were screened in an electrophoretic mobility shift assay (EMSA) at 100 µM concentrations for their abilities to disrupt c-Myc–Max(S)/DNA complexes. The structures of these compounds are presented in Table 1 along with the EMSA screening results, where “0%” indicates complete disruption of the c-Myc–Max/DNA complex relative to the DMSO vehicle control (“100%”). Encouragingly, two compounds, 4aa and 4da (“4da”), from the initial batch of 10 derivatives of 4 demonstrated potent activity in this assay. In addition to 4aa and 4da, the next most active compound, 4bb, also possesses an ionizable carboxylic acid. The remaining seven compounds are methyl ester derivatives and were largely inactive, thus indicating the important contribution of the carboxylic acid.
Table 1.
EMSA screening analysis of abilities of synthetic α-helix mimetics to disrupt the c-Myc–Max/DNA complex at 100 µM (n = 1). For these experiments, we used the 151 residue isoform of Max, which we term Max(S), which binds DNA only in heterodimeric association with c-Myc and not as a homodimer.60 The last column refers to the amount of DNA binding obtained in the presence of the compound relative to that obtained in its absence. See Experimental Section for further information.
Given the promising EMSA screening data in Table 1, we expanded our library of c-Myc–Max disruptors. Although two methyl esters (14ba and 14ca) demonstrated modest inhibitory activities, they exhibited poor solubilities. Hence, we focused largely on the preparation of only carboxylic acid-functionalized compounds; their structures are shown in Tables 2 and 3. All compounds in Table 2 exhibit a nitro group at the N-terminus, and either a carboxylic acid or a methyl ester at the C-terminus. In Table 3, these functional groups were varied somewhat. Also, the scaffold benzene rings were changed to pyridines and the positions of the side-chains was investigated. We again used an EMSA-based screening assay to evaluate the second group of helix mimetic-based putative c-Myc inhibitors (full structures in SI). From a total of 31 tested compounds, we identified an additional four inhibitors (4ca, 15, 16 and 21) that produced complete disruption of the c-Myc–Max/DNA binding at 100 µM (Tables 2 and 3). Although this screen was performed only once, it is tempting to speculate that the nature and position of the hydrophobic side-chains are critical to activity (compare the data for 4ac, 4ca and 4jj, and 4ca with 4ga, and also 4aa with 32–34). A carboxylic acid appears important at the C-terminus (for example, compare 4aa and 14aa), as does a nitro group at the N-terminus (for example, compare 4aa with 17 and 19, where the nitro group has been replaced with a hydrogen atom or an amino group, respectively). Finally, replacement of the scaffold benzene rings with more hydrophilic pyridines seems to be tolerated (compare 4aa with 15 and 16).
Table 2.
EMSA-based screening results of the disruption of c-Myc–Max(S)/DNA binding at 100 µM of synthetic α-helix mimetic (n = 1).
Table 3.
EMSA-based screening results of the disruption of c-Myc–Max(S)/DNA binding at 100 µM of synthetic α-helix mimetic (n = 1). R4 and R5 groups located at the 3-positions of the aryl rings unless otherwise stated with a (2), indicating the group is at the 2-position.
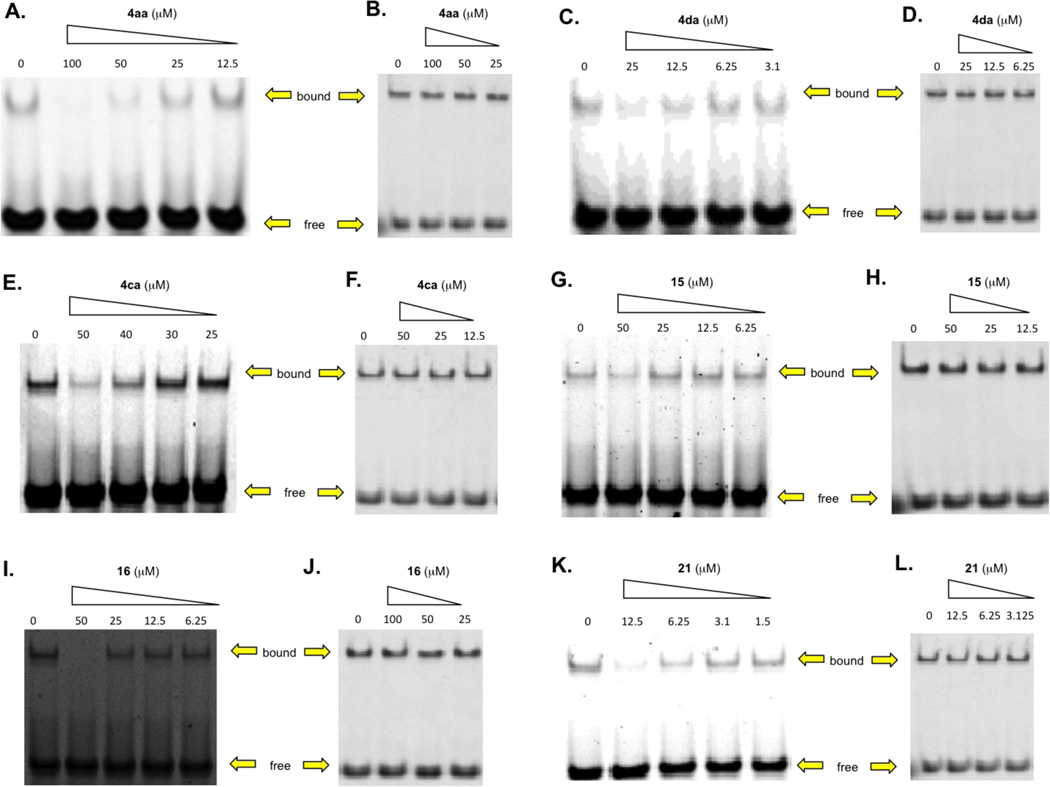
With the above screening data in hand, 4aa, 4da, 4ca, 15, 16 and 21 were evaluated in further detail by confirmatory EMSA analysis. Each compound demonstrated a dose-dependent inhibition of the c-Myc–Max(S)/DNA ternary complex with IC50s < 50 µM (Figure 2, Table 4). The most potent inhibitor 21 (IC50 = 5.6 µM) displays an ethyl aminooxalate, indicating the nitro group in our original design of generic compound 4 is not critical for activity, provided a suitable bioisostere is present. To examine the specificity of these compounds, we also tested their abilities to disrupt DNA binding by Max homodimers in otherwise identical EMSA assays (Figure 2, Table 4). The foregoing experiments capitalized on the fact that the 151 residue “short” isoform of Max [Max(S)] only binds DNA as a heterodimer in association with c-Myc. In contrast, the 160 reside “Max(L)” isoform also binds DNA as a homodimer and can therefore be used to assess the specificity of direct Myc inhibitors.59–61 In each case, the six helix mimetics exhibited a two-fold or greater selectivity for c-Myc–Max(S)/DNA complexes over Max(L)–Max(L)/DNA complexes thereby suggesting that these compounds recognize c-Myc rather than Max (Table 4).
Figure 2.
A, C, E, G, I, K: c-Myc–Max(S) EMSA assays for synthetic α-helix mimetics. Recombinant c-Myc353–437 and full-length Max(S), which does not bind DNA as homodimers,59–61 were purified to homogeneity from E. coli and used at a final concentration of 30 nM; B, D, F, H, J, L: control experiments showed that none of the compounds significantly affected DNA binding by Max(L)–Max(L) homodimers, which bind DNA well. Each figure is representative of three independent trials (Supplementary Figure 1).
Table 4.
EMSA-determined disruption of ternary c-Myc–Max(S)/DNA and Max(L)–Max(L)/DNA complexes by synthetic α-helix mimetics. Each IC50 and standard error were calculated based on three independent EMSAs performed for each compound concentration tested (Supplementary Figure 1).
| Code Number | Inhibition (IC50, µM) | |
|---|---|---|
| c-Myc-Max(S)/DNA | Max(L)-Max(L)/DNA | |
| 4aa | 20.2 ± 1.3 | 84.9 ± 12.0 |
| 4da | 11.6 ± 2.3 | >20 |
| 4ca | 43.0 ± 1.7 | 79.0 ± 8.5 |
| 15 | 43.8 ± 2.9 | >100 |
| 16 | 23.8 ± 2.7 | 68.7 ± 4.5 |
| 21 | 5.6 ± 0.7 | >12.5 |
4da recognizes the α-helical structure of c-Myc in the c-Myc–Max(S) heterodimer
We next performed NMR experiments to further our understanding of the mechanism of 4da’s inhibition at an atomic level. For this we utilized homogeneously pure, recombinant 15N-labeled or unlabeled c-Myc and Max(S) proteins. 15N-HSQC spectroscopy allows the observation of interactions between small molecules and a protein of interest over a broad range of affinities. Furthermore, even without residue-specific assignment, the technique provides insight into changes in secondary structures and potentially allows the elucidation of affinities. First, we examined the ability of 4da to interact with the Max(S) homodimer or the c-Myc monomer (Figure 3A and B). Titration of the compound up to 100 µM failed to reveal any observable chemical shift perturbations, thereby indicating that this compound does not bind either of the individual proteins. We then formed the c-Myc–15N-Max(S) heterodimer through the addition of an equimolar amount of unlabeled c-Myc to 15N-Max(S). This led to the expected substantial change in the HSQC spectrum of 15N-Max(S) (Figure 3C). Finally, addition of 4da to the heterodimer led to further and substantial chemical shift perturbations, clearly indicating that the heterodimer is needed to provide a suitable binding site for this inhibitor and that, following 4da addition, the final configuration attained is unlike that of either of the individual proteins alone (Figure 3D). Careful analysis of four isolated peaks suggested a Kd ~ 13 µM for 4da (Figure 3E).
Figure 3.
A) 15N-Max(S) HSQC spectra with (red) and without (black) the addition of 100 µM 4da; B) 15N-c-Myc HSQC spectra with (red) and without (black) the addition of 100 µM 4da; C) heterodimeric complex of 15N-Max(S) (black alone) and with c-Myc (red); D) the c-Myc–15N-Max(S) binary complex (black) with the addition of 100 µM of 4da (red); E) dose response curves of four isolated NMR cross-peak intensities (108.98/8.35, 109.37/8.02, 123.96/7.63, 133.38/7.992 [15N ppm / 1H ppm]). (Analysis was performed based on Equation 1 (NMR Experimental Section)).
This study conclusively shows that 4da is unable to bind these proteins in either of their alternate conformations by themselves (c-Myc monomer or Max(S) homodimers), but clearly interacts with the c-Myc–Max heterodimer. This is particularly important in the case of the Max(S) homodimer, which has an α-helical structure that closely resembles that of the c-Myc–Max heterodimer.11 The binding of 4da to specific epitopes dictated by this heterodimer alters its structure in a manner that does not restore either of the spectra generated by the individual protein partners. Rather, the novel structure indicates that it remains in a heterodimeric form that is unable to bind DNA, as shown in Figure 3C.
Surface plasmon resonance (SPR) indicates 4da binds the c-Myc–Max(S) heterodimer
We next utilized SPR to determine whether 4da behaves in a manner distinct from that of previously described direct c-Myc inhibitors such as 10058-F4 and 2 which interfere with c-Myc–Max dimerization and DNA binding as a consequence of their efficient interaction with the unstructured c-Myc monomer, rendering it incapable of interacting with Max.34 However, they are relatively inefficient at disrupting pre-formed dimers due to the high free energy of protein–protein association.34–38,45–49 An experimental outcome consistent with this previously proposed model is shown in Figure 4. After first establishing conditions under which c-Myc–Max(S) heterodimer binding to an E-box-containing oligonucleotide could be quantified (Figure 4A), we showed that 2 prevented c-Myc–Max(S) oligonucleotide binding significantly better if it was pre-incubated with c-Myc monomer prior to the addition of Max(S) (Figure 4B vs. 4C). In contrast, while 4da was also somewhat better at inhibiting DNA binding if added to c-Myc and Max(S) proteins prior to their heterodimerization, it was much more effective than 2 against pre-formed c-Myc–Max(S) heterodimers (Figure 4D and 4C). In light of our other findings reported herein, particularly those depicted in Figure 3, these results are most consistent with the idea that 4da, rather than binding to the c-Myc monomer, preferentially interacts with the c-Myc–Max(S) heterodimer, alters its structure and renders it incapable of DNA binding without actually promoting its dissociation into c-Myc and Max(S) monomers.
Figure 4.
SPR analysis of 2 and 4da. Concentrations refer to (A) c-Myc–Max(S) dimer or (B)–(E) small-molecule ligand. (A). Concentration-dependent binding of c-Myc–Max(S) heterodimers to a biotin-tagged E-box-containing double-stranded oligonucleotide immobilized to a streptavidin-coated biosensor chip. Binding of the oligonucleotide itself was associated with a response of 700–800 units. After resetting this value to baseline, equimolar concentrations of c-Myc and Max(S) were allowed to dimerize for 30 min and then passed over the biosensor chip. Note the concentration-dependent increase in response. (B). Prevention of heterodimerization by 2. The indicated concentrations of 2 were incubated with 20 nM of c-Myc for 20 min followed by the addition of 20 nM Max(S). Following an additional 30 min incubation, the mixture was applied to the E-Box-containing biosensor chip. (C). DNA binding by pre-formed c-Myc–Max(S) heterodimers is relatively resistant to 2. Pre-formed c-Myc–Max(S) heterodimers (20 nM of each protein) were incubated for 30 min. with the indicated concentrations of 2 and then applied to the E-Box-containing biosensor chip. (D). Disruption of DNA binding by 4da. The experiment was performed as described in (B) except that the indicated concentrations of 4da were incubated with c-Myc monomer prior to the addition of Max(S). (E). The experiment was performed as described in (D) except that pre-formed c-Myc–Max(S) heterodimers were incubated with the indicated concentrations of 4da. Blue bars indicate the period during which protein-compound mixtures were being injected. Note that the relative response in each case has been normalized to 100 to denote the maximal response for each set of curves in order to allow inter-group comparisons. Similar results for each panel were obtained on at least two additional occasions.
4da inhibits c-Myc-overexpressing cells
The six active compounds described above were next investigated for their abilities to inhibit the proliferation of HL60 human promyelocytic leukaemia cells and Daudi Burkitt’s lymphoma cells, both of which over-express c-Myc and are highly susceptible to various c-Myc inhibitors.37,38,45,48,49 As shown in Table 5, all compounds except 21 inhibited the proliferation of both cell lines with IC50s below 50 µM. Indeed, the HL60 inhibition data closely mirrored the in vitro data for inhibition of the c-Myc–Max(S)/DNA ternary complex. 4da proved the most potent inhibitor of cell proliferation across both cell lines. It is unclear at this stage why compound 21 did not exhibit cell activity. Standard hydrolysis of the ethyl ester of 21 by cellular esterase will furnish the inactive compound 25, whilst amide hydrolysis by peptidases will afford inactive compound 19. Therefore, we hypothesize that metabolism of 21 to either 19 or 25 might explain the lack of cell activity.
Table 5.
Viability of HL60 and Daudi cells in the presence of synthetic α-helix mimetics after 3 days. MTT cell proliferation assays performed as previously described.37,38,45,48,49 The results shown represent the means and standard deviations of quadruplicate experiments.
| Code Number | Inhibition (IC50, µM) | |
|---|---|---|
| HL60 Cells | Daudi Cells | |
| 4aa | 39.9 ± 5.3 | 22.9 ± 2.5 |
| 4da | 19.9 ± 1.6 | 9.5 ± 0.7 |
| 4ca | 44.8 ± 0.9 | 25.6 ± 1.4 |
| 15 | 21.5 ± 2.7 | 11.2 ± 1.4 |
| 16 | 30.3 ± 4.6 | 11.1 ± 1.1 |
| 21 | >50 | >50 |
Having established that 4da was the most potent of the above compounds in vitro and in cells, we tested it further against several other cell lines (Table 6), including those representing epithelial cancers and several human multiple myeloma lines, the latter having previously been shown to be moderately sensitive to the growth inhibitory effects of the highly specific direct c-Myc inhibitor 10058-F437,62 (IC50s = 52–90 µM. One of these myeloma lines, U266, expresses L-Myc instead of c-Myc and had previously been shown to be the least sensitive of all myeloma cell lines to growth inhibition by 10058-F4 (IC50 ~100 µM62). We also tested 4da against rat fibroblasts with a homozygous deletion of the endogenous myc gene (HO15.19 cells) as well as HO15.19 cells in which Myc expression had been restored by stable transduction with a c-Myc-expressing retroviral vector (HO15.19-wt-Myc cells).63,64 Consistent with the idea that 4da is a specific c-Myc inhibitor, we found all c-Myc-expressing cell lines to be sensitive to relatively low concentrations of the compounds. However, U266 myeloma cells were the most resistant of the myeloma cell lines tested and HO15.19 fibroblasts were more resistant to 4da than HO15.19-Myc cells. That both transformed and non-transformed cells were growth-inhibited by 4da is consistent with the idea that c-Myc expression is necessary for the proliferation of virtually all cells and that c-Myc over-expression and/or de-regulation is not a pre-requisite for sensitivity to this class of inhibitors or to other modes of pharmacologic or genetically-mediated Myc inhibition in general.21,65 The residual susceptibilities of U266 and myeloma cells and HO15.19 suggests that 4da may have additional targets that are necessary to support proliferation and/or non-specific toxicities.
Table 6.
Growth inhibition studies were performed as described for Table 3. Cells were incubated in serial dilutions of 4da for 3 days before determining viable cell numbers relative to untreated cells.
| Cell Line or Strain |
Cell Type | Inhibition (IC50, µM) |
|---|---|---|
| H460 | lung cancer | 15.9 ± 1.1 |
| HeLa | uterine cervical cancer | 10.8 ± 0.8 |
| HEK-293 | embryonic kidney | 10.4 ± 1.3 |
| JJN3 | myeloma | 15.7 ± 0.9 |
| INA-6 | myeloma | 18.1 ± 0.8 |
| IH-1 | myeloma | 20.6 ± 1.1 |
| ANBL-6 | myeloma | 35.6 ± 2.6 |
| KJON | myeloma | 23.0 ± 2.6 |
| U266 | myeloma | 46.0 ± 3.2 |
| HO15.19 (myc−/−) | rat fibroblasts | 20.6 ± 1.0 |
| HO15.19-wt-Myc | rat fibroblasts | 14.1 ± 1.2 |
4da promotes cell cycle arrest and accumulation of neutral lipids
The inhibition of c-Myc in proliferating cells, either in vitro or in vivo, leads to cell cycle arrest and eventual apoptosis.66,67 This induced quiescence is likely related to changes in cellular energy metabolism given the fact that c-Myc is intimately involved in supporting mitochondrial biogenesis and glycolysis.68–70 Indeed, c-Myc-depleted cells possess atrophic mitochondria and defective electron transport chain complexes as well as low levels of oxidative phosphorylation, glycolysis and ATP.68–70 Recently, Zirath et al. have shown that N-Myc-amplified neuroblastoma cells accumulate stores of neutral lipid following their exposure to the small-molecule c-Myc inhibitor 10058-F4,28 and we have made similar findings in myc−/− fibroblasts.71 We have shown this to result from an increase in the transport of exogenous very long chain fatty acids that represents a compensatory attempt to fuel the TCA cycle with substrates other than glucose or glutamine, whose utilization by the TCA cycle is compromised in c-Myc-depleted cells. However, because of the defective mitochondrial structure and function, the uptake of these fatty acids exceeds their utilization and the difference is stored as neutral lipid.
To determine whether our synthetic α-helix mimetics imitated the above-described phenotypes associated with c-Myc depletion, we treated HL60 promyelocytic leukemia cells and H460 large cell lung cancer cells with 4da, and then evaluated the cells for evidence of proliferative arrest and neutral lipid accumulation, respectively. As positive controls for these studies, parallel cultures were treated with the previously well-characterized direct Myc inhibitors 10058-F4 or 2.37 As seen in Figure 5, both 10058-F4 and 4da promoted a G0/G1 arrest in HL60 cells, with the latter compound being both more efficient and potent. Repeat experiments performed on different occasions yielded very similar results (Supplementary Table 1). Similarly, treatment of H460 cells with both 2 and 4da led to an increase in neutral lipid stores, with 4da again being more effective (Figure 6). From these studies, we conclude that these structurally unrelated c-Myc inhibitors exert similar effects on proliferation and cellular energetics.
Figure 5.
4da promotes cell cycle arrest. Logarithmically growing HL60 promyelocytic leukemia cells were incubated with the indicated concentrations of 4da or the previously described c-Myc inhibitor 10058-F437 for 24 h. Cells were then collected and stained with propidium iodide as previously described.71 Typical results are shown. Repeat experiments performed in triplicate on a separate occasion yielded very similar findings (Supplementary Table 1).
Figure 6.
4da causes neutral lipid accumulation. H460 large cell lung cancer cells were incubated in the indicated concentrations of 2 or 4da for 48 h and then stained with the neutral lipid-specific fluorescent dye BODIPY-459/503.71 Control, untreated cells were stained in parallel. Curves depict the fluorescence distribution of at least 104 individual events from each group. The numbers in the upper left corner depict the ratio of mean fluorescence intensity of inhibitor-treated cells to control cells, based on the average of biological triplicates stained in parallel ± one standard error. Typical flow diagrams are indicated in each panel.
4da specifically inhibits a c-Myc-dependent reporter
To determine whether, as previously described for 10058-F4,72 4da specifically inhibited c-Myc-dependent genes, we stably transfected HeLa cells with a vector encoding a highly labile luciferase protein. The minimal promoter of this vector was engineered to contain tandemly triplicated binding sites for c-Myc (wt-Myc) or NF-κB. Control vectors contained either no binding sites or mutant c-Myc binding sites (mut-Myc) (CTCGAG rather than the canonical CACGTG). Relative to the latter control lines whose expression of luciferase was indistinguishable from the background of untransfected cells, the luciferase activities in cells expressing the wt-c-Myc and NF-kB vectors were 10–20 times and 100–200 times, respectively, higher than the background. As seen in Figure 7, both 4da and the control direct c-Myc inhibitor showed a dose-dependent inhibition of luciferase in cells expressing the wt-c-Myc vector, whereas no inhibition of luciferase was observed in cells expressing the NF-kB vector. From these studies, we conclude that 2 and 4da selectively suppress the expression of genes containing c-Myc binding sites, with the latter compound again being more potent.
Figure 7.
4da specifically inhibits a c-Myc reporter vector. HeLa cells stably expressing luciferase vectors under the control of a minimal promoter bearing c-Myc or NF-kB binding sites were exposed to the indicated concentrations of 4da or the previously well-characterized c-Myc inhibitor 2 for 6 h. Cells were assayed for luciferase activity as described in the Experimental Section. The results depicted represent the mean of triplicate determinations +/− 1 standard error with the value for untreated cells arbitrarily being set to 100%.
4da does not disrupt c-Myc–Max heterodimers in cells
The foregoing studies, particularly those shown in Figure 3 suggested that, while 4da was specific for c-Myc, its mechanism of action was distinct from that of previously described direct Myc inhibitors which prevent/disrupt Myc–Max association.34 To further confirm that 4da did not promote c-Myc–Max dissociation, we exposed HL60 cells to concentrations of the compound shown in the above studies to inhibit cell cycle progression, proliferation and Myc-specific gene expression and then performed c-Myc–Max co-IP studies. As a positive control, a parallel culture of cells was exposed to the compound 10058-F4, at a concentration previously shown to disrupt c-Myc–Max heterodimers.38 As shown in Figure 8, 10058-F4 treatment abrogated c-Myc’s association with Max, in a dose-dependent manner, without significantly affecting the overall level of the former protein. In contrast, c-Myc–Max heterodimers remain largely intact following exposure to even the highest levels of 4da. Taken together with the studies shown in Figure 2, these findings provide independent confirmation that 4da, while inhibiting a variety of c-Myc-regulated functions, does so without promoting c-Myc–Max dissociation.
Figure 8.
4da fails to promote c-Myc–Max dissociation in situ. HL60 promyelocytic leukemia cells were exposed for 6 h the indicated concentrations of 10058-F4 or 4da or to DMSO vehicle alone. Total cell lysates were then prepared as previously described48 and Max proteins were immunoprecipitated (IP) followed by immunoblotting (IB) of the precipitate with an anti-c-Myc antibody. The bottom portion of each panel shows the total amount of c-Myc protein in lysates prior to IP.
Discussion
Synthetic α-helix mimetics have been successfully utilized in the past to interrogate and disrupt aberrant helix-mediated PPIs.51–58 The transcriptionally active c-Myc–Max coiled coil presents itself as a potential target that might be disrupted or otherwise perturbed by such agents. We designed biphenyl-based α-helix mimetics that, along with a hydrophobic core and electron-rich peripheries, were intended to recognize a hydrophobic domain of helical c-Myc flanked by arginines and other polar residues, specifically the region R366RNELKRSFFALR378, that ensures the formation of a rigid tertiary structure upon dimerizing with Max. Importantly, this would represent an unprecedented strategy to inhibit the oncogenic activity of c-Myc since those direct inhibitors that have been fully characterized to date operate through binding and stabilizing the unstructured c-Myc monomer, rendering it incapable of interacting with Max.35
Several of the compounds reported herein impaired the ability of the c-Myc–Max(S) dimer to recognize its DNA-binding sequence, as shown by EMSA. To delineate the mode of inhibition of the synthetic helix mimetics, we recruited an array of biophysical techniques. HSQC NMR spectroscopy employing 15N-labeled Max(S) indicated that the helix mimetic 4da interacted only with c-Myc–Max(S) heterodimers and not with Max(S)–Max(S) homodimers or c-Myc monomers. This allowed us to infer that the biological target is the c-Myc–Max(S) heterodimer exclusively. Furthermore, according to NMR titrations, 4da bound helical c-Myc with a Kd of approximately 13 µM. These findings were further corroborated by SPR, which was consistent with the idea that 4da binds the c-Myc–Max heterodimer and alters its structure such that DNA binding is impaired. Indeed, the Kd for this interaction, ca. 10 µM (Figure 4D), is in excellent agreement with that obtained by NMR (Kd~13 µM (Figure 2E)). The somewhat higher Kd obtained under conditions in which c-Myc–Max(S) heterodimers were allowed to pre-form before the addition of 4da (Figure 4E) suggests that the heterodimer is more stable under these conditions. This could be explained by assuming that, in the initial stages of heterodimerization, the helix 1 regions of c-Myc and Max(S) may assemble into a structure that is recognized by 4da, as originally proposed in the design section, before the remaining regions of the proteins have the opportunity to fully assemble into their final and most stable conformations. Such a partially formed heterodimer might be more readily distorted by 4da.
SPR alone cannot directly discriminate between a mechanism that distorts the c-Myc–Max(S) heterodimer as proposed here for 4da and one that prevents c-Myc–Max(S) interaction. However, the markedly different behaviors in response to 2 and 4da (Figure 5), together with the results of our NMR studies (Figure 3) argue strongly in favor of the former model to explain 4da’s mechanism of action. Moreover, they are consistent with our inability to demonstrate disruption of c-Myc–Max heterodimers with 4da by in situ co-immunoprecipitation under conditions where it can be readily demonstrated by inhibitors that directly bind the c-Myc monomer, such as 10058-F4, 2 or their analogues.37,38,47–49 The helix mimetics reported herein inhibited the proliferation of c-Myc-overexpressing cells in a manner that reflected their abilities to disrupt DNA binding by the c-Myc–Max heterodimer in vitro. Finally, the induction of cell cycle arrest, the accumulation of neutral lipids and the inhibition of c-Myc-dependent gene expression, are all consistent with c-Myc being the main cellular target of 4da and other such compounds. Nonetheless, 4da may possess some non-specific toxicities or recognize other targets that are needed to drive proliferation as evidenced by the inhibition of some cell lines that do not express c-Myc (Table 6). It will be important to minimize such properties in future attempts to optimize this class of compounds. It is tempting to speculate that compounds such as 4da might be particularly effective in conjunction with other direct inhibitors such as 10058-F4 and 2 by preventing or reversing DNA binding by any residual c-Myc–Max(S) heterodimers that formed in the presence of the latter compounds.
Conclusions
Novel, direct inhibitors of c-Myc were designed based on a synthetic α-helix mimetic strategy to recognize and perturb the structure of the coiled coil c-Myc–Max heterodimer. Our most potent compound, 4da, demonstrated this activity in vitro with IC50 values in the low micromolar range. Several techniques were enlisted to delineate the mechanism of inhibition of our compounds. The outcomes of these studies are consistent with the idea that, as designed, these compounds bind helical c-Myc, and impair the c-Myc–Max heterodimer’s ability to bind DNA without causing the dissociation of c-Myc–Max heterodimers. The in vitro results were mirrored by studies showing that under conditions where 4da inhibited cell growth, c-Myc–Max heterodimers remained intact in situ. Current work is focused on determining the metabolic stability of 4da, since the c-Myc inhibitor 2 exhibits a short half-life due to metabolism of the nitro group to the toxic hydroxylamino derivative and the inactive amino-derivative.73 Hence, in addition to contributing to a short half-life, the nitro group in 4da may also be responsible for the general toxicity of the compound. Thus, towards the further optimization of our helix mimetics, efforts will be made to seek out nitro group bioisosteres, of which it is already known there exists at least one (ethyl aminooxalate in compound 21). More generally, the identification of a new mechanism of action by which direct c-Myc inhibitors may function should provide ample opportunities for the design of novel analogues of compounds such as those described here that are intended to promote more efficient cellular uptake and PPI disruption.
EXPERIMENTAL
Chemistry: General
Unless otherwise stated, all reactions were performed under an inert (N2) atmosphere. Reagents and solvents were reagent grade and purchased from Sigma-Aldrich, Alfa Aesar, Oakwood and TCI America. Anhydrous solvents were purchased from Sigma-Aldrich and used as provided. Reactions were monitored by TLC, visualizing with a UV lamp and/or KMnO4 stain. Silica gel 60 (70–230 mesh, Merck) was used for flash column chromatography. 1H and 13C NMR spectra were recorded on Varian INOVA 400 MHz NMR spectrometers at 25 °C. Chemical shifts are reported in parts per million (ppm). Data for 1H NMR are reported as follows: chemical shift (δ ppm) (integration, multiplicity, coupling constant (Hz), identity). Multiplicities are reported as follows: s = singlet, d = doublet, t = triplet, q = quartet, hep = heptet, dd = doublet of doublets, m = multiplet. Data for 13C are reported in terms of chemical shifts (δ ppm). The residual solvent peak was used as an internal reference. The mass spectra were obtained on an Electrospray TOF (ESI-TOF) mass spectrometer (BrukerAmaZon X). Target molecules that were evaluated beyond the initial EMSA screen (4aa, 4da, 4ca, 15, 16 and 21) exhibited purities of >95% as determined by CHN elemental combustion analysis.
General procedure A: Nucleophilic aromatic substitution (SNAr)
NaH (3 eq) was suspended in anhydrous THF (0.1 M), and the requisite alcohol (1.3 eq) was added at 0 °C under an inert (N2) atmosphere. After 15 min, 3-fluoro-4-nitrobenzoic acid (1 eq) was added. The reaction mixture was stirred for 15 min at 0 °C, then at RT for 2 h. TLC indicated the reaction was complete. The reaction was quenched by adding sat. NH4Cl, then partitioned with EtOAc. The aqueous layer was put to one side, and then the organic layer was washed twice with 0.1 M HCl. The organic layer was dried over Na2SO4, filtered, concentrated and purified by flash column chromatography over silica gel using an eluent of Hex/EtOAc/AcOH, 1:1:0.1 to provide the title compound.
General Procedure B: Acid chloride synthesis
To a solution of the carboxylic acid in CH2Cl2 or THF (0.1 M) at 0 °C was added dropwise oxalyl chloride followed by one drop of DMF. The reaction was stirred under an inert atmosphere at room temperature for 2 h (typically, all reactions were complete after this time), and then concentrated in vacuo and dried for at least 5 h under high vacuum to afford the acid chloride which was used without further purification.
General Procedure C: Reduction of nitro group with Pd/C
10% Pd/C (20wt%) was carefully added to an evacuated flask of the nitro compound in EtOH (0.1 M). H2 was bubbled through the reaction mixture for 10 min, then the reaction was stirred under 1 atm of H2 (balloon) for 1 – 5 h. TLC confirmed the reaction was complete. The reaction mixture was filtered over Celite, washing with MeOH. The filtrate was concentrated in vacuo, then dried under high vacuum to furnish the title compound.
General Procedure D: Reduction of nitro group with SnCl2.2H2O
The nitro compound (1 eq) was dissolved in EtOAc (0.1 M), and then SnCl2•2H2O (5 eq) was added. The reaction mixture was stirred for 3 h at 50 °C, by which time TLC confirmed the reaction was complete. The reaction mixture was partitioned between EtOAc and sat. NaHCO3. The organic layer was collected and the aqueous layer was extracted with further EtOAc (×2). The organic layers were combined, washed with sat. NaHCO3, brine, dried (Na2SO4), filtered and concentrated. The residue was purified by silica gel flash column chromatography using a gradient of EtOAc in Hex as eluent to furnish the corresponding aniline.
General procedure E: Methyl ester hydrolysis
NaOH (4 eq) was added to a solution of the methyl ester (1 eq) in a 3:1:1 mixture of THF/MeOH/H2O (0.08 M). The reaction was stirred at room temperature until complete consumption of the starting material (1 h – 4 d). The reaction mixture was diluted with further water, acidified to pH 5 with 1N HCl and then extracted into EtOAc (×5). The organic layers were combined, dried over Na2SO4, filtered and concentrated to give the carboxylic acid which did not require further purification.
General procedure F: Bis-arylamide synthesis
To a solution of the aniline (1 eq) and N,N-dimethylaniline in anhydrous acetone (0.2 M) at 0 °C was added dropwise a solution of the acid chloride of the appropriate 4-nitrobenzoic acid (1 eq) in anhydrous acetone (0.2 M). The resulting mixture was allowed to warm to room temperature overnight. The white precipitate was collected by vacuum filtration, washed with acetone, 2N HCl, and water until the filtrate was neutral, and then dried in vacuo at 40 °C overnight.
3-Isobutoxy-4-(3-isobutoxy-4-nitrobenzamido)benzoic acid (4aa)
3-Isobutoxy-4-nitrobenzoic acid74 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.33 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid74 (8a) according to General Procedure F to deliver the title compound as a pale yellow solid (80 mg, 56%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.98 (1H, s, CO2H), 9.79 (1H, s, NH), 8.03 (1H, d, J=8.0 Hz, Ar), 7.97 (1H, d, J=8.0 Hz, Ar), 7.79 (1H, s, Ar), 7.62 (2H, d, J=8.0 Hz, Ar), 7.57 (1H, s, Ar), 4.03 (2H, d, J=6.4 Hz, OCH2), 3.89 (2H, d, J=6.4 Hz, OCH2), 2.08 (2H, m, 2xCH(CH3)2), 0.99 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 163.9, 151.4, 150.8, 141.5, 139.7, 131.2, 128.5, 125.5, 123.7, 122.3, 119.9, 114.2, 112.9, 75.6, 74.9, 28.2, 28.0, 19.4, 19.1; MS (ESI) m/z Calcd for C22H26N2O7 (M+): 430.2, Found: 431.1 (M+H+); Anal. Calcd for C22H26N2O7: C, 61.39; H, 6.09; N, 6.51. Found: C, 61.11; H, 6.07; N, 6.37.
3-(Benzyloxy)-4-(3-isobutoxy-4-nitrobenzamido)benzoic acid (4ac)
3-Isobutoxy-4-nitrobenzoic acid74 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.19 mmol scale, then coupled to 4-amino-3-benzyloxybenzoic acid75 (8c) according to General Procedure F to deliver the title compound as a pale yellow solid (46 mg, 52%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.96 (1H, s, CO2H), 9.91 (1H, s, NH), 7.99 (1H, d, J=8.0 Hz, Ar), 7.91 (1H, d, J=8.0 Hz, Ar), 7.74 (1H, s, Ar), 7.65 (1H, s, Ar), 7.59 (2H, t, J=8.4 Hz, Ar), 7.50 (2H, d, J=6.8 Hz, Ar), 7.34 (2H, t, J=6.8 Hz, Ar), 7.30 (1H, d, J=6.8 Hz, Ar), 5.24 (2H, s, OCH2), 3.96 (2H, d, J=5.6 Hz, OCH2), 2.01 (1H, m, CH(CH3)2), 0.95 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.2, 164.1, 151.5, 150.6, 141.5, 139.7, 137.1, 131.4, 128.8, 128.6, 128.2, 127.6, 125.4, 124.3, 122.6, 120.0, 114.3, 113.7, 75.6, 70.3, 28.0, 19.1; MS (ESI) m/z Calcd for C25H24N2O7 (M+): 464.2, Found: 465.1 (M+H+).
4-(3-Isobutoxy-4-nitrobenzamido)-3-(pyridin-2-ylmethoxy)benzoic acid (4ag)
3-Isobutoxy-4-nitrobenzoic acid74 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.24 mmol scale, then coupled to 4-amino-3-(pyridin-2-ylmethoxy)benzoic acid (8g) according to General Procedure F to deliver the title compound as a white solid (55 mg, 49%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.99 (1H, s, CO2H), 10.19 (1H, s, NH), 8.06 (1H, d, J=8.0 Hz, Ar), 8.02 (1H, d, J=8.0 Hz, Ar), 7.82 (2H, m, Py), 7.68 (3H, m, Ar+Py), 7.61 (1H, d, J=8.0 Hz, Ar), 7.35 (1H, t, J=4.8 Hz, Py), 5.37 (2H, s, OCH2), 4.02 (2H, d, J=6.4 Hz, OCH2), 2.05 (1H, m, CH(CH3)2), 0.99 (6H, d, J=6.0 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.1, 164.1, 156.7, 151.5, 150.2, 149.4, 141.6, 139.7, 137.4, 131.9, 128.4, 125.5, 123.9, 123.5, 123.2, 121.8, 119.9, 114.7, 114.5, 75.6, 71.8, 28.0, 19.1; MS (ESI) m/z Calcd for C24H23N3O7 (M+): 465.2, Found: 466.1 (M+H+).
4-(3-Isobutoxy-4-nitrobenzamido)-3-(pyridin-3-ylmethoxy)benzoic acid (4ah)
3-Isobutoxy-4-nitrobenzoic acid74 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.37 mmol scale, then coupled to 4-amino-3-(pyridin-3-ylmethoxy)benzoic acid (8h) according to General Procedure F to deliver the title compound as a white solid (120 mg, 70%): 1H-NMR (DMSO-d6, 400 MHz) δ 13.01 (1H, bs, CO2H), 10.01 (1H, s, NH), 8.94 (1H, s, Py), 8.71 (1H, d, J=4.0 Hz, Py), 8.35 (1H, d, J=8.0 Hz, Ar), 8.00 (1H, d, J=8.4 Hz, Ar), 7.91 (1H, d, J=8.0 Hz, Py), 7.77 (2H, m, Ar+Py), 7.69 (1H, s, Ar), 7.65 (1H, d, J=8.0 Hz, Ar), 7.61 (1H, d, J=8.4 Hz, Ar), 5.41 (2H, s, OCH2), 3.98 (2H, d, J=6.4 Hz, OCH2), 2.02 (1H, m, CH(CH3)2), 0.96 (6H, d, J=6.8 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.1, 164.2, 151.5, 150.3, 145.4, 144.7, 141.5, 140.5, 139.6, 135.2, 131.4, 128.7, 125.7, 125.4, 124.7, 123.0, 120.1, 114.5, 113.7, 75.6, 67.4, 28.0, 19.1; MS (ESI) m/z Calcd for C24H23N3O7 (M+): 465.2, Found: 466.1 (M+H+).
4-(3-Isobutoxy-4-nitrobenzamido)-3-(pyridin-4-ylmethoxy)benzoic acid (4ai)
3-Isobutoxy-4-nitrobenzoic acid74 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.28 mmol scale, then coupled to 4-amino-3-(pyridin-4-ylmethoxy)benzoic acid (8i) according to General Procedure F to deliver the title compound as a white solid (94 mg, 72%): 1H-NMR (DMSO-d6, 400 MHz) δ 13.00 (1H, bs, CO2H), 10.27 (1H, s, NH), 8.87 (2H, d, J=5.6 Hz, Py), 8.06 (2H, d, J=5.6 Hz, Py), 8.02 (1H, d, J=8.8 Hz, Ar), 7.87 (2H, d, J=8.4 Hz, Ar), 7.66 (3H, m, Ar), 5.58 (2H, s, OCH2), 4.01 (2H, d, J=5.6 Hz, OCH2), 2.02 (1H, m, CH(CH3)2), 0.96 (6H, d, J=6.0 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 166.9, 164.1, 155.6, 151.4, 150.1, 143.4, 141.5, 139.4, 131.2, 128.8, 125.3, 125.2, 123.9, 123.1, 120.0, 114.5, 113.6, 75.6, 68.1, 27.9, 19.0; MS (ESI) m/z Calcd for C24H23N3O7 (M+): 465.2, Found: 466.1 (M+H+).
4-(3-Isobutoxy-4-nitrobenzamido)benzoic acid (4aj)
3-Isobutoxy-4-nitrobenzoic acid74 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.31 mmol scale, then coupled to 4-aminobenzoic acid (8j) according to General Procedure F to deliver the title compound as a pale yellow solid (76 mg, 69%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.78 (1H, s, CO2H), 10.65 (1H, s, NH), 8.00 (1H, d, J=8.8 Hz, Ar), 7.94 (2H, d, J=8.8 Hz, Ar), 7.87 (2H, d, J=8.8 Hz, Ar), 7.76 (1H, s, Ar), 7.61 (1H, d, J=8.0 Hz, Ar), 4.02 (2H, d, J=6.0 Hz, OCH2), 2.03 (1H, m, CH(CH3)2), 0.97 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.7, 151.4, 143.1, 141.5, 140.1, 130.7, 126.4, 125.3, 120.1, 114.7, 75.7, 28.1, 19.1; MS (ESI) m/z Calcd for C18H18N2O6 (M+): 358.1, Found: 359.1 (M+H+).
3-Isopropoxy-4-(3-isopropoxy-4-nitrobenzamido)benzoic acid (4bb)
3-Isopropoxy-4-nitrobenzoic acid53 (6b) was converted to its corresponding acid chloride according to General Procedure B on a 0.31 mmol scale, then coupled to 4-amino-3-isopropoxybenzoic acid74 (8b) according to General Procedure F to deliver the title compound as a pale yellow solid (38 mg, 31%): 1H-NMR (CDCl3+CD3OD, 400 MHz) δ 8.44 (1H, s, NH), 7.80 (2H, d, J=8.0 Hz, Ar), 7.59 (2H, s, 2xAr-1H), 7.27 (2H, d, J=8.0 Hz, Ar), 4.73 (2H, m, 2xOCH(CH3)2), 1.38 (12H, d, J=5.2 Hz, 2xOCH(CH3)2; 13C-NMR (CDCl3+CD3OD, 100 MHz) δ 163.0, 151.4, 145.7, 142.7, 139.3, 131.7, 125.7, 118.5, 117.1, 115.2, 73.0, 71.6, 29.6, 22.0, 21.7; MS (ESI) m/z Calcd for C20H22N2O7 (M+): 402.1, Found: 403.0 (M+H+).
3-Isobutoxy-4-(3-benzyloxy-4-nitrobenzamido)benzoic acid (4ca)
3-Benzyloxy-4-nitrobenzoic acid75 (6c) was converted to its corresponding acid chloride according to General Procedure B on a 0.37 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid75 (8a) according to General Procedure F to deliver the title compound as a pale yellow solid (68 mg, 40%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.98 (1H, s, CO2H), 9.82 (1H, s, NH), 8.07 (1H, d, J=8.0 Hz, Ar), 7.94 (2H, m, Ar), 7.66 (1H, d, J=8.0 Hz, Ar), 7.62 (1H, d, J=8.0 Hz, Ar), 7.57 (1H, s, Ar), 7.46-7.41 (4H, m, Ar), 7.38 (1H, d, J=6.4 Hz, Ar), 5.40 (2H, s, OCH2), 3.89 (2H, d, J=5.6 Hz, OCH2), 2.07 (1H, m, CH(CH3)2), 0.99 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 163.9, 151.0, 150.9, 141.8, 139.7, 136.1, 131.1, 129.0, 128.6, 127.8, 125.6, 123.9, 122.3, 120.2, 115.0, 113.0, 74.9, 71.2, 28.1, 19.4; MS (ESI) m/z Calcd for C25H24N2O7 (M+): 464.2, Found: 465.1 (M+H+); Anal. Calcd for C25H24N2O7: C, 64.65; H, 5.21; N, 6.03. Found: C, 64.86; H, 5.23; N, 6.06.
3-Isobutoxy-4-(3-(naphthalen-1-ylmethoxy)-4-nitrobenzamido)benzoic acid (4da (“JKY-2-169”))
3-Naphthalen-1-ylmethoxy-4-nitrobenzoic acid75 (6d) was converted to its corresponding acid chloride according to General Procedure B on a 0.31 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid74 (8a) according to General Procedure F to deliver the title compound as a pale yellow solid (134 mg, 84%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.96 (1H, s, CO2H), 9.80 (1H, s, NH), 8.14 (1H, s, Ar), 8.07 (1H, d, J=7.2 Hz, Ar), 8.04 (1H, d, J=8.0 Hz, Ar), 7.96-7.92 (3H, m, Ar), 7.69 (1H, d, J=7.2 Hz, Ar), 7.65 (1H, d, J=8.0 Hz, Ar), 7.60-7.50 (5H, m, Ar), 5.82 (2H, s, OCH2), 3.85 (2H, d, J=5.6 Hz, OCH2), 2.04 (1H, m, CH(CH3)2), 0.95 (6H, d, J=6.0 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.0, 151.1, 150.8, 141.8, 139.8, 133.6, 131.6, 131.2, 129.4, 128.9, 128.5, 126.9, 126.8, 126.5, 125.8, 125.6, 124.1, 123.8, 122.3, 120.3, 115.0, 112.9, 74.9, 69.8, 28.1, 19.4; MS (ESI) m/z Calcd for C29H26N2O7 (M+): 514.2, Found: 515.1 (M+H+); Anal. Calcd for C29H26N2O7: C, 67.70; H, 5.09; N, 5.44. Found: C, 67.41; H, 4.94; N, 5.35.
4-(3-((2,3-Dihydro-1H-inden-2-yl)oxy)-4-nitrobenzamido)-3-isobutoxybenzoic acid (4ea)
3-(2,3-dihydro-1H-inden-2-yl)oxy)-4-nitrobenzoic acid (6e) was converted to its corresponding acid chloride according to General Procedure B on a 0.66 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid75 (8a) according to General Procedure F to deliver the title compound as a pale yellow solid (252 mg, 78%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.95 (1H, s, CO2H), 9.79 (1H, s, NH), 7.98 (1H, d, J=8.4 Hz, Ar), 7.95 (1H, d, J=8.4 Hz, Ar), 7.87 (1H, s, Ar), 7.59 (2H, m, Ar), 7.54 (1H, s, Ar), 7.23 (2H, m, Ar), 7.16 (2H, m, Ar), 5.50 (1H, m, OCH), 3.86 (2H, d, J=6.4 Hz, OCH2), 3.45 (2H, dd, J=6.0 Hz, 17.2 Hz, CHCH2), 3.08 (2H, d, J=17.2 Hz, CHCH2), 2.06 (1H, m, CH(CH3)2), 0.97 (6H, d, J=6.0 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.0, 150.9, 150.2, 142.3, 140.5, 139.6, 131.2, 128.5, 127.1, 125.7, 125.0, 123.9, 122.3, 120.2, 115.1, 112.9, 80.3, 74.9, 28.3, 19.4; MS (ESI) m/z Calcd for C27H26N2O7 (M+): 490.2, Found: 491.1 (M+H+).
3-Methoxy-4-(3-methoxy-4-nitrobenzamido)benzoic acid (4ff)
3-Methoxy-4-nitrobenzoic acid (6f) was converted to its corresponding acid chloride according to General Procedure B on a 0.49 mmol scale, then coupled to 4-amino-3-methoxybenzoic acid (8f) according to General Procedure F to deliver the title compound as a pale yellow solid (129 mg, 76%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.99 (1H, s, CO2H), 9.92 (1H, s, NH), 8.02 (1H, d, J=8.0 Hz, Ar), 7.95 (1H, d, J=8.0 Hz, Ar), 7.83 (1H, s, Ar), 7.65-7.60 (3H, m, Ar), 4.02 (3H, s, OCH3), 3.91 (3H, s, OCH3); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.2, 152.0, 151.3, 141.4, 139.8, 131.0, 128.4, 125.4, 124.0, 122.3, 120.1, 114.0, 112.1, 57.3, 56.3; MS (ESI) m/z Calcd for C16H14N2O7 (M+): 346.1, Found: 347.0 (M+H+).
3-Isobutoxy-4-(4-nitro-3-(pyridin-2-ylmethoxy)benzamido)benzoic acid (4ga)
4-Nitro-3-(pyridin-2-ylmethoxy)benzoic acid (6g) was converted to its corresponding acid chloride according to General Procedure B on a 0.20 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid75 (8a) according to General Procedure F to deliver the title compound as a white solid (72 mg, 77%): 1H-NMR (DMSO-d6, 400 MHz) δ 13.00 (1H, s, CO2H), 9.85 (1H, s, NH), 8.61 (1H, s, Ar), 8.11 (1H, d, J=8.0 Hz, Ar), 7.97 (1H, s, Ar), 7.93-7.89 (2H, m, Ar+Py), 7.69 (1H, d, J=8.4 Hz, Ar), 7.63 (1H, d, J=8.4 Hz, Ar), 7.58 (2H, m, Py), 7.39 (1H, t, J=6.0 Hz, Py), 5.49 (2H, s, OCH2), 3.89 (2H, d, J=5.2 Hz, OCH2), 2.07 (1H, m, CH(CH3)2), 0.99 (6H, d, J=6.0 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 163.8, 155.8, 151.0, 150.9, 149.6, 141.6, 139.8, 137.6, 131.1, 128.6, 125.8, 124.1, 123.6, 122.3, 121.7, 120.5, 114.9, 113.0, 74.9, 71.8, 28.1, 19.4; MS (ESI) m/z Calcd for C24H23N3O7 (M+): 465.2, Found: 466.1 (M+H+).
3-Isobutoxy-4-(4-nitrobenzamido)benzoic acid (4ja)
4-Nitrobenzoyl chloride (7j; 0.45 mmol) was coupled to 4-amino-3-isobutyoxybenzoic acid74 (8a) according to General Procedure F to deliver the title compound as a white solid (154 mg, 96%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.94 (1H, s, CO2H), 9.85 (1H, s, NH), 8.35 (2H, d, J=8.4 Hz, Ar), 8.13 (2H, d, J=8.4 Hz, Ar), 7.88 (1H, d, J=7.6 Hz, Ar), 7.58 (1H, d, J=7.6 Hz, Ar), 7.53 (1H, s, Ar), 3.85 (2H, d, J=6.4 Hz, OCH2), 2.03 (1H, m, CH(CH3)2), 0.95 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.0, 151.0, 149.7, 140.4, 131.2, 129.4, 128.6, 124.2, 124.1, 122.2, 113.0, 74.9, 28.1, 19.4; MS (ESI) m/z Calcd for C18H18N2O6 (M+): 358.1, Found: 359.1 (M+H+).
4-(4-Nitrobenzamido)benzoic acid (4jj)
4-Nitrobenzoyl chloride (7j; 0.693 mmol) was coupled to 4-aminobenzoic acid according to General Procedure F to deliver the title compound as a pale yellow solid (180 mg, 91%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.81 (1H, s, CO2H), 10.85 (1H, s, NH), 8.40 (2H, d, J=8.4 Hz, Ar), 8.21 (2H, d, J=8.4 Hz, Ar), 7.99 (2H, d, J=8.4 Hz, Ar), 7.93 (2H, d, J=8.4 Hz, Ar); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.7, 149.7, 143.2, 140.6, 130.7, 129.7, 126.4, 124.0, 120.1; MS (ESI) m/z Calcd for C14H10N2O5 (M+): 286.1, Found: 287.0 (M+H+).
Methyl 3-isobutoxy-4-(3-isobutoxy-4-nitrobenzamido)benzoate (14aa)
3-Isobutoxy-4-nitrobenzoic acid74 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.18 mmol scale, then coupled to methyl 4-amino-3-isobutoxybenzoate74 (9a) according to General Procedure F to deliver the title compound as a pale yellow solid (51 mg, 64%): 1H-NMR (CDCl3, 400 MHz) δ 8.78 (1H, s, NH), 8.60 (1H, d, J=8.4 Hz, Ar), 7.93 (1H, d, J=8.4 Hz, Ar), 7.75 (1H, d, J=8.4 Hz, Ar), 7.64 (1H, s, Ar), 7.58 (1H, s, Ar), 7.39 (1H, d, J=8.4 Hz, Ar), 3.95 (4H, m, 2xOCH2), 3.92 (3H, s, OCH3), 2.18 (2H, m, 2xCH(CH3)2, 1.11 (6H, d, J=6.4Hz, CH(CH3)2, 1.07 (6H, d, J=6.4Hz, CH(CH3)2; 13C-NMR (CDCl3, 100 MHz) δ 166.5, 163.0, 152.8, 147.0, 141.6, 139.7, 131.4, 125.9, 125.8, 123.3, 118.6, 117.2, 113.7, 111.5, 76.0, 75.0, 52.1, 28.2, 28.1, 19.3, 18.9; MS (ESI) m/z Calcd for C23H28N2O7 (M+): 444.2, Found: 445.1 (M+H+).
Methyl 3-isobutoxy-4-(3-isopropoxy-4-nitrobenzamido)benzoate (14ba)
3-Isopropoxy-4-nitrobenzoic acid53 (6b) was converted to its corresponding acid chloride according to General Procedure B on a 0.18 mmol scale, then coupled to methyl 4-amino-3-isobutoxybenzoate74 (9a) according to General Procedure F to deliver the title compound as a pale yellow solid (77 mg, 100%): 1H-NMR (CDCl3, 400 MHz) δ 8.76 (1H, s, NH), 8.59 (1H, d, J=8.4 Hz, Ar), 7.86 (1H, d, J=8.4 Hz, Ar), 7.75 (1H, d, J=8.4 Hz, Ar), 7.67 (1H, s, Ar), 7.57 (1H, s, Ar), 7.36 (1H, d, J=8.4 Hz, Ar), 4.80 (1H, m, OCH(CH3)2), 3.94-3.91 (5H, m, OCH3+OCH2CH), 2.19 (1H, m, CH2CH(CH3)2), 1.43 (6H, d, J=5.6 Hz, OCH(CH3)2), 1.10 (6H, d, J=7.2 Hz, CH2CH(CH3)2); 13C-NMR (CDCl3, 100 MHz) δ 166.5, 163.1, 151.5, 147.0, 142.8, 139.4, 131.4, 125.7, 123.3, 118.6, 117.0, 115.3, 111.5, 75.1, 73.0, 52.1, 28.2, 21.7, 19.3; MS (ESI) m/z Calcd for C22H26N2O7 (M+): 430.2, Found: 431.1 (M+H+).
Methyl 3-(benzyloxy)-4-(3-isopropoxy-4-nitrobenzamido)benzoate (14bc)
3-Isopropoxy-4-nitrobenzoic acid53 (6b) was converted to its corresponding acid chloride according to General Procedure B on a 0.16 mmol scale, then coupled to methyl 4-amino-3-benzyloxybenzoate75 (9c) according to General Procedure F to deliver the title compound as a pale yellow solid (71 mg, 96%): 1H-NMR (CDCl3, 400 MHz) δ 8.72 (1H, s, NH), 8.60 (1H, d, J=8.4 Hz, Ar), 7.78 (1H, d, J=8.4 Hz, Ar), 7.74 (1H, d, J=8.4 Hz, Ar), 7.71 (1H, s, Ar), 7.54 (1H, s, Ar), 7.42-7.40 (5H, m, Ar), 7.23 (1H, d, J=8.4 Hz, Ar), 5.19 (2H, s, OCH2), 4.63 (1H, m, OCH), 3.90 (3H, s, OCH3), 1.34 (6H, d, J=5.6 Hz, CH(CH3)2); 13C-NMR (CDCl3, 100 MHz) δ 166.7, 163.3, 151.6, 147.2, 143.0, 139.3, 135.9, 131.9, 129.1, 129.0, 128.2, 125.9, 124.1, 119.0, 117.7, 115.3, 112.6, 73.2, 71.8, 52.4, 21.9; MS (ESI) m/z Calcd for C25H24N2O7 (M+): 464.2, Found: 465.1 (M+H+).
Methyl 3-benzyloxy-4-(3-isobutoxy-4-nitrobenzamido)benzoate (14ca)
3-Benzyloxy-4-nitrobenzoic acid75 (6c) was converted to its corresponding acid chloride according to General Procedure B on a 0.18 mmol scale, then coupled to methyl 4-amino-3-isobutoxybenzoate74 (9a) according to General Procedure F to deliver the title compound as a pale yellow solid (86 mg, 100%): 1H-NMR (CDCl3, 400 MHz) δ 8.77 (1H, s, NH), 8.54 (1H, d, J=8.4 Hz, Ar), 7.94 (1H, d, J=8.4 Hz, Ar), 7.61 (1H, s, Ar), 7.73 (1H, d, J=8.4 Hz, Ar), 7.56 (1H, s, Ar), 7.46-7.30 (5H, m, Ar), 5.27 (2H, s, OCH2), 3.92-3.89 (5H, m, OCH3+OCH2CH), 2.18 (1H, m, CH2CH(CH3)2), 1.08 (6H, d, J=7.2 Hz, CH2CH(CH3)2); 13C-NMR (CDCl3, 100 MHz) δ 166.6, 162.9, 152.1, 147.1, 141.9, 139.7, 134.8, 131.3, 128.7, 128.4, 127.0, 126.0, 125.8, 123.2, 118.7, 117.6, 114.6, 111.6, 75.1, 71.3, 52.2, 28.2, 19.2; MS (ESI) m/z Calcd for C26H26N2O7 (M+): 478.2, Found: 479.1 (M+H+).
Methyl 4-(3-((2,3-dihydro-1H-inden-2-yl)oxy)-4-nitrobenzamido)-3-isobutoxybenzoate (14ea)
3-((2,3-dihydro-1H-inden-2-yl)oxy)-4-nitrobenzoic acid (6e) was converted to its corresponding acid chloride according to General Procedure B on a 0.18 mmol scale, then coupled to methyl 4-amino-3-isobutoxybenzoate74 (9a) according to General Procedure F to deliver the title compound as a yellow solid (85 mg, 94%): 1H-NMR (CDCl3, 400 MHz) δ 8.80 (1H, s, NH), 8.61 (1H, d, J=8.4 Hz, Ar), 7.90 (1H, d, J=8.4 Hz, Ar), 7.77 (2H, d, J=10.0 Hz, Ar), 7.59 (1H, s, Ar), 7.40 (1H, d, J=8.4 Hz, Ar), 7.26-7.19 (4H, m, Ar), 5.38 (1H, m, OCH), 3.95 (2H, d, J=6.4 Hz, OCH2CH), 3.92 (3H, s, OCH3), 3.52 (2H, dd, J=6.4 Hz, 16.8 Hz, CHCH2), 3.30 (2H, dd, J=6.4 Hz, 16.8 Hz, CHCH2), 2.21 (1H, m, CH2CH(CH3)2), 1.10 (6H, d, J=6.4 Hz, CH2CH(CH3)2); 13C-NMR (CDCl3, 100 MHz) δ 166.5, 162.9, 151.5, 147.0, 142.4, 139.6, 139.5, 131.3, 127.0, 126.0, 125.8, 124.6, 123.3, 118.6, 117.2, 115.0, 111.6, 80.2, 75.1, 52.2, 39.5, 28.2, 19.3; MS (ESI) m/z Calcd for C28H28N2O7 (M+): 504.2, Found: 505.1 (M+H+).
Methyl 3-(benzyloxy)-4-(3-benzyloxy-4-nitrobenzamido)benzoate (14cc)
3-Benzyloxy-4-nitrobenzoic acid75 (6c) was converted to its corresponding acid chloride according to General Procedure B on a 0.16 mmol scale, then coupled to methyl 4-amino-3-benzyloxy-benzoate75 (9c) according to General Procedure F to deliver the title compound as a pale yellow solid (67 mg, 82%): 1H-NMR (CDCl3, 400 MHz) δ 8.73 (1H, s, NH), 8.58 (1H, d, J=8.4 Hz, Ar), 7.83 (1H, d, J=8.4 Hz, Ar), 7.77 (1H, d, J=8.4 Hz, Ar), 7.71 (1H, s, Ar), 7.64 (1H, s, Ar), 7.44-7.32 (10H, m, Ar), 7.25 (1H, d, J=8.4 Hz, Ar), 5.19 (2H, s, OCH2), 5.16 (2H, s, OCH2), 3.91 (3H, s, OCH3); 13C-NMR (CDCl3, 100 MHz) δ 166.7, 163.0, 152.3, 147.2, 142.2, 139.6, 135.9, 135.0, 131.9, 129.2, 129.0, 128.7, 128.1, 127.4, 126.1, 124.1, 119.1, 118.3, 114.6, 112.7, 71.8, 71.5, 52.5; MS (ESI) m/z Calcd for C29H24N2O7 (M+): 512.2, Found: 513.1 (M+H+).
Methyl 4-(3-(benzyloxy)-4-nitrobenzamido)-3-((2,3-dihydro-1H-inden-2-yl)oxy)benzoate (14ce)
3-Benzyloxy-4-nitrobenzoic acid75 (6c) was converted to its corresponding acid chloride according to General Procedure B on a 0.14 mmol scale, then coupled to methyl 4-amino-3-((2,3-dihydro-1H-inden-2-yl)oxy)-benzoic acid (9e) according to General Procedure F to deliver the title compound as a pale yellow solid (69 mg, 92%): 1H-NMR (CDCl3, 400 MHz) δ 8.54 (1H, d, J=8.0 Hz, Ar), 8.42 (1H, d, NH), 7.77 (2H, m, Ar), 7.62 (2H, m, Ar), 7.43 (2H, d, J=8.0 Hz, Ar), 7.37 (2H, d, J=8.0 Hz, Ar), 7.32 (1H, d, J=8.0 Hz, Ar), 7.29 (2H, m, Ar), 7.20 (2H, m, Ar), 6.41 (1H, dd, J=1.6 Hz, 8.4 Hz, Ar), 5.40 (1H, t, J=5.2 Hz, OCH), 5.21 (2H, s, OCH2), 3.92 (3H, s, OCH3), 3.34 (2H, dd, J=5.2 Hz, 16.8 Hz, CHCH2), 3.17 (2H, d, J=16.8 Hz, CHCH2); 13C-NMR (CDCl3, 100 MHz) δ 166.4, 162.7, 151.9, 145.1, 141.7, 140.2, 139.2, 134.8, 133.5, 128.7, 128.4, 127.1, 125.8, 125.0, 124.3, 118.8, 117.2, 115.3, 114.6, 80.8, 71.2, 52.2, 39.5; MS (ESI) m/z Calcd for C31H26N2O7 (M+): 538.2, Found: 539.1 (M+H+).
Methyl 3-isobutoxy-4-(3-(naphthalen-1-ylmethoxy)-4-nitrobenzamido)benzoate (14da)
3-Naphthalen-1-ylmethoxy-4-nitrobenzoic acid75 (6d) was converted to its corresponding acid chloride according to General Procedure B on a 0.18 mmol scale, then coupled to methyl 4-amino-3-isobutoxybenzoate74 (9a) according to General Procedure F to deliver the title compound as a pale yellow solid (95 mg, 100%): 1H-NMR (CDCl3, 400 MHz) δ 8.79 (1H, s, NH), 8.60 (1H, d, J=8.4 Hz, Ar), 8.05 (1H, d, J=8.4 Hz, Ar), 7.95 (2H, t, J=8.4 Hz, Ar), 7.88 (2H, t, J=9.2 Hz, Ar), 7.76 (1H, d, J=9.2 Hz, Ar), 7.71 (1H, d, J=6.8 Hz, Ar), 7.60-7.47 (4H, m, Ar), 7.41 (1H, d, J=6.8 Hz, Ar), 5.75 (2H, s, OCH2), 3.94-3.92 (5H, m, OCH3+OCH2CH), 2.19 (1H, m, CH2CH(CH3)2), 1.09 (6H, d, J=6.4 Hz, CH2CH(CH3)2); 13C-NMR (CDCl3, 100 MHz) δ 166.5, 162.8, 152.2, 147.0, 142.0, 139.7, 133.6, 131.3, 130.9, 130.1, 129.4, 128.7, 126.7, 126.3, 126.1, 125.8, 125.3, 123.3, 123.1, 118.6, 117.6, 114.7, 111.6, 75.1, 70.1, 52.2, 28.2, 19.3; MS (ESI) m/z Calcd for C30H28N2O7 (M+): 528.2, Found: 529.1 (M+H+).
Methyl 3-(benzyloxy)-4-(3-(naphthalen-1-ylmethoxy)-4-nitrobenzamido)benzoate (14dc)
3-(Naphthalen-1-ylmethoxy)-4-nitrobenzoic acid (6d) was converted to its corresponding acid chloride according to General Procedure B on a 0.16 mmol scale, then coupled to methyl 4-amino-3-benzyloxy-benzoic acid75 (9c) according to General Procedure F to deliver the title compound as a pale yellow solid (40 mg, 45%): 1H-NMR (CDCl3, 400 MHz) δ 8.76 (1H, s, NH), 8.56 (1H, d, J=8.4 Hz, Ar), 7.97 (1H, d, J=8.0 Hz, Ar), 7.86-7.95 (4H, m, Ar), 7.76 (1H, d, J=8.4 Hz, Ar), 7.69 (1H, s, Ar), 7.60 (1H, d, J=6.8 Hz, Ar), 7.50 (2H, d, J=8.4 Hz, Ar), 7.45 (1H, d, J=8.4 Hz, Ar), 7.40 (2H, d, J=6.8 Hz, Ar), 7.32 (2H, t, J=8.4 Hz, Ar), 7.24 (2H, t, J=6.4 Hz, Ar), 5.52 (2H, s, OCH2), 5.16 (2H, s, OCH2), 3.89 (3H, s, OCH3); 13C-NMR (CDCl3, 100 MHz) δ 166.8, 163.2, 163.1, 152.3, 147.2, 142.2, 139.6, 135.8, 133.9, 131.8, 131.2, 130.3, 129.7, 129.1, 128.9, 128.1, 126.9, 126.8, 126.3, 126.1, 125.5, 124.1, 123.5, 119.2, 119.1, 118.3, 114.6, 112.7, 71.8, 70.3, 52.5; MS (ESI) m/z Calcd for C33H26N2O7 (M+): 562.2, Found: 563.1 (M+H+).
Methyl 3-((2,3-dihydro-1H-inden-2-yl)oxy)-4-(3-(naphthalen-1-ylmethoxy)-4-nitrobenzamido)benzoate (14de)
3-(Naphthalen-1-ylmethoxy)-4-nitrobenzoic acid75 (6d) was converted to its corresponding acid chloride according to General Procedure B on a 0.14 mmol scale, then coupled to methyl 4-amino-3-((2,3-dihydro-1H-inden-2-yl)oxy)-benzoic acid (9e) according to General Procedure F to deliver the title compound as a pale yellow solid (60 mg, 73%): 1H-NMR (CDCl3, 400 MHz) δ 8.56 (1H, d, J=8.4 Hz, Ar), 8.44 (1H, s, NH), 8.02 (1H, d, J=8.4 Hz, Ar), 7.87 (1H, d, J=8.0 Hz, Ar), 7.84 (1H, d, J=8.0 Hz, Ar), 7.79 (3H, m, Ar), 7.63 (2H, t, J=8.4 Hz, Ar), 7.57 (1H, t, J=8.4 Hz, Ar), 7.50 (1H, d, J=8.4 Hz, Ar), 7.46 (1H, d, J=8.0 Hz, Ar), 7.24 (2H, m, Ar), 7.15 (2H, m, Ar), 6.42 (1H, dd, J=1.6 Hz, 8.4 Hz, Ar), 5.65 (2H, s, OCH2), 5.40 (1H, t, J=5.2 Hz, OCH), 3.92 (3H, s, OCH3), 3.33 (2H, dd, J=4.8 Hz, 17.2 Hz, CHCH2), 3.17 (2H, d, J=17.2 Hz, CHCH2); 13C-NMR (CDCl3, 100 MHz) δ 166.4, 162.7, 151.9, 145.1, 141.8, 140.2, 139.2, 133.6, 133.5, 130.9, 130.2, 129.4, 128.7, 127.1, 126.6, 126.4, 126.1, 125.9, 125.2, 125.0, 124.3, 123.2, 118.8, 117.3, 115.3, 114.7, 80.8, 70.0, 52.2, 39.5; MS (ESI) m/z Calcd for C35H28N2O7 (M+): 588.2, Found: 589.1 (M+H+).
Methyl 3-((2,3-dihydro-1H-inden-2-yl)oxy)-4-(3-isopropoxy-4-nitrobenzamido)benzoate (14eb)
3-Isopropoxy-4-nitrobenzoic acid53 (6b) was converted to its corresponding acid chloride according to General Procedure B on a 0.14 mmol scale, then coupled to methyl 4-amino-3-((2,3-dihydro-1H-inden-2-yl)oxy)-benzoic acid (9e) according to General Procedure F to deliver the title compound as a pale yellow solid (60 mg, 87%): 1H-NMR (CDCl3, 400 MHz) δ 8.55 (1H, d, J=8.0 Hz, Ar), 8.43 (1H, s, NH), 7.77 (2H, m, Ar), 7.53 (2H, m, Ar), 7.28-7.25 (2H, m, Ar), 7.22-7.19 (2H, m, Ar), 6.38 (1H, dd, J=6.8 Hz, 0.8 Hz, Ar), 5.40 (1H, t, J=5.2 Hz, OCH), 4.69 (1H, m, OCH), 3.92 (3H, s, OCH3), 3.35 (2H, dd, J=5.2 Hz, 16.8 Hz, CHCH2), 3.18 (2H, d, J=16.8 Hz, CHCH2), 1.37 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (CDCl3, 100 MHz) δ 166.4, 163.0, 151.3, 145.1, 142.6, 140.2, 138.8, 133.5, 127.1, 125.7, 125.5, 125.0, 124.2, 118.8, 116.6, 115.3, 115.1, 80.6, 72.8, 52.2, 39.5, 21.7; MS (ESI) m/z Calcd for C27H26N2O7 (M+): 490.2, Found: 491.1 (M+H+).
Methyl 3-(benzyloxy)-4-(3-((2,3-dihydro-1H-inden-2-yl)oxy)-4-nitrobenzamido)benzoate (14ec)
3-((2,3-Dihydro-1H-inden-2-yl)oxy-4-nitrobenzoic acid (6e) was converted to its corresponding acid chloride according to General Procedure B on a 0.14 mmol scale, then coupled to methyl 4-amino-3-benzyloxy)-benzoic acid75 (9c) according to General Procedure F to deliver the title compound as a pale yellow solid (67 mg, 78%): 1H-NMR (CDCl3, 400 MHz) δ 8.77 (1H, s, NH), 8.61 (1H, d, J=7.6 Hz, Ar), 7.78 (2H, d, J=8.4 Hz, Ar), 7.72 (1H, s, Ar), 7.60 (1H, s, Ar), 7.42 (2H, d, J=7.6 Hz, Ar), 7.34 (3H, m, Ar), 7.25 (1H, d, J=7.6 Hz, Ar), 7.22 (3H, m, Ar), 5.19-5.17 (3H, m, OCH2+OCH), 3.91 (3H, s, OCH3), 3.38 (2H, dd, J=6.8 Hz, 17.2 Hz, CHCH2), 3.21 (2H, dd, J=2.4 Hz, 17 Hz, CHCH2); 13C-NMR (CDCl3, 100 MHz) δ 166.7, 163.1, 151.6, 147.2, 142.6, 139.9, 139.4, 135.8, 131.9, 129.1, 128.2, 127.2, 126.2, 124.9, 124.1, 119.1, 118.0, 114.8, 112.6, 80.4, 71.8, 52.5, 39.8; MS (ESI) m/z Calcd for C31H26N2O7 (M+): 538.2, Found: 539.1 (M+H+).
Methyl 3-((2,3-dihydro-1H-inden-2-yl)oxy)-4-(3-((2,3-dihydro-1H-inden-2-yl)oxy)-4-nitrobenzamido)benzoate (14ee)
3-((2,3-Dihydro-1H-inden-2-yl)oxy)-benzoic acid (6e) was converted to its corresponding acid chloride according to General Procedure B on a 0.14 mmol scale, then coupled to methyl 4-amino-3-((2,3-dihydro-1H-inden-2-yl)oxy)-benzoic acid (9e) according to General Procedure F to deliver the title compound as a pale yellow solid (30 mg, 40%): 1H-NMR (CDCl3, 400 MHz) δ 8.57 (1H, d, J=8.4 Hz, Ar), 8.47 (1H, s, NH), 7.78 (2H, d, J=8.0 Hz, Ar), 7.62 (1H, s, Ar), 7.58 (1H, d, J=8.0 Hz, Ar), 7.27 (2H, m, Ar), 7.20-7.16 (6H, m, Ar), 6.41 (1H, dd, J=1.6 Hz, 8.8 Hz, Ar), 5.42 (1H, m, OCH), 5.27 (1H, m, OCH), 3.92 (3H, s, OCH3), 3.43 (2H, dd, J=6.8 Hz, 16.8 Hz, CHCH2), 3.19 (2H, dd, J=6.8 Hz, 16.8 Hz, CHCH2), 3.24-3.14 (4H, m, 2xCHCH2); 13C-NMR (CDCl3, 100 MHz) δ 166.4, 162.8, 151.2, 145.1, 142.2, 140.2, 139.6, 138.9, 133.5, 127.1, 126.9, 125.8, 125.0, 124.6, 124.3, 118.8, 116.9, 115.1, 115.0, 80.6, 80.1, 52.2, 39.5; MS (ESI) m/z Calcd for C33H28N2O7 (M+): 564.2, Found: 565.1 (M+H+).
3-Isobutoxy-4-(6-isobutoxy-5-nitropicolinamido)benzoic acid (15)
6-Isobutoxy-5-nitropicolinic acid (11) was converted to its corresponding acid chloride according to General Procedure B on a 0.41 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid75 (6a) according to General Procedure F to deliver the title compound as a pale yellow solid (156 mg, 88%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.94 (1H, s, CO2H), 10.22 (1H, s, NH), 8.67 (1H, d, J=8.8 Hz, Py), 8.54 (1H, d, J=8.8 Hz, Py), 7.95 (1H, d, J=8.4 Hz, Ar), 7.65 (1H, d, J=8.4 Hz, Ar), 7.58 (1H, s, Ar), 4.35 (2H, d, J=5.6 Hz, OCH2), 3.98 (2H, d, J=6.4 Hz, OCH2), 2.12 (2H, m, 2xCH(CH3)2), 1.04 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 166.7, 159.8, 154.5, 149.5, 147.4, 137.3, 136.2, 130.3, 126.7, 122.5, 118.4, 115.8, 111.9, 74.7, 73.3, 27.8, 27.5, 19.1, 18.8; MS (ESI) m/z Calcd for C21H25N3O7 (M+): 431.2, Found: 432.1 (M+H+)+); Anal. Calcd for C21H25N3O7: C, 58.46; H, 5.84; N, 9.74. Found: C, 58.37; H, 5.84; N, 9.58.
6-Isobutoxy-5-(6-isobutoxy-5-nitropicolinamido)picolinic acid (16)
6-Isobutoxy-5-nitropicolinic acid (11) was converted to its corresponding acid chloride according to General Procedure B on a 0.41 mmol scale, then coupled to 6-isobutoxy-5-aminopicolinic acid (13) according to General Procedure F to deliver the title compound as a pale yellow solid (115 mg, 65%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.97 (1H, s, CO2H), 10.03 (1H, s, NH), 8.71 (1H, d, J=7.6 Hz, Py), 8.62 (1H, d, J=7.6 Hz, Py), 7.88 (1H, d, J=7.6 Hz, Py), 7.73 (1H, d, J=7.6 Hz, Py), 4.29 (2H, d, J=5.2 Hz, OCH2), 4.19 (2H, d, J=6.8 Hz, OCH2), 2.09 (2H, m, 2xCH(CH3)2), 1.00 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 165.7, 160.8, 154.9, 152.6, 149.3, 139.8, 137.8, 136.7, 126.1, 125.0, 120.0, 116.2, 73.8, 73.1, 27.9, 19.6, 19.2; MS (ESI) m/z Calcd for C20H24N4O7 (M+): 432.2, Found: 433.1 (M+H+)+); Anal. Calcd for C20H24N4O7: C, 55.55; H, 5.59; N, 12.96. Found: C, 55.72; H, 5.49; N, 12.97.
3-Isobutoxy-4-(3-isobutoxybenzamido)benzoic acid (17)
3-Isobutoxy-benzoic acid was converted to its corresponding acid chloride according to General Procedure B on a 0.26 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid75 (8a) according to General Procedure F to deliver the title compound as a white solid (43 mg, 43%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.97 (1H, s, CO2H), 9.41 (1H, s, NH), 8.08 (1H, d, J=8.0 Hz, Ar), 7.61 (1H, d, J=8.0 Hz, Ar), 7.55 (1H, s, Ar), 7.50-7.43 (3H, m, Ar), 7.18 (1H, d, J=6.8 Hz, Ar), 3.90 (2H, d, J=6.4 Hz, OCH2), 3.83 (2H, d, J=6.4 Hz, OCH2), 2.12-2.03 (2H, m, CH(CH3)2), 1.03-0.99 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 165.0, 159.2, 149.9, 136.1, 131.7, 130.3, 127.7, 122.5, 122.4, 119.9, 118.9, 113.1, 112.6, 74.8, 74.3, 28.2, 28.1, 19.4; MS (ESI) m/z Calcd for C22H27NO5 (M+): 385.2, Found: 386.1 (M+H+).
5-(5-Amino-6-isobutoxypicolinamido)-6-isobutoxypicolinic acid (18)
6-Isobutoxy-5-(6-isobutoxy-5-nitropicolinamido)picolinic acid (16; 40 mg, 0.093 mmol) was reduced according to General Procedure C: pale yellow solid (30 mg, 81%): 1H-NMR (DMSO-d6, 400 MHz) δ 9.96 (1H, s, NH), 8.70 (1H, d, J=7.6 Hz, Py), 7.64 (1H, d, J=7.6 Hz, Py), 7.58 (1H, d, J=7.6 Hz, Py), 6.96 (1H, d, J=7.6 Hz, Py), 5.87 (2H, s, NH2), 4.20 (2H, d, J=6.4 Hz, OCH2), 4.12 (2H, d, J=6.4 Hz, OCH2), 2.11 (2H, m, 2xCH(CH3)2), 1.03 (6H, d, J=6.4 Hz, CH(CH3)2), 1.00 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 166.8, 163.1, 152.0, 150.0, 137.7, 131.7, 124.8, 119.2, 118.8, 117.7, 72.7, 71.9, 28.1, 28.0, 19.6; MS (ESI) m/z Calcd for C20H26N4O5 (M+): 402.2, Found: 403.1 (M+H+).
4-(4-Amino-3-isobutoxybenzamido)-3-isobutoxybenzoic acid (19)
3-Isobutoxy-4-(3-isobutoxy-4-nitrobenzamido)benzoic acid (4aa; 200 mg, 0.46 mmol) was reduced according to General Procedure C: pale yellow solid (184 mg, 100%): 1H-NMR (DMSO-d6, 400 MHz) δ 8.93 (1H, s, NH), 8.18 (1H, d, J=8.4 Hz, Ar), 7.55 (1H, d, J=8.4 Hz, Ar), 7.50 (1H, s, Ar), 7.32 (1H, d, J=8.4 Hz, Ar), 7.26 (1H, s, Ar), 6.68 (1H, d, J=7.6 Hz, Ar), 5.44 (2H, s, NH2), 3.88 (2H, d, J=6.4 Hz, OCH2), 3.76 (2H, d, J=6.4 Hz, OCH2), 2.10 (2H, m, 2xCH(CH3)2), 1.00 (12H, m, 2x CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.6, 164.8, 148.5, 145.1, 142.5, 132.0, 122.6, 121.7, 121.2, 120.3, 112.7, 112.3, 110.2, 74.8, 74.4, 28.2, 19.6, 19.5; MS (ESI) m/z Calcd for C22H28N2O5 (M+): 400.2, Found: 401.1 (M+H+).
4-(4-Amino-3-((2,3-dihydro-1H-inden-2-yl)oxy)benzamido)-3-isobutoxybenzoic acid (20)
4-(3-((2,3-Dihydro-1H-inden-2-yl)oxy)-4-nitrobenzamido)-3-isobutoxybenzoic acid (4ea; 60 mg, 0.12 mmol) was reduced according to General Procedure C: pale yellow solid (55 mg, 100%): 1H-NMR (DMSO-d6, 400 MHz) δ 8.97 (1H, s, NH), 8.17 (1H, d, J=8.0 Hz, Ar), 7.56 (1H, d, J=8.0 Hz, Ar), 7.51 (1H, s, Ar), 7.40 (1H, s, Ar), 7.33 (1H, d, J=8.0 Hz, Ar), 7.23 (2H, m, Ar), 7.15 (2H, m, Ar), 6.67 (1H, d, J=8.4 Hz, Ar), 5.38 (2H, s, NH2), 5.22 (1H, m, OCH), 3.87 (2H, d, J=6.0 Hz, OCH2), 3.40 (2H, dd, J=6.0 Hz, 17.2 Hz, CHCH2), 3.12 (2H, d, J=16.4 Hz, CHCH2), 2.10 (1H, m, CH(CH3)2), 0.99 (6H, d, J=7.2 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 164.9, 148.8, 143.6, 143.2, 141.0, 126.9, 125.0, 122.6, 121.8, 121.2, 120.7, 113.0, 112.4, 111.8, 79.6, 78.2, 74.8, 28.2, 19.4; MS (ESI) m/z Calcd for C27H28N2O5 (M+): 460.2, Found: 461.1 (M+H+).
4-(4-(2-Ethoxy-2-oxoacetamido)-3-isobutoxybenzamido)-3-isobutoxybenzoic acid (21)
To an ice-cooled solution of 4-(4-amino-3-isobutoxybenzamido)-3-isobutoxybenzoic acid (19; 60 mg, 0.15 mmol, 1 eq) and pyridine (24 µL, 0.30 mmol, 2 eq) in CH2Cl2 (7 mL) was added dropwise a solution of ethyl chlorooxoacetate (18 µL, 0.16 mmol, 1.1 eq) in CH2Cl2 (1 mL). The reaction mixture was allowed to warm to room temperature and stirred overnight. The resulting precipitate was collected by vacuum filtration, and the product was washed with cold 1 M HCl, cold EtOAc, then dried under high vacuum at 40 °C overnight to furnish the title compound as a white solid (57 mg, 76%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.94 (1H, bs, CO2H), 9.73 (1H, s, NH), 9.43 (1H, s, NH), 8.83 (1H, d, J=3.6 Hz, Ar), 8.23 (1H, d, J=8.0 Hz, Ar), 8.04 (1H, d, J=8.0 Hz, Ar), 7.56 (3H, m, Ar), 7.52 (1H, s, Ar), 4.30 (2H, q, J=6.8 Hz, OCH2CH3), 3.95 (2H, d, J=5.2 Hz, OCH2), 3.87 (2H, d, J=5.6 Hz, OCH2), 2.10 (2H, m, 2xCH(CH3)2), 1.29 (3H, t, J=6.8 Hz, OCH2CH3), 1.03 (6H, d, J=6.4 Hz, CH(CH3)2), 0.99 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.5, 160.3, 154.7, 150.0, 148.2, 144.2, 131.7, 131.3, 129.4, 127.6, 126.8, 122.5, 120.8, 119.5, 112.7, 111.1, 75.0, 74.9, 63.3, 28.2, 19.5, 19.3, 14.2; MS (ESI) m/z Calcd for C26H32N2O8 (M+): 500.2, Found: 501.1 (M+H+); Anal. Calcd for C29H26N2O7: C, 62.39; H, 6.44; N, 5.60. Found: C, 62.45; H, 6.25; N, 5.54.
4-(4-(2-(Carboxymethoxy)acetamido)-3-isobutoxybenzamido)-3-isobutoxybenzoic acid (22)
To an ice-cooled solution of 4-(4-amino-3-isobutoxybenzamido)-3-isobutoxybenzoic acid (19; 60 mg, 0.15 mmol, 1 eq) and Et3N (42 µL, 0.30 mmol, 2 eq) in anhydrous THF (7 mL) was added diglycolic anhydride (21 mg, 0.18 mmol, 1.2 eq). The reaction mixture was heated at 50 °C overnight. The resulting white precipitate was collected by vacuum filtration, washing with cold 1 M HCl, cold EtOAc, then dried under high vacuum at 40 °C overnight to deliver the title compound (22 mg, 28%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.92 (2H, bs, 2xCO2H), 9.38 (1H, s, NH), 9.35 (1H, s, NH), 8.34 (1H, d, J=8.0 Hz, Ar), 8.06 (1H, d, J=8.0 Hz, Ar), 7.57-7.51 (4H, m, Ar), 4.18 (4H, s, 2xOCH2CO), 3.90 (4H, m, 2xOCH2), 2.08 (2H, m, 2xCH(CH3)2), 0.99 (12H, d, J=6.4 Hz, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 171.5, 168.0, 167.3, 164.5, 149.8, 147.6, 131.8, 130.5, 129.9, 127.5, 122.5, 122.2, 120.7, 118.6, 112.6, 110.8, 75.0, 74.9, 70.7, 68.3, 28.2, 28.1, 19.5, 19.3; MS (ESI) m/z Calcd for C26H32N2O9 (M+): 516.2, Found: 517.1 (M+H+).
4-(4-(Carboxyformamido)-3-isobutoxybenzamido)-3-isobutoxybenzoic acid (23)
4-(4-(2-Ethoxy-2-oxoacetamido)-3-isobutoxybenzamido)-3-isobutoxybenzoic acid (21; 33 mg, 0.066 mmol, 1 eq) was dissolved in a 3:1:1 mixture of THF/MeOH/H2O. LiOH.H2O (7 mg, 0.17 mmol, 2.5 eq) was added to the reaction mixture, which was stirred at room temperature for 24 h. The reaction was acidified to pH 1–2 with 1N HCl, which generated a white precipitate that was collected by vacuum filtration. The precipitate was washed on the filter with 0.1N HCl, and dried under high vacuum to afford the title compound as a white solid (18 mg, 58%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.89 (1H, s, CO2H), 9.78 (1H, s, NH), 9.41 (1H, s, NH), 8.26 (1H, d, J=8.8 Hz, Ar), 8.04 (1H, d, J=8.8 Hz, Ar), 7.59-7.56 (3H, m, Ar), 7.52 (1H, s, Ar), 3.94 (2H, d, J=6.4 Hz, OCH2), 3.87 (2H, d, J=6.0 Hz, OCH2), 2.09 (2H, m, 2xCH(CH3)2), 1.02 (6H, d, J=6.0 Hz, CH(CH3)2), 0.99 (6H, d, J=6.4 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.3, 164.5, 161.8, 156.1, 150.0, 148.1, 131.8, 131.0, 129.7, 127.6, 122.5, 120.8, 119.2, 112.7, 111.1, 75.0, 74.9, 28.2, 28.1, 19.5, 19.3; MS (ESI) m/z Calcd for C24H28N2O8 (M+): 472.2, Found: 473.1 (M+H+).
2-(3-Isobutoxy-4-(3-isobutoxy-4-nitrobenzamido)benzamido)acetic acid (26)
24 (28 mg, 0.052 mmol) was dissolved in 20% TFA/CH2Cl2 (2 mL). After stirring at room temperature for 3 h, TLC indicated the reaction was complete. All solvents were removed in vacuo and residual TFA was removed by repeated azeotroping with CHCl3. The residue was dried under high vacuum to deliver the title compound as a sticky, white solid (25 mg, 100%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.58 (1H, s, CO2H), 9.76 (1H, s, NH), 8.83 (1H, s, NH), 7.99 (1H, d, J=8.0 Hz, Ar), 7.85 (1H, d, J=8.8 Hz, Ar), 7.76 (1H, s, Ar), 7.58 (1H, d, J=8.8 Hz, Ar), 7.54 (1H, s, Ar), 7.51 (1H, d, J=8.0 Hz, Ar), 4.00 (2H, d, J=6.4 Hz, OCH2), 3.91 (2H, d, J=5.6 Hz, NHCH2), 3.85 (2H, d, J=6.4 Hz, OCH2), 2.05 (2H, m, 2xCH(CH3)2), 0.96 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 171.8, 166.2, 163.9, 151.4, 151.1, 141.5, 139.8, 131.8, 129.7, 125.5, 124.1, 120.0, 119.9, 114.2, 111.5, 75.6, 74.9, 41.6, 28.2, 28.0, 19.5, 19.1; MS (ESI) m/z Calcd for C24H29N3O8 (M+): 487.2, Found: 488.1 (M+H+).
2,2'-((3-Isobutoxy-4-(3-isobutoxy-4-nitrobenzamido)benzoyl)azanediyl)diacetic acid (27)
25 (36 mg, 0.055 mmol) was dissolved in 20% TFA/CH2Cl2 (2 mL). After stirring at room temperature for 3 h, TLC indicated the reaction was complete. All solvents were removed in vacuo and residual TFA was removed by repeated azeotroping with CHCl3. The residue was dried under high vacuum to deliver the title compound as a sticky, white solid (30 mg, 100%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.95 (2H, bs, 2xCO2H), 9.75 (1H, s, NH), 7.99 (1H, d, J=8.0 Hz, Ar), 7.75 (2H, m, Ar), 7.57 (1H, d, J=8.0 Hz, Ar), 6.93 (2H, m, Ar), 3.99 (4H, m, 2xNCH2CO), 3.85 (2H, d, J=6.4 Hz, OCH2), 3.76 (2H, d, J=6.4 Hz, OCH2), 2.01 (2H, m, 2xCH(CH3)2), 0.94 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 171.4, 171.0, 170.6, 168.3, 163.9, 151.4, 141.5, 139.8, 133.3, 128.3, 125.5, 125.0, 119.9, 118.9, 114.2, 110.8, 75.6, 74.8, 66.7, 52.0, 48.3, 46.9, 28.1, 28.0, 19.4, 19.1; MS (ESI) m/z Calcd for C26H31N3O10 (M+): 545.2, Found: 546.1 (M+H+).
2-Isobutoxy-4-(3-isobutoxy-4-nitrobenzamido)benzoic acid (31)
3-Isobutoxy-4-nitrobenzoic acid75 (6a) was converted to its corresponding acid chloride according to General Procedure B on a 0.42 mmol scale, then coupled to 4-amino-2-isobutyoxybenzoic acid (30) according to General Procedure F to deliver the title compound as an off-white solid (120 mg, 66%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.31 (1H, s, CO2H), 10.55 (1H, s, NH), 8.00 (1H, d, J=8.8 Hz, Ar), 7.75 (1H, s, Ar), 7.69 (1H, d, J=8.8 Hz, Ar), 7.62 (2H, m, Ar), 7.38 (1H, d, J=8.0 Hz, Ar), 4.02 (2H, d, J=6.0 Hz, OCH2), 3.76 (2H, d, J=6.4 Hz, OCH2), 2.03 (2H, m, 2xCH(CH3)2), 0.97 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.0, 164.6, 159.0, 151.4, 143.6, 141.6, 140.0, 132.4, 125.3, 120.0, 116.6, 114.7, 111.8, 105.1, 75.7, 74.8, 28.2, 28.1, 19.4, 19.1; MS (ESI) m/z Calcd for C22H26N2O7 (M+): 430.2, Found: 431.1(M+H+).
2-Isobutoxy-4-(2-isobutoxy-4-nitrobenzamido)benzoic acid (32)
Compound 29 was converted to its corresponding acid chloride according to General Procedure B on a 0.42 mmol scale, then coupled to 4-amino-2-isobutyoxybenzoic acid (30) according to General Procedure F to deliver the title compound as an off-white solid (102 mg, 57%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.28 (1H, s, CO2H), 10.51 (1H, s, NH), 7.87 (2H, m, Ar), 7.75 (1H, d, J=8.0 Hz, Ar), 7.67 (1H, d, J=8.8 Hz, Ar), 7.51 (1H, s, Ar), 7.25 (1H, d, J=8.0 Hz, Ar), 3.97 (2H, d, J=6.0 Hz, OCH2), 3.73 (2H, d, J=6.4 Hz, OCH2), 2.01 (2H, m, 2xCH(CH3)2), 0.97 (6H, d, J=6.4 Hz, CH(CH3)2), 0.92 (6H, d, J=6.0 Hz, CH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.0, 164.2, 159.1, 156.6, 149.8, 143.5, 132.6, 132.3, 130.5, 116.3, 115.8, 110.9, 107.9, 104.2, 75.4, 74.7, 28.1, 28.0, 19.3, 19.2; MS (ESI) m/z Calcd for C22H26N2O7 (M+): 430.2, Found: 431.1 (M+H+).
3-Isobutoxy-4-(2-isobutoxy-4-nitrobenzamido)benzoic acid (33)
Compound 29 was converted to its corresponding acid chloride according to General Procedure B on a 0.42 mmol scale, then coupled to 4-amino-3-isobutyoxybenzoic acid75 (8a) according to General Procedure F to deliver the title compound as an off-white solid (106 mg, 59%): 1H-NMR (DMSO-d6, 400 MHz) δ 12.83 (1H, bs, CO2H), 10.00 (1H, s, NH), 8.40 (1H, d, J=8.0 Hz, Ar), 8.09 (1H, d, J=8.8 Hz, Ar), 7.96 (1H, s, Ar), 7.92 (1H, d, J=8.0 Hz, Ar), 7.59 (1H, d, J=8.8 Hz, Ar), 7.54 (1H, s, Ar), 4.14 (2H, d, J=6.8 Hz, OCH2), 3.91 (2H, d, J=6.4 Hz, OCH2), 2.07 (2H, m, 2xCH(CH3)2), 0.92 (12H, m, 2xCH(CH3)2); 13C-NMR (DMSO-d6, 100 MHz) δ 167.2, 162.4, 156.9, 150.5, 148.2, 132.6, 131.6, 128.9, 127.1, 122.8, 120.5, 116.2, 112.8, 109.1, 76.4, 75.3, 27.9, 27.7, 19.3, 19.1; MS (ESI) m/z Calcd for C22H26N2O7 (M+): 430.2, Found: 431.1 (M+H+).
Recombinant proteins and EMSAs
The human c-Myc bHLH-ZIP domain (residues 353–437) and full-length human Max(L) (160 residues) and Max(S) (151 residues) were used for all studies requiring purified proteins. Max(L) and Max(S) arise by alternate splicing and differ only by virtue of a nine amino acid deletion near the N-terminus of the latter that prevents it from binding DNA as a homodimer while still being able to associate with c-Myc and bind DNA as a heterodimer.59,60 Recombinant c-Myc bHLH-ZIP, Max(L) and Max(S) were expressed in the E. coli strain BL21DE3(plysS)38,45,46 as N-terminal His6-tagged proteins in the pET151-D-TOPO vector, Bacteria were grown at 37C in L-Broth to an A600 ≈ 0.8 and then for an additional 16–18 h in the presence of 1 mM isopropyl-L-thio-B-D-galactopyranoside to induce expression of the recombinant proteins. Cultures were harvested, pelleted by centrifugation as 5,000 × g for 10 min, washed in ice cold PBS and lysed in a buffer containing 8 M urea, 100 mM NaH2PO4 and 10 mM Tris-HCl, pH 8.0. Clarified lysates were then applied to Ni-NTA-Agarose affinity chromatography columns (Qiagen, Inc. Chatsworth, CA), which were washed exhaustively in lysis buffer and then developed with a pH gradient according to the directions of the supplier. Fractions containing the highest concentrations of protein were then pooled, dialyzed against 150 mM NaCl, Tris-HCl pH 6.7 and cleaved with TEV protease at 25°C as previously described.11,45,75 For larger amounts of protein, the TEV protease:His6-tagged protein molar ratios were increased to 1:50 and digestions were allowed to proceed for up to 72 h. The cleaved residues containing the His6 tag were then removed by an additional round of Ni-NTA-Aagarose chromatography. Proteins were dialyzed against storage buffer comprised of TrisHCl 50 mM pH 6.3, NaCl 150 mM and 30% glycerol and were then stored in small aliquots at −80°C. SDS-PAGE and Coomassie Blue staining of all purified proteins was performed to verify that purity exceeded 95% is all cases.
EMSA assays were performed as previously described37,38,45–49 using 30 nM of each of the above-described purified proteins and 30 nM of a 6-carboxy-2',4,4',5',7,7'-hexachloro-fluorescein (HEX)-tagged double-stranded oligonucleotide containing a c-Myc binding site (IDT, Coralville, IA). Binding reactions contained 1 × PBS (pH 7.3), 1 mmol/L EDTA, 0.1% NP40, 5% glycerol, 1 mmol/L DTT, and 400 µg/mL bovine serum albumin. Protein dimerization was allowed to proceed at room temperature for 90 min prior to the addition of the oligonucleotide for an additional 15 min. The entire reaction was subjected to electrophoresis in an 8% polyacrylamide/bis-acrylamide (80:1) gel in 0.5 × Tris-borate EDTA at 20 °C.
NMR Samples
For NMR studies, which utilized 15N-labeled proteins, proteins were propagated and purified as described above except that the growth medium contained 1 g/L 15NH4Cl. Uniformly 15N labeled Max(S) and c-Myc (353–437) were dialyzed at 4°C against an identical buffer comprised of 50 mM Tris pH 7.5, 150 mM NaCl, 0.01% sodium azide. The dilute samples of Max(S) and c-Myc were concentrated to 0.43 mM and 0.23 mM, respectively by stirred cell concentration (Millipore MWCO 3000). An NMR sample of Max(S) was formed by combining the 15N labeled Max(S) protein with a buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.01% sodium azide) to a final concentration of 0.153 mM with 10% D2O. NMR samples of 15N labeled c-Myc–Max(S) were prepared by combining equal molar amounts of the proteins to a final concentration of 0.153 mM with 10% D2O. Titration using compound 4da was conducted with addition of small volumes of a stock solution of 4da in DMSO, reaching a maximum endpoint DMSO concentration of 8%.
NMR Spectroscopy
NMR Experiments were performed on a Bruker Avance 600 MHz spectrometer. A standard 1H/15N fast HSQC pulse sequence was used for experiments with the Max/c-Myc proteins at 298K. The chemical shifts were referenced to the 1H2O signal at 4.87 ppm. The NMR data were processed using NMRPipe.77 Peak heights were measured using a modified version of Sparky.78
Calculation of Dissociation Constants (Kd)
The dissociation constants, Kd, for the binding of 4da to the c-Myc–Max(S) complex were determined by fitting the change in intensity of the bound and unbound amide crosspeaks to a simple bimolecular association model.79
In this expression C0 and CT are the intensities at the initial and final points of the titration. PT is the total concentration of the c-Myc–Max(S) complex, and MT is the total concentration of 4da at each particular point in the titration. Fitting of the binding data was carried out using Matlab (MathWorks, USA).
Surface plasmon resonance (SPR)
Recombinant, N-terminal His6-tagged c-Myc and Max(S) and Max(L) proteins were expressed in the pET151/D-Topo bacterial expression vectors and purified to >90% homogeneity using nickel-agarose gel chromatography as previously described.38,45,48,49 In the case of c-Myc, only an 85 residue bHLH-ZIP domain was expressed whereas, in the case of Max(S) and Max(L), the full-length proteins were expressed. His6 tags were cleaved with TEV protease and removed with an additional nickel-agarose chromatographic step. The proteins were then dialyzed against PBS (pH 6.5) concentrated by Centriprep Ultracel YM-3 filtration (Millipore, Carrigtwohill, Co. Cork, Ireland) and stored at −80C in small aliquots. Preliminary studies with these proteins showed them to be capable of functioning in EMSA experiments as both c-Myc–Max(S) heterodimers and Max(L) homodimers (not shown).
SPR was performed with a Biacore Model 3000 instrument (Biacor, Uppsala, Sweden) employing a streptavadin-coated SA biosensor chip (BR-1000-32) (GE Healthcare Bio-Sciences AB, Pittsburgh, PA). The following biotinylated E-box-containing oligonucleotide (5’-biotin-[TGAAGCAGACCACGTGGTCGTCTTCA] was annealed with its unlabeled opposite strand oligonucleotide, diluted in HBS-EP running buffer (0.5M NaCl; 10 mMM HEPES, pH 7.4; 3 mM EDTA and 0.005% v/v Surfactant P20) and immobilized on the flow cell to allow the attachment of ~700 RU. A second flow cell without any bound DNA was used as a blank. DNA binding experiments were carried out with running buffer (0.15 M NaCl; 10 mM HEPES, pH 7.4; 3 mM EDTA, 0.005% v/v Surfactant P20 and 5% DMSO) at a flow rate of 60 µL min−1.
Binding of pre-formed c-Myc–Max(S) or Max(L)–Max(L) dimers was achieved by first allowing dimerization to occur at a concentration of 20 nM of each protein HBS-EP buffer for 30 min. Dimers were then introduced onto the oligonucleotide-containing chip at a flow rate of 60 µL min−1. In each case, the amount of c-Myc–Max(S) heterodimer or Max(L) homodimer binding was found to be linearly proportional to concentration and reached equilibrium at the highest concentration, indicating that binding was both specific and saturable. In all cases, protein concentrations were chosen so as to provide an additional 250 relative response units upon binding to the sensor chip. Under similar conditions, no binding to the immobilized oligonucleotide was seen when c-Myc monomer alone was tested (not shown)).
Small molecule interaction studies were performed either by pre-incubating the molecules at the indicated concentrations for 30 min with c-Myc (20 nM) followed by the addition of Max(S) at the above-noted concentrations. In other experiments, designed to determine whether compounds could perturb the binding of pre-formed c-Myc–Max heterodimers, c-Myc and Max(S) proteins were first allowed to heterodimerize at the above concentrations followed by the addition of compounds to the final stated concentrations for 30 min. The mixture was then passed over the oligonucleotide-bound chip at a rate of 60 µL min−1 while maintaining a constant concentration of compound.
Binding constants and graphing were performed with BIAevaluation 4.1.1 software (University of Reading, UK), all response units were normalized to 0–100 RU. More detailed descriptions of these procedures will be published elsewhere (Wang et al, in preparation)
Cell viability assays
These were performed as previously described.37,38,47,48,62 Briefly, cells at >90% viability were seeded into 96 well plates (2×103 cells/well) in fresh growth medium containing 10-point serial dilutions of the compounds of interest. Incubations were allowed to proceed for three days at which time cell viability was assed using the MTT assay as previously described.37,47,48 Each point was assessed in at least triplicate determinations.
Cell cycle analyses
Cell cycle analyses were performed as previously described.37 Briefly, after treating with the indicated c-Myc inhibitors, cells were washed twice in cold PBS and re-suspended in 1 ml of 10 mM NaCl, 10 mM Tris-HCl, (pH 8.0), 0.1% NP40, 10 µg/ml RNase A, and 15 µg/ml propidium iodide (Sigma-Aldrich, Inc. St. Louis, MO). Stained nuclei were analyzed on a Becton Dickinson FACStar fluorescence-activated cell sorter and cell cycle quantification was performed using ModFit LT 3.0 software (Verity Software House, Topsham ME).
Neutral lipid quantification
H460 large cell lung cancer cells were incubated at sub-confluent densities in 6 well plates with the indicated concentrations of c-Myc inhibitors for 48 h. The cells were then washed twice in PBS, trypsinized and again washed twice in PBS before being re-suspended in 0.1 ml of PBS. 0.9 ml of PBS containing 50 µg/ml of BODIPY (4,4-difluoro-3a,4adiaza-s-indacene) 459/503 (Life Technologies, Carlsbad, CA), which is specific for neutral lipid, was then added and the re-suspended cells were incubated for 30 min at ambient temperature. Cells were then washed three additional times in PBS before being re-suspended in 0.5 ml of PBS and analyzed by flow cytometry. Fluorescence quantification was performed using Cell Quest Pro software (BD Biosciences, Franklin Lakes, NJ) and mean fluorescence of each sample was determined.
Luciferase assays
The luciferase expression vector pGL4.28[luc2CP/minP/Hygro] (Promega, Inc. Madison, WI) contains a minimal mammalian promoter (minP) and a luciferase2CP expression cassette that imparts additional protein instability by fusing hCL1 and PEST sequences. A large number of additional mutations were also introduced throughout the vector to minimize the non-specific binding of mammalian transcription factors and otherwise reduce background expression. Into a unique XhoI site of the polylinker adjacent to the minP sequence, we ligated one of three individual oligonucleotides containing tandemly triplicated c-Myc binding sites (5’-TCGAGCCACGTGGCCACGTGGCCACGTGGC-3’) (Wt-c-Myc), mutant Myc binding sites (5’-TCGAGCCTCGAGGCCTCGAGGCCTCGAGGC-3’), Mut-c-Myc) and NF-κB binding sites (5’-TCGAGGGACTTTCCATGGGACTTTCCATGGGACTTTCC-3’) (NF-κB) where underlined bases indicate the relevant transcription factor binding sites. The identities of all constructs were determined by automated Sanger DNA sequencing. Each vector was then combined with a 5-fold excess of the vector pFR400, which encodes a mutant mammalian dihydrofolate reductase with a lower Km for the cytotoxic drug methotrexate78 and stably transfected into HeLa cells using Superfect (Qiagen, Valencia, CA). Following selection in hygromycin, stably transfected clones were pooled and subjected to at least two rounds of selection in 4-fold stepwise increasing concentrations of methotrexate (Sigma-Aldrich, St Louis, MO) as previously described to allow for amplification of the adjacent vector sequences.78 Cells were cultured continuously in methotrexate until the day of plating for luciferase assays so as to ensure maintenance of the amplified plasmid sequences.
To perform luciferase assays, 106 cells were seeded into 6 well plates. The following day, the medium was removed and fresh medium containing the indicated concentration of c-Myc inhibitor was added for 6 h. Cells were then trypsinized, washed twice in PBS and pelleted by low-speed centrifugation. An aliquot of each sample was used for protein determinations against which subsequent luciferase determinations were normalized. The remaining cell pellets were lysed and assayed in triplicate in 96 well plates using a ONE-Glo™ Luciferase Assay luciferase assay kit according to the directions of the supplier (Promega). Readings were obtained on a Wallac 1420 VICTOR2 (PerkinElmer Life and Analytical Sciences). Under these conditions, background luciferase activity in cells expressing the Mut-c-Myc vector was indistinguishable from that of untransefected cells whereas the expression of luciferase in control lysates of cells transfected with the Wt-c-Myc and NF-kB vectors was typically 50–100-fold higher. Background values were subtracted from all experimental determinations. All results obtained from cells exposed to c-Myc inhibitors were plotted as mean values +/− 1 standard error and expressed relative to those of untreated cells, which were arbitrarily set at 100%.
Co-immunoprecipitation (co-IP) studies
Co-IP assays were performed as previously described.48 Briefly, approximately 5 × 106 HL60 cells were exposed for 6 hr to the stated concentration of c-Myc inhibitor or to the DMSO vehicle only, which served as a negative control. Cells were collected by centrifugation at 500 × g for 10 min, washed twice in ice-cold PBS and lysed in IP Buffer.48,60 An aliquot of each cleared lysate was saved prior to co-IP to allow a comparison of input c-Myc protein levels. 300 µg of the cleared lysate in 1 ml of IP buffer was then precipitated at 4C overnight with a 1:200 dilution of anti-Max antibody60 followed by precipitation with protein G-Sepharose using conditions suggested by the supplier (Santa Cruz Biotechnology, Inc. Santa Cruz, CA). The precipitate was then washed three times in IP buffer and subjected to 10% SDS-PAGE and immunoblotting with a 1:1000 dilution of anti-Myc monoclonal antibody (9E10, Santa Cruz Biotechnology, Santa Cruz, CA). Blots were developed using an enhanced chemiluminescence kit following the directions recommended by the supplier (Pierce ECL Plus, Thermo-Fisher, Pittsburgh, PA).
Supplementary Material
Chart 1.
Structures of some inhibitors that bind monomeric c-Myc.
BRIEFS.
Synthetic α-helix mimetics perturb the c-Myc–Max dimer, abrogating its ability to bind DNA.
Acknowledgments
Financial support of this work was provided by an American Cancer Society Young Investigator Award (S.F.), the University of Maryland School of Pharmacy (S.F.), an American Chemical Society Pre-Doctoral Medicinal Chemistry Fellowship and American Foundation for Pharmaceutical Education Fellowship (both to J.L.Y), and the National Institutes of Health (R01CA140624 to E.V.P.).
ABBREVIATIONS
- BET
bromodomain and extraterminal domain
- bHLH-ZIP
basic helix-loop-helix leucine zipper
- EMSA
electrophoretic mobility shift assay
- HSQC
heteronuclear single quantum coherence
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PPI
protein–protein interaction
- SPR
surface plasmon resonance
- TCA
tricarboxylic acid cycle
Footnotes
SUPPORTING INFORMATION AVAILABLE: Synthesis of intermediates 3e, 3g, 8, 5g–5i, 6e, 10, 21, 22, 26–28, EMSA dose-response plots for 1aa, 4da, 1ca, 12, 13 and 18, and cell cycle analysis data for 4da and 10058-F4. This information if available free of charge via the internet at http://pubs.acs.org
REFERENCES
- 1.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 3.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 5.Eisenman RN. Deconstructing myc. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 6.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 7.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 8.Pelengaris S, Khan M. The many faces of c-MYC. Arch. Biochem. Biophys. 2003;416:129–136. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 9.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 10.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 11.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 12.Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 13.Berns EM, Klijn JG, van Putten WL, van Staveren IL, Portengen H, Foekens JA. c-myc amplification is a better prognostic factor than HER2/neu amplification in primary breast cancer. Cancer Res. 1992;52:1107–1113. [PubMed] [Google Scholar]
- 14.Buchholz M, Schatz A, Wagner M, Michl P, Linhart T, Adler G, Gress TM, Ellenrieder V. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 2006;25:3714–3724. doi: 10.1038/sj.emboj.7601246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitani S, Kamata H, Fujiwara M, Aoki N, Tango T, Fukuchi K, Oka T. Analysis of c-myc DNA amplification in non-small cell lung carcinoma in comparison with small cell lung carcinoma using polymerase chain reaction. Clin. Exp. Med. 2001;1:105–111. doi: 10.1007/s10238-001-8020-5. [DOI] [PubMed] [Google Scholar]
- 16.Erisman MD, Rothberg PG, Diehl RE, Morse CC, Spandorfer JM, Astrin SM. Mol. Cell. Biol. 1985;5:1969–1976. doi: 10.1128/mcb.5.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado MD, Leon J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–616. doi: 10.1177/1947601910377495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Eyss B, Eilers M. Addicted to Myc--but why? Genes Dev. 2011;25:895–897. doi: 10.1101/gad.2053311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, Gudkov AV, Prochownik EV, Nikiforov MA. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene. 2008;27:1905–1915. doi: 10.1038/sj.onc.1210823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev. Mol. Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prinjha RK, Witherington J, Lee K. Place your BETs: the therapeutic potential of bromodomains. Trends Pharmacol. Sci. 2012;33:146–153. doi: 10.1016/j.tips.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Hewings DS, Rooney TP, Jennings LE, Hay DA, Schofield CJ, Brennan PE, Knapp S, Conway SJ. Progress in the development and application of small molecule inhibitors of bromodomain-acetyl-lysine interactions. J. Med. Chem. 2012;55:9393–9413. doi: 10.1021/jm300915b. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez R, Meslamani J, Zhou MM. The bromodomain: from epigenome reader to druggable target. Biochim. Biophys. Acta. 2014;1839:676–685. doi: 10.1016/j.bbagrm.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zirath H, Frenzel A, Oliynyk G, Segerström L, Westermark UK, Larsson K, Munksgaard Persson M, Hultenby K, Lehtiö J, Einvik C, Påhlman S, Kogner P, Jakobsson PJ, Henriksson MA. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. U S A. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soucek L, Nasi S, Evan GI. Omomyc expression in skin prevents Myc-induced papillomatosis. Cell Death. Differ. 2004;11:1038–1045. doi: 10.1038/sj.cdd.4401443. [DOI] [PubMed] [Google Scholar]
- 29.Annibali D, Whitfield JR, Favuzzi E, Jauset T, Serrano E, Cuartas I, Redondo-Campos S, Folch G, Gonzàlez-Juncà A, Sodir NM, Massó-Vallés D, Beaulieu ME, Swigart LB, McGee MM, Somma MP, Nasi S, Seoane J, Evan GI, Soucek L. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soucek L, Evan GI. The ups and downs of Myc biology. Curr. Opin. Genet. Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fletcher S, Prochownik EV. Small-molecule inhibitors of the Myc oncoprotein. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbagrm.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap JL, Chauhan J, Jung K-Y, Chen L, Prochownik EV, Fletcher S. Small-molecule inhibitors of dimeric transcription factors: Antagonism of protein–protein and protein–DNA interactions. Med Chem Comm. 2012;3:541–551. [Google Scholar]
- 35.Prochownik EV, Vogt PK. Therapeutic Targeting of Myc. Genes Cancer. 2010;1:650–659. doi: 10.1177/1947601910377494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, Prochownik EV. Improved low molecular weight Myc-Max inhibitors. Mol. Cancer Ther. 2007;6:2399–2408. doi: 10.1158/1535-7163.MCT-07-0005. [DOI] [PubMed] [Google Scholar]
- 38.Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. USA. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi J, Strover JS, Whitby LR, Vogt PK, Boger DL. Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg. Med. Chem. Lett. 2009;19:6038–6041. doi: 10.1016/j.bmcl.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, Janda KD. A credit-card library approach for disrupting protein-protein interactions. Bioorg. Med. Chem. 2006;14:2660–2673. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 41.Kiessling A, Sperl B, Hollis A, Eick D, Berg T. Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem. Biol. 2006;13:745–751. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Kiessling A, Wiesinger R, Sperl B, Berg T. Selective inhibition of c-Myc/Max dimerization by a pyrazolo[1,5-a]pyrimidine. Chem Med Chem. 2007;2:627–630. doi: 10.1002/cmdc.200600294. [DOI] [PubMed] [Google Scholar]
- 43.Follis AV, Hammoudeh DI, Wang HB, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 2008;15:1149–1155. doi: 10.1016/j.chembiol.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 2009;131:7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- 45.Yap JL, Wang H, Hu A, Chauhan J, Jung K-Y, Gharavi RB, Prochownik EV, Fletcher S. Pharmacophore identification of c-Myc inhibitor 2. Bioorg. Med. Chem. Lett. 2013;23:370–374. doi: 10.1016/j.bmcl.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, Chauhan J, Hu A, Pendleton K, Yap JL, Sabato PE, Jones JW, Perri M, Yu J, Cione E, Kane MA, Fletcher S, Prochownik EV. Disruption of Myc-Max heterodimerization with improved cell-penetrating analogs of the small molecule 2. Oncotarget. 2013;4:936–947. doi: 10.18632/oncotarget.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chauhan J, Wang H, Yap JL, Sabato PE, Hu A, Prochownik EV, Fletcher S. Discovery of methyl 4'-methyl-5-(7-nitrobenzo[c][1,2,5]oxadiazol-4-yl)-[1,1'-biphenyl]-3-carboxylate, an improved small-molecule inhibitor of c-Myc-Max dimerization. Chem Med Chem. 2014;9:2274–2285. doi: 10.1002/cmdc.201402189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fieber W, Schneider ML, Matt T, Kräutler B, Konrat R, Bister K. Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J. Mol. Biol. 2001;307:1395–1410. doi: 10.1006/jmbi.2001.4537. [DOI] [PubMed] [Google Scholar]
- 49.Ernst JT, Kutzki O, Debnath AK, Jiang S, Lu H, Hamilton AD. Design of a protein surface antagonist based on alpha-helix mimicry: inhibition of gp41 assembly and viral fusion. Angew. Chem. Int. Ed. Engl. 2002;41:278–281. doi: 10.1002/1521-3773(20020118)41:2<278::aid-anie278>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 50.Saraogi I, Hebda JA, Becerril J, Estroff LA, Miranker AD, Hamilton AD. Synthetic alpha-helix mimetics as agonists and antagonists of islet amyloid polypeptide aggregation. Angew. Chem. Int. Ed. Engl. 2010;49:736–739. doi: 10.1002/anie.200901694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yap JL, Cao X, Vanommeslaeghe K, Jung K-Y, Wilder P, Nan A, MacKerell AD, Jr, Smythe WR, Fletcher S. Relaxation of the rigid backbone of an oligoamide-foldamer-based α-helix mimetic: identification of potent Bcl-xL inhibitors. Org. Biomol. Chem. 2012;10:2928–2933. doi: 10.1039/c2ob07125h. [DOI] [PubMed] [Google Scholar]
- 52.Cao X, Yap JL, Newell-Rogers MK, Peddaboina C, Hua J, Papaconstantinou HT, Jupitor D, Rai A, Jung K-Y, Tubin RP, Yu W, Vanommeslaeghe K, Wilder PT, MacKerell AD, Jr, Fletcher S, Smythe WR. The novel BH3 α-helix mimetic JY-1-106 induces apoptosis in a subset of cancer cells (lung cancer, colon cancer and mesothelioma) by disrupting Bcl-xL and Mcl-1 protein-protein interactions with Bak. Mol. Cancer. 2013;12:42. doi: 10.1186/1476-4598-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung K-Y, Vanommeslaeghe K, Lanning ME, Yap JL, Gordon C, Wilder PT, MacKerell AD, Jr, Fletcher S. Amphipathic α-helix mimetics based on a 1,2-diphenylacetylene scaffold. Org. Lett. 2013;15:3234–3237. doi: 10.1021/ol401197n. [DOI] [PubMed] [Google Scholar]
- 54.Orner BP, Ernst JT, Hamilton AD. Toward proteomimetics: Terphenyl derivatives as structural and functional mimics of extended regions of an α-helix. J. Am. Chem. Soc. 2001;123:5382–5383. doi: 10.1021/ja0025548. [DOI] [PubMed] [Google Scholar]
- 55.Plante JP, Burnley T, Malkova B, Webb ME, Warriner SL, Edwards TA, Wilson AJ. Oligobenzamide proteomimetic inhibitors of the p53-hDM2 protein-protein interaction. Chem. Commun. 2009:5091–5093. doi: 10.1039/b908207g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lanning ME, Fletcher S. Recapitulating the α-helix: nonpeptidic, low-molecular-weight ligands for the modulation of helix-mediated protein–protein interactions. Future Med. Chem. 2013;5:2157–2174. doi: 10.4155/fmc.13.176. [DOI] [PubMed] [Google Scholar]
- 57.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H, Fan S, Prochownik EV. Distinct roles for MAX protein isoforms in proliferation and apoptosis. J. Biol. Chem. 1997;272:17416–17424. doi: 10.1074/jbc.272.28.17416. [DOI] [PubMed] [Google Scholar]
- 59.Prochownik EV, VanAntwerp ME. Differential patterns of DNA binding by myc and max proteins. Proc. Natl. Acad. Sci. U. S. A. 1993;90:960–964. doi: 10.1073/pnas.90.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holien T, Vatsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120:2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 61.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibrobasta isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 62.Graves JA, Rothermund K, Wang T, Qian W, Van Houten B, Prochownik EV. Point mutations in c-Myc uncouple neoplastic transformation from multiple other phenotypes in rat fibroblasts. PLoS One. 2010;5(10):e13717. doi: 10.1371/journal.pone.0013717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bennett MR, Anglin S, McEwan JR, Jagoe R, Newby AC, Evan GI. Inhibition of vascular smooth muscle cell proliferation in vitro and in vivo by c-myc antisense oligodeoxynucleotides. J. Clin. Invest. 1994;93:820–828. doi: 10.1172/JCI117036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin. Cancer Biol. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, Gudkov AV, Prochownik EV, Nikiforov MA. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene. 2008;27:1905–1915. doi: 10.1038/sj.onc.1210823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dang CV. Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011;76:369–374. doi: 10.1101/sqb.2011.76.011296. [DOI] [PubMed] [Google Scholar]
- 67.Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, Prochownik EV. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS One. 2012;7:e37699. doi: 10.1371/journal.pone.0037699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edmunds LR, Sharma L, Kang A, Lu J, Vockley J, Basu S, Uppala R, Goetzman ES, Beck ME, Scott D, Prochownik EV. c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J. Biol. Chem. 2014 doi: 10.1074/jbc.M114.580662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clausen DM, Guo J, Parise RA, Beumer JH, Egorin MJ, Lazo JS, Prochownik EV, Eiseman JL. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics, and metabolism of 2, a novel small-molecule inhibitor of c-Myc/Max dimerization. J. Pharmacol. Exp. Ther. 2010;335:715–727. doi: 10.1124/jpet.110.170555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy NS, Prabhakaran P, Azzarito V, Plante JP, Hardie MJ, Kilner CA, Warriner SL, Wilson AJ. Solid-phase methodology for synthesis of O-alkylated aromatic oligoamide inhibitors of α-helix-mediated protein-protein interactions. Chem. Eur. J. 2013;19:5546–5550. doi: 10.1002/chem.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plante J, Campbell F, Malkova B, Kilner C, Warriner SL, Wilson AJ. Synthesis of functionalised aromatic oligamide rods. Org. Biomol. Chem. 2008;6:138–146. doi: 10.1039/b712606a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferre-D'Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 75.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 76.Goddard TD, Kneller DG. SPARKY3. San Francisco, CA: University of California; 2001. [Google Scholar]
- 77.Lian LY, Barsukov IL, Sutcliffe MJ, Sze KH, Roberts GC. Protein-ligand interactions: exchange processes and determination of ligand conformation and protein-ligand contacts. Meth. Enzymol. 1994;239:657–700. doi: 10.1016/s0076-6879(94)39025-8. [DOI] [PubMed] [Google Scholar]
- 78.Prochownik EV, Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. Nature. 1986;322:848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.