Abstract
Background:
This was a meta-analysis and systematic review to determine the global prevalence of the mitochondrially encoded 12S RNA (MT-RNR1) genetic mutation in order to assess the need for neonatal screening prior to aminoglycoside therapy.
Materials and Methods:
A comprehensive search of MEDLINE, EMBASE, Ovid, Database of Abstracts of Reviews of Effect, Cochrane Library, Clinical Evidence and Cochrane Central Register of Trials was performed including cross-referencing independently by 2 assessors. Selections were restricted to human studies in English. Meta-analysis was done with MetaXL 2013.
Results:
Forty-five papers out of 295 met the criteria. Pooled prevalence in the general population for MT-RNR1 gene mutations (A1555G, C1494T, A7445G) was 2% (1–4%) at 99%.
Conclusion:
Routine screening for MT-RNR1 mutations in the general population prior to treatment with aminoglycosides appear desirable but poorly supported by the weak level of evidence available in the literature. Routine screening in high-risk (Chinese and Spanish) populations appear justified.
Key words: Aminoglycoside, mitochondrial genetics, mutation, ribosomal ribonucleic acid, sensorineural hearing loss
INTRODUCTION
The presence of gene variants in mitochondrial encoded MT-RNR1 (12S RNA) genes is associated with aminoglycoside-induced sensorineural hearing loss (SNHL).[1,2,3,4,5,6,7] Global reports have shown variations in the prevalence/incidence of MT-RNR1; however, the A1555G mutation (the first identified homoplasmic variant)[2] is frequent among families in Africa, Asia, Europe and America.[6,7,8,9,10,11,12,13] This mutation has a matrilineal inheritance and in one study a reported propensity in more than 50% of those affected to develop SNHL following exposure to aminoglycosides.[14] Sporadic development of hearing loss in those who carry this mutation without aminoglycoside exposure is rare.[15]
Despite the risk of ototoxicity, aminoglycosides are widely used. Gentamicin is the most commonly used aminoglycoside in the treatment of neonatal sepsis due to bacterial sensitivity and cost effectiveness compared to other groups of antibiotics. Minimizing the risk of ototoxic hearing loss may require screening of all patients especially newborns for MT-RNR1 mutant genes prior to aminoglycoside therapy if MT-RNR1 mutations are common in the exposed population and if the risk of ototoxicity in the affected patients is significant.
Rapid screening tests with the ability to detect the most common variants are available.[16,17] The cost effectiveness of such screenings are yet to be done, but would need to consider both the upfront costs associated with screening for the mutation versus the lifetime cost of habilitation of a deaf child, either of which can be significant. Mohr et al. in a societal study put the lifetime cost of habilitation of children with severe-to-profound hearing loss at one million dollars USD per individual.[18]
The objective of this systematic review was to determine the prevalence/incidence of MT-RNR1 genetic mutations in the general population as a first step toward determining the cost-effectiveness of neonatal screening for MT-RNR1 mutations prior to gentamicin therapy.
MATERIALS AND METHODS
A comprehensive search of MEDLINE (from 1947 to April 2014), EMBASE (from 1974 to April 2014), Publius Ovidius Naso (Ovoid), Database of Abstracts of Reviews of Effects and the Cochrane Central Register of Controlled Trials in issue 3, 2014 of the Cochrane Library was performed. The studies were restricted to humans and English language. The search strategy used the exploded Medical Subject Headings term “hearing screening” “hearing loss” combined with the second set obtained using the exploded terms “Genetic screening gene” or “susceptible gene for newborn” or “aminoglycosides” OR “MT-RNR1” OR “A1555G” OR “A7445G” OR “C1494T”. Reference lists and citation indexes of identified manuscripts were cross-referenced to identify further relevant literature. Short-listing of titles and abstracts on the basis of relevancy and subsequent data extraction were undertaken independently by two authors. Differences were resolved by mutual consensus.
Inclusion criteria
Studies investigating the prevalence/incidence of MT-RNR1 gene mutations (A1555G, C1494T, A7445G) in the population.
Relevant human studies on screening for MT-RNR1.
Studies describing the risk of SNHL in MT-RNR1 mutation.
Exclusion criteria
Letters to the editor, conference proceedings and editorials.
Non-English literature and animal studies.
Case reports from which incidence/prevalence data could not be extracted.
Studies that did not meet the inclusion criteria, though considered relevant to the body of evidence were reviewed as appropriate, including review of references to ensure inclusion of all relevant literature.
The 2011 Centre for Evidence Based Medicine Oxford criteria was used to assess/classify the quality designs of studies.[19]
Results/Meta-analysis
This was performed on all the prevalence recorded globally using the MetaXL 2013 random effect package and method.[20]
The preliminary search yielded 295 articles. Sixty-nine full text articles were selected following review of the titles and abstracts. Cross-referencing of these articles provided a further 25 studies bringing the cumulative total to 91 of which 45 were found to meet our inclusion criteria [Figure 1], representing cohorts, case–control studies and case series only. All papers were independently reviewed for study design and data extraction by at least two investigators [Table 1].
Figure 1.
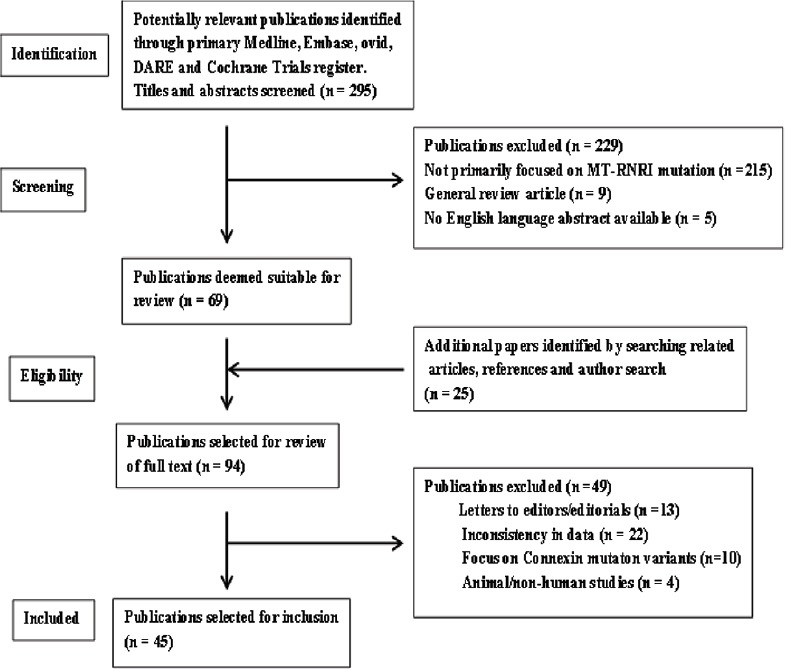
Illustrates the search and selection processes for the articles utilised in the mitochondrially encoded 12S RNA systematic review and meta-analysis
Table 1.
This contains the summary of the contents of the articles selected for inclusion in this study including the level of evidence, article type, race studied, results and outcome
| Paper | Level of evidence | Type of paper | What is the focus | Study population | Race | Drug | Mutation | Outcome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Postal et al. (2009) | Level III | Case control | Incidence | NICU (high-risk) | Latin American (Brazil) | Aminoglycoside | C1494T | No mutation seen | |||||||||
| A=HL + A (n=20) | |||||||||||||||||
| B=NHL + A (n=20) | |||||||||||||||||
| C=HL − A (n=20) | |||||||||||||||||
| Han et al. (2007) | Level IV | Cross-section | Incidence/effect | Adults | Chinese | Aminoglycoside (n=2/66) | C1494T | 15/66 mutation seen | |||||||||
| HL (n=13/66) | |||||||||||||||||
| Wang et al. (2011) | Level III | Cross-section | Prevalence | Neonatal | Chinese | None | 12S rRNA, A1555G | 18/14913 mutation seen | |||||||||
| Johnson et al. (2010) | Level III | Case control | Prevalence/effect | NICU-LBW | Caucasian (USA) | Gentamicin | 12S sRNA, T961C, T1494C, A1555G | 4/436, 1/4 immediate HL post exposure | |||||||||
| Tang et al. (2002) | Level III | Cross-section | Prevalence | NICU | Mixed (USA) | None | A15555G | 1/1161 (0.09%) | |||||||||
| De Moraes (2009) | Case control | Prevalence | NICU (high-risk)-A, B, C Adults-D | Latin America (Argentina) | Aminoglycoside | A1555G, C1494T, A827G | 19/85 | ||||||||||
| Diverse data, unable to have clear outcome | A=HL + A (n=25) | ||||||||||||||||
| B=NH + A (n=25) | |||||||||||||||||
| C=HL − A (n=25) | |||||||||||||||||
| D=HL − A (n=10) | |||||||||||||||||
| Rydzanicz (2009) | Level IV | Cross-section (n=500) | Prevalence | General population | Polish (Caucasians) | None | A827G, T961C, A1555G | A827G (1/500), T961C (1/250), A1555G (1/250) | |||||||||
| Shen et al. (2011) | Level III | Case (n=440), control (n=449) | Prevalence | Paediatric subjects with HL | Chinese | None | A1555G, T1494C, T1095>C | Case: A1555G (33/440, 7.5%), T1494C (2/440), T1095c (4/440), no clear control details | |||||||||
| Zhu (2009) | Level IV | Cross-section | Prevalence | HL in Chinese | Chinese | Aminoglycoside | C1494T | 13/3133 | |||||||||
| Rodriguez-Ballesteros (2006) | Level III | Case (n=1340) Control (n=894) | Relative risk | General population (1340 + HL), 894 no HL | Caucasian (Spanish) | None | C1494T | Case: 20/1340 Control: 0 | |||||||||
| Estivill et al. 1998 | Level III | Case (n=429 out of which Bil NSHL=214 Control (n=200) | Prevalence Relative risk | General population + NSNHL (at risk gp) | Caucasian (Spanish) | Aminoglycoside | A155G | Case: Control: 0 Relative risk postaminogly=X2.3 | |||||||||
| Del Castillo et al. (2003) | Level IV | Case (n=649 families with HL) | Prevalence | General population | Caucasian (Spanish) | None | A1555G | 105/649 | |||||||||
| Wu et al. (2007) | Level IV | Cases (n=families 3 15, 620 patient with NSHL) | Prevalence | General population | Hans-Chinese | None | A1555G | 10/315 families, penetration depends on mitochondrial haplotype | |||||||||
| Maniglia et al. (2008) | Level 1V | Cross-section Case (n=27) Control (n=100) | Prevalence | General population | Latin Americans (Brazil) | Aminoglycoside | A1555G | 0 for case and control | |||||||||
| Ealy (2001) | Level III | Cases: 703 (NICU) Control: 1473 Total=2176 | Prevalence | NICU and general population | USA (Iowa) | None | A1555G, C1494T, T1095C, A827G | NICU 1.85%, genral population 1.83% (A827G, most common, next A1555G) | |||||||||
| Lu et al. (2010) | Level III | Paed cases: 1642 (HL) Control: 449 | Prevalence | Paediatric and general population | Han-Chinese | Aminoglycoside | A1555G, T1494C, T1095C | A1555G 3.9%, T1494C (0.18%), T1095C (0.61%), T 961 (1.7%) Control: no mutation, no details of the effect of aminoglycoside | |||||||||
| Saunders et al. (2009) | Level IV | Cases (n=31) | Prevalence | Paediatrics=31 | Latin America (Nicaragua) | Aminoglycoside | A1555G, T1494C, T1095C | No mutant detection | |||||||||
| Fischel-Ghodsian et al. (1997) | Level IV | Cases (n=41) | Incidence in high-risk group | General population (NSHL) | Mixed races (USA) | Aminoglycoside | A1555G | 17% incidence | |||||||||
| Gravina et al. (2007) | Level IV | Blood donors=712 + dried blood spots=330 new born | Prevalence | General population | Latin America (Argentina) | None | A1555G, T1291C | 1/1042 for T1291C, none for A1555G | |||||||||
| Bai et al. (2008) | Level IV | Pedigree with mutation A1555G | To assess HL and frequency of mutation | Single family pedigree | Chinese | Aminoglycoside | A1555G | 19/27 positive for A1555G, 4/27 have normal hearing with mutation (2nd and 3rd decade) | |||||||||
| Kato et al. (2010) | Level IV | Case series n=373 (SNHL) | Assess accuracy and of screening test (1 h), prevalence | Adult HL | Japanese | None | A1555G, A3243G, A8348G, G1177A | A1555G 11/373 (2.9%, A 3243G 9/373 (2.7%), A8348G 1/373, G1177A 1/373. Test was accurate in 99.8% | |||||||||
| Human et al. (2010) | Level IV | MDR-TB cases=115 General population n=439 (Afrikaner 93, black 112, Caucasian 104 and mixed ancestry 130) | Prevalence | Adult TB patients | South Africans (Afrikaner, Black, Caucasian and mixed ancestry) | Aminoglycoside (Strepto, Kana, Amikacin) | A1555G, A827G, C1494T, T1095C, T1291C | A827G 1/115, no other mutation Control: A1555G 1/112 black, 1/93 Afrikaner. A827G 1/93 Afrikaner | |||||||||
| Bardien et al. (2009) | Level IV | General population (black 106, mixed ancestry 98) | Prevalence | General population | South Africans | None | A1555G, A827G, C1494T, T1095C, T1291C | A1555G 1/106 black, no other mutation identified | |||||||||
| Wan et al. (2013) | Level III | Case control Meniere’s disease population (Caucassian 33) | Prevalence | Meniere’s disease population | Canadians | None | A1555G, A827G, C1494T | No mutation 0/33 | |||||||||
| Huang et al. (2013) | Level III | Case control deaf population (Chinese) | Prevalence | Deaf | Chinese | None | A1555G, A827G, C1494T, T1095C, T1291C | 6/6000, 0.1% | |||||||||
| Guaran et al. (2013) | Level III | Cohort study | Prevalence | Deaf | Italian | None | A1555G | 4/169, 2.3% | |||||||||
| Zhu et al. (2014) | Level IV | Case series | Penetrance | Single family pedigree | Chinese | Aminoglycoside | A1555G | Penetrance varies=10-52%; risk of deafness−~ heteroplasmy levels | |||||||||
| Liang et al. (2013) | Level III | Case series | Penetrance | Single family pedigree | Chinese | tRNAlle | A1555G, A4317G | Penetrance high 66.7% and 81% | |||||||||
| Yang et al. (2013) | Level III | Case control | Prevalence | General population | N/W China (Tibet, Tu and Mongolians) | None | A1555G, C1494T | 13/189, 6.9% | |||||||||
| Zhang et al. (2013) | Level III | Cohort | Prevalence | Deaf | Chinese | None | A1555G, C1494T | 25/215, 11.6% | |||||||||
| Yao et al. (2013) | Level III | Case control | Prevalence | Deaf and general population | Chinese | None | A1555G, C1494T | Case 6/227=2.64% Control 0/200 | |||||||||
| Wang et al. (2011) | Level III | Cohort | Prevalence | Deaf | Chinese | None | A1555G, C1494T | 29/1448, 1.8% | |||||||||
| Stefanovska et al. (2012) | Level III | Cohort | Prevalence | Deaf | Macedonia | Aminoglycoside | A1555G, C1494T | 0/130, 0% | |||||||||
| Danilenko et al. (2012) | Level III | Case control | Prevalence | General | Belarius | None | A1555G, C1494T | 2/391, 0.55% | |||||||||
| Zohour et al. (2012) | Level III | Cohort | Prevalence | Prelingual deaf | Iranian | None | A1555G, C1494T, A7445G | 8/2000, 0.4% | |||||||||
| Cai et al. | Level III | Cohort | Prevalence | General neonantal | China | None | A1555G | 2/1000, 0.2% | |||||||||
| Zhong et al. (2013) | Level III | Cohort | Prevalence | General neonatal | China | None | A1555G, C1494T, A7445G | 3225/58, 97, 5.52% | |||||||||
| Nahil et al. (2010) | Level III | Case control | Prevalence | General | Morocco | None | A1555G | 6/164, 3.6% | |||||||||
TB: Tuberculosis; HL: Hearing loss; NICU: Neonatal intensive care unit; LBW: Low birth weight; NSHL: Nonsyndromic hearing loss; MDR: Multi drug resistant; SNHL: Sensorineural hearing loss
Forty-two articles[3,6,7,9,10,11,12,13,16,17,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36] (the others are as shown in Figure 2) addressed the prevalence/incidence of MT-RNR1 mutation in the populations with SNHL for which a range of 0% in Brazilian[30] and Argentine[32] subjects to 100% in 3 Spanish families[31] with aminoglycoside-induced hearing loss were presented. The modal prevalence was 3.2% and only A1555G and/or C1494T variants were assessed. Fourteen papers[9,10,16,21,30,32,33,34] [rest as seen in Figure 2] described prevalence/incidence in the general population/normal controls; these included low birth weight neonates, newborns and neonatal intensive care unit (NICU) patients. A range of 0% to 1.8% was presented. Three papers documented ototoxicity in patients with no observed MT-RNR1 mutations[9,21,24] while two papers had no hearing impairment among patients with the mutation with or without the gentamicin exposure.[26,33] Eight papers[7,17,22,26,29,35,36,37] recorded a wide phenotypic variance of hearing impairment, age-of-onset and audiometric configurations in patients with the mutation and hearing impairment.
Figure 2.
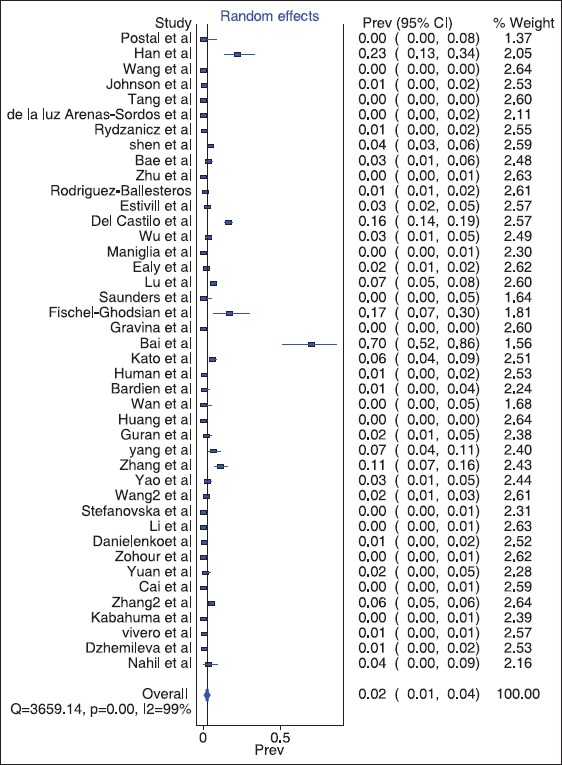
The Forest plot of the meta-analysis of the mitochondrially encoded 12S RNA pooled global prevalence using the MetaXL 2013
Two articles[22,31] described the adverse effects/relative risks on hearing following aminoglycoside exposure in patients carrying the mutation. Both articles focused on Spanish populations and reported a relative increase in development of SNHL following aminoglycoside administration among the genetic mutation carriers, as high as 2.3 times greater in one of the studies[22] compared to those without the mutation.
A noteworthy article[16] focused on the accuracy and rapidity of a screening test for common MT-RNR1 gene mutations. The screening test was found to be highly accurate (up to 99.8%), and results were rapidly available (within 60 min). Other innovative methods for screening MT-RNR1 include MassArray (Sequenom Inc, USA)(that combines a highly specific polymerase chain reaction [PCR] and the accuracy of mass spectrometry)[38] and the “on/off switch” that is combined with PCR.[39] Base quenched probe technique in a PCR assay[40] and the multiplex primer extension methods[41] were described as the most appropriate for mass screening of MT-RNR1. The denaturing high-performance liquid chromatography[42] method was found to be simple, accurate and cost effective whereas, Li et al.[43] also described the Real-time quantitative PCR as suitable for a rapid screening for MT-RNR1 in the deaf population.
The meta-analysis result gave a global pooled prevalence 0.02 (confidence interval 0.01; 0.04) at 99% of the global population. This represents approximately 2% (1-4%) of the general global population. The forest plot is as below in Figure 2.
Mutations in the MT-RNR1 gene result in increased susceptibility to ototoxic hearing loss following treatment with aminoglycoside antibiotics. Mutations are globally prevalent across all races[9,10,16,18,19,20,21,24,33,44] with variable susceptibility and penetrance,[42,45] affecting both sexes and all age groups.[46] Before now, there is no acceptable global prevalence because of the skewed distribution in the inheritance pattern of this gene. This necessitated the systematic review and metanlysis on all documented prevalences within the general population to give insight into this lacuna. A pooled global prevalence of 0.02 recorded represents 2% of the world's population, and this suggests that up to 0.14 billion people are at risk of MT-RNR1 mutation.
DISCUSSION
The prevalence estimates of MT-RNR1 mutations vary widely across different populations, perhaps artificially influenced by the screening method used and the accuracy of available epidemiologic data. In African countries,[6,9,16] estimated prevalence ranged from 0.9% to 2%, in Asia[7,16,21,25,28,34,35,36] 0.09-17%, in the Polish[28] population 0.4% and in America[10,12] 1.85%. In a recent report[13] no patients with the mutation were found among Mexicans whereas prevalence estimates ranged as high as 17% in Chinese and Spanish populations.[24,29,32,35] A limited comparative study conducted in Iowa (USA) suggested no significant difference in the prevalence of MT-RNR1 mutations between NICU babies and the general population (1.85% vs. 1.83%).[9] Exposure to aminoglycosides could increase the relative risk of developing SNHL among the mutant gene carriers by 2.3 times.[31,36] However, this could be confounded by low birth weight[33] prematurity, variations in duration of drug exposure[33] and race[21,34] which were not controlled for in the available limited studies.
The penetrance of the effects of the MT-RNR1 gene A1555G variant mutation (hearing loss following aminoglycoside exposure) among Koreans[26] was estimated between 28.6% and 75%, with an average of 60.8%. This is similar to that recorded for a large Arab-Israeli population (65.4%)[47] and compares with the Spanish population (54.1%).[25] It differs significantly in the Chinese population with a relatively low penetrance ranging from 4% to 18%,[23,37,48,49] a factor to consider when deciding on the merits of population screening. Other factors worthy of consideration that may explain variations in clinical expression include the total dose of the aminoglycoside administered, other potentiating medications given simultaneously as well as the age of the patient. Extremes of ages appear to be more vulnerable.[26]
Mitochondrially encoded 12S RNA gene mutation on its own is insufficient to produce significant hearing loss and, therefore, aminoglycoside exposure is necessary for full expression of the mutation and for hearing loss to occur.[7,49,50,51,52]
Globally, it is estimated that 10-20% of patients with aminoglycoside–induced ototoxicity carry the MT-RNR1 mutation.[35,53] This suggests that a significant percentage of non-MT-RNR1 gene carriers may also develop SNHL resulting from aminoglycoside ototoxicity. Some studies showed no correlation between aminoglycoside blood levels and development of ototoxicity[54,55,56,57,58] in contrast to an earlier finding that high serum levels of amikacin were significantly associated with the development of cochlear toxicity.[59] In addition, some idiosyncratic reactions (resulting in profound hearing losses) following exposure to an otherwise “low-dose” of an aminoglycoside in some individuals have been reported.[56] Under-reporting of MT-RNR1 genes due to non-screening or incomplete assessment of the patients for all variations of mutations of the MT-RNR1 genes may be responsible. However, 38.5% of the documentations on prevalence of MT-RNR1 in our study recorded 0 (0%) prevalence [Figure 2]. which hypothetically support the notion that the occurrence, spectrum and distribution of the gene are narrow. Hence, there is a need for the identification of susceptible populations for the genetic screening rather than the general population.
Mechanism of action
The exact biochemical mechanism of ototoxicity related to the various MT-RNR1 mutations is unclear. There is evidence to suggest the most common mutations (A1555G and C1494T) tend to reduce the accuracy of the translation in the mitochondria to render the ribosome decoding site hyper-susceptible to aminoglycoside.[60,61] The mutations locate to the penultimate helix of the 12S rRNA, which is a component of the aminoacyl-tRNA necessary for the decoding of the mRNA acceptor site (the A-site). The A-site is the target site of the aminoglycoside in bacteria.[15] Therefore, a defect in this mitochondrial site in humans is thought to result in defective metabolism and elimination of the aminoglycosides and/or their by-products resulting in the exaggerated concentration of metabolites within the system and hence toxicity to susceptible organs.
Screening methods: Feasibility and cost-effectiveness
The universal application of aminoglycosides in the management of neonatal sepsis/childhood infections emphasises the clinical relevance of knowledge of the prevalence of these mutations. In the USA alone, over 5 million patients are treated with aminoglycoside antibiotics annually.[51,53] The worldwide re-emergence of tuberculosis (TB) and the growing incidence of the multi- and extreme-drug resistant (MDR & XDR) TB that are only amenable to aminoglycosides (kanamycin and amikacin) are further cause for concern.
At least five different methods are employed in the (PCR) screening for the MT-RNR1 mutation. These include: Allele specific PCR testing, DNA sequencing, PCR-restriction fragment chain length polymorphism (PCR-RFLP) analysis and allele specific oligonucleotide hybridisation and SNaPshot analysis technique. The SNaPshot analysis technique developed at the Stellenbosch University of Cape Town (2009) possesses the advantage of multiplexing and, therefore, is capable of screening for all five common genetic variants. The quoted costs per screening an individual sample including the cost of DNA extraction is USD $16 for SNaPshot compared to USD $30 for PCR methods.[16]
With over 5 million patients in the USA treated with aminoglycoside antibiotics annually and with the cost of screening using the SNaPshot technique at $16 each, an estimated $80 million would be incurred in screening all patients receiving aminoglycoside therapy in the USA annually. Assuming the prevalence rate of 1.85% in the USA for the MT-RNR1 mutant gene, an estimated 92,500 patients are at risk of developing ototoxic hearing loss each year. If the cost of habilitation of a severe-to-profound prelingually deaf child is truly one million dollars screening for MT-RNR1 mutations warrants further consideration, including prospective consecutive cohorts screened for the prevalence of a relevant mutation and followed prospectively for development of aminoglycoside-induced hearing loss. This would allow better estimates of cost-effectiveness of screening programs.
Hypothetically, the cost of screening the entire world population will be about $22.4 Billion USD against the cost of full habilitation of the potential 0.14 billion at risk that is estimated at about $140 trillion USD. The above are quantifiable costs and exclude the non-quantifiable costs. It is true that the above figures for just MT-RNR1 may be unrealistic considering the world's limited resources chasing several global health concerns and disasters; however, it makes economic sense to prevent possible complications that might emanate from this genetic anomaly. On the other hand, effort toward reducing this occurrence appear achievable through the screening of all patients before receiving the first dose of aminoglycosides and/or the screening of every newborn within the at risk population group.
LIMITATIONS
At the onset of the study, the quality and number of the available studies for review evidence were suboptimal and hence the delay in the reporting 2 years after first presentation at the American Academy of Otorhinolaryngology Head and Neck Surgery Conference in Boston USA. This delay was necessary to enable us recruit more papers. No studies identified were felt to offer level 1 evidence (local and current random sample surveys [or censuses]). Our systematic review represents an attempt at obtaining level 2 evidence (systemic reviews and meta-analysis of surveys that allow matching to local circumstances). Many identified studies suffered poor design/sample selection and or ambiguous eligibility criteria for inclusion/exclusion of patients.
In conclusion, the global pooled prevalence of MT-RNR1 mutations in the general population appears significant with a racial bias in Chinese and Spanish populations. Patients with MT-RNR1 mutations are susceptible to aminoglycoside-induced ototoxicity, but with variable penetrance. Routine screening for MT-RNR1 mutations in the general population prior to treatment with aminoglycosides appear desirable, but currently poorly supported by the weak level of evidence available in the literature, but warrants further consideration as more data become available. Routine screening in high-risk (Chinese and Spanish populations) appear justified.
ACKNOWLEDGMENTS
The Librarian of the Children's Hospital Library of the University of British Columbia Vancouver for assisting with paper searches and retrievals.
Footnotes
Source of Support: The Global Surgery Foundation of the University of British Columbia supported with training grant during this study and a travel grant to the AAO-HNSF annual meeting where the paper was first presented in 2012.
Conflict of Interest: None declared.
REFERENCES
- 1.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames. org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–9. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–94. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 3.Li R, Greinwald JH, Jr, Yang L, Choo DI, Wenstrup RJ, Guan MX. Molecular analysis of the mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with non-syndromic hearing loss. J Med Genet. 2004;41:615–20. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lévêque M, Marlin S, Jonard L, Procaccio V, Reynier P, Amati-Bonneau P, et al. Whole mitochondrial genome screening in maternally inherited non-syndromic hearing impairment using a microarray resequencing mitochondrial DNA chip. Eur J Hum Genet. 2007;15:1145–55. doi: 10.1038/sj.ejhg.5201891. [DOI] [PubMed] [Google Scholar]
- 5.Konings A, Van Camp G, Goethals A, Van Eyken E, Vandevelde A, Ben Azza J, et al. Mutation analysis of mitochondrial DNA 12SrRNA and tRNASer(UCN) genes in non-syndromic hearing loss patients. Mitochondrion. 2008;8:377–82. doi: 10.1016/j.mito.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Nahili H, Charif M, Boulouiz R, Bounaceur S, Benrahma H, Abidi O, et al. Prevalence of the mitochondrial A 1555G mutation in Moroccan patients with non-syndromic hearing loss. Int J Pediatr Otorhinolaryngol. 2010;74:1071–4. doi: 10.1016/j.ijporl.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Li Z, Zhu Y, Yang A, Li R, Zheng J, et al. Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion. 2010;10:380–90. doi: 10.1016/j.mito.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs HT, Hutchin TP, Käppi T, Gillies G, Minkkinen K, Walker J, et al. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur J Hum Genet. 2005;13:26–33. doi: 10.1038/sj.ejhg.5201250. [DOI] [PubMed] [Google Scholar]
- 9.Human H, Hagen CM, de Jong G, Harris T, Lombard D, Christiansen M, et al. Investigation of mitochondrial sequence variants associated with aminoglycoside-induced ototoxicity in South African TB patients on aminoglycosides. Biochem Biophys Res Commun. 2010;393:751–6. doi: 10.1016/j.bbrc.2010.02.075. [DOI] [PubMed] [Google Scholar]
- 10.Ealy M, Lynch KA, Meyer NC, Smith RJ. The prevalence of mitochondrial mutations associated with aminoglycoside-induced sensorineural hearing loss in an NICU population. Laryngoscope. 2011;121:1184–6. doi: 10.1002/lary.21778. [DOI] [PubMed] [Google Scholar]
- 11.Dzhemileva LU, Posukh OL, Tazetdinov AM, Barashkov NA, Zhuravskii SG, Ponidelko SN, et al. Analysis of mitochondrial 12S rRNA and tRNA(Ser(UCN)) genes in patients with nonsyndromic sensorineural hearing loss from various regions of Russia. Genetika. 2009;45:982–91. [PubMed] [Google Scholar]
- 12.Vivero RJ, Ouyang X, Yan D, Angeli S, Liu XZ. Mitochondrial DNA mutation screening in South Florida. Laryngoscope. 2010;120(Suppl 3):S92. [Google Scholar]
- 13.de la Luz Arenas-Sordo M, Menendez I, Hernández-Zamora E, Sirmaci A, Gutiérrez-Tinajero D, McGetrick M, et al. Unique spectrum of GJB2 mutations in Mexico. Int J Pediatr Otorhinolaryngol. 2012;76:1678–80. doi: 10.1016/j.ijporl.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Chinnery PF, Elliott C, Green GR, Rees A, Coulthard A, Turnbull DM, et al. The spectrum of hearing loss due to mitochondrial DNA defects. Brain. 2000;123(Pt 1):82–92. doi: 10.1093/brain/123.1.82. [DOI] [PubMed] [Google Scholar]
- 15.Hobbie SN, Bruell CM, Akshay S, Kalapala SK, Shcherbakov D, Böttger EC. Mitochondrial deafness alleles confer misreading of the genetic code. Proc Natl Acad Sci U S A. 2008;105:3244–9. doi: 10.1073/pnas.0707265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardien S, Human H, Harris T, Hefke G, Veikondis R, Schaaf HS, et al. A rapid method for detection of five known mutations associated with aminoglycoside-induced deafness. BMC Med Genet. 2009;10:2. doi: 10.1186/1471-2350-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato T, Nishigaki Y, Noguchi Y, Ueno H, Hosoya H, Ito T, et al. Extensive and rapid screening for major mitochondrial DNA point mutations in patients with hereditary hearing loss. J Hum Genet. 2010;55:147–54. doi: 10.1038/jhg.2009.143. [DOI] [PubMed] [Google Scholar]
- 18.Mohr PE, Feldman JJ, Dunbar JL, McConkey-Robbins A, Niparko JK, Rittenhouse RK, et al. The societal costs of severe to profound hearing loss in the United States. Int J Technol Assess Health Care. 2000;16:1120–35. doi: 10.1017/s0266462300103162. [DOI] [PubMed] [Google Scholar]
- 19.Group OLoEW. The Oxford 2011 Levels of Evidence. UK Oxford Centre for Evidence-Based Medicine; 2011. [Google Scholar]
- 20.Barendregt JJ, Doi SA, Lee YY, Norman RE, Vos T. Meta-analysis of prevalence. J Epidemiol Community Health. 2013;67:974–8. doi: 10.1136/jech-2013-203104. [DOI] [PubMed] [Google Scholar]
- 21.Postal M, Palodeto B, Sartorato EL, Oliveira CA. C1494T mitochondrial DNA mutation, hearing loss, and aminoglycosides antibiotics. Braz J Otorhinolaryngol. 2009;75:884–7. doi: 10.1016/S1808-8694(15)30554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estivill X, Govea N, Barceló E, Badenas C, Romero E, Moral L, et al. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X, Yang L, Zhu Y, Liao Z, Wang J, Qian Y, et al. Very low penetrance of hearing loss in seven Han Chinese pedigrees carrying the deafness-associated 12S rRNA A1555G mutation. Gene. 2007;393:11–9. doi: 10.1016/j.gene.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Saunders JE, Greinwald JH, Vaz S, Guo Y. Aminoglycoside ototoxicity in Nicaraguan children: Patient risk factors and mitochondrial DNA results. Otolaryngol Head Neck Surg. 2009;140:103–7. doi: 10.1016/j.otohns.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y, Li Q, Chen Z, Kun Y, Liu L, Liu X, et al. Mitochondrial haplotype and phenotype of 13 Chinese families may suggest multi-original evolution of mitochondrial C1494T mutation. Mitochondrion. 2009;9:418–28. doi: 10.1016/j.mito.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Bae JW, Kim DB, Choi JY, Park HJ, Lee JD, Hur DG, et al. Molecular and clinical characterization of the variable phenotype in Korean families with hearing loss associated with the mitochondrial A1555G mutation. PLoS One. 2012;7:e42463. doi: 10.1371/journal.pone.0042463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu CC, Chiu YH, Chen PJ, Hsu CJ. Prevalence and clinical features of the mitochondrial m.1555A>G mutation in Taiwanese patients with idiopathic sensorineural hearing loss and association of haplogroup F with low penetrance in three families. Ear Hear. 2007;28:332–42. doi: 10.1097/AUD.0b013e318047941e. [DOI] [PubMed] [Google Scholar]
- 28.Rydzanicz M, Wróbel M, Cywinska K, Froehlich D, Gawecki W, Szyfter W, et al. Screening of the general Polish population for deafness-associated mutations in mitochondrial 12S rRNA and tRNA Ser(UCN) genes. Genet Test Mol Biomarkers. 2009;13:167–72. doi: 10.1089/gtmb.2008.0098. [DOI] [PubMed] [Google Scholar]
- 29.del Castillo FJ, Rodríguez-Ballesteros M, Martín Y, Arellano B, Gallo-Terán J, Morales-Angulo C, et al. Heteroplasmy for the 1555A>G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet. 2003;40:632–6. doi: 10.1136/jmg.40.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniglia LP, Moreira BC, Silva MA, Piatto VB, Maniglia JV. Screening of the mitochondrial A1555G mutation in patients with sensorineural hearing loss. Braz J Otorhinolaryngol. 2008;74:731–6. doi: 10.1016/S1808-8694(15)31384-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Ballesteros M, Olarte M, Aguirre LA, Galán F, Galán R, Vallejo LA, et al. Molecular and clinical characterisation of three Spanish families with maternally inherited non-syndromic hearing loss caused by the 1494C->T mutation in the mitochondrial 12S rRNA gene. J Med Genet. 2006;43:e54. doi: 10.1136/jmg.2006.042440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gravina LP, Foncuberta ME, Estrada RC, Barreiro C, Chertkoff L. Carrier frequency of the 35delG and A1555G deafness mutations in the Argentinean population. Impact on the newborn hearing screening. Int J Pediatr Otorhinolaryngol. 2007;71:639–43. doi: 10.1016/j.ijporl.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Johnson RF, Cohen AP, Guo Y, Schibler K, Greinwald JH. Genetic mutations and aminoglycoside-induced ototoxicity in neonates. Otolaryngol Head Neck Surg. 2010;142:704–7. doi: 10.1016/j.otohns.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 34.Chen G, Wang X, Fu S. Prevalence of A1555G mitochondrial mutation in Chinese newborns and the correlation with neonatal hearing screening. Int J Pediatr Otorhinolaryngol. 2011;75:532–4. doi: 10.1016/j.ijporl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Bai YH, Ren CC, Gong XR, Meng LP. A maternal hereditary deafness pedigree of the A1555G mitochondrial mutation, causing aminoglycoside ototoxicity predisposition. J Laryngol Otol. 2008;122:1037–41. doi: 10.1017/S0022215107001648. [DOI] [PubMed] [Google Scholar]
- 36.Wang QJ, Zhao YL, Rao SQ, Guo YF, He Y, Lan L, et al. Newborn hearing concurrent gene screening can improve care for hearing loss: A study on 14,913 Chinese newborns. Int J Pediatr Otorhinolaryngol. 2011;75:535–42. doi: 10.1016/j.ijporl.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Dai P, Liu X, Han D, Qian Y, Huang D, Yuan H, et al. Extremely low penetrance of deafness associated with the mitochondrial 12S rRNA mutation in 16 Chinese families: Implication for early detection and prevention of deafness. Biochem Biophys Res Commun. 2006;340:194–9. doi: 10.1016/j.bbrc.2005.11.156. [DOI] [PubMed] [Google Scholar]
- 38.Yao GD, Li SX, Chen DL, Feng HQ, Zhao SB, Liu YJ, et al. Combination of hearing screening and genetic screening for deafness-susceptibility genes in newborns. Exp Ther Med. 2014;7:218–22. doi: 10.3892/etm.2013.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xuan GP, Guo ZF, Chen FF, Dong WL. Efficient discrimination of three hotspot mutations associated with aminoglycoside antibiotics induced deafness in the mitochondrial 12S rRNA by on/off switch. Acta Pharmacol Sin. 2013;34:157. [Google Scholar]
- 40.Zheng L, Luo GH, Zhang J, Zhang XY, Xu N. A novel method of detecting mitochondrial C1494T and A1555G mutations by using the base-quenched probe technique in a single PCR assay. Chin J Otorhinolaryngol Head Neck Surg. 2012;47:326–9. [PubMed] [Google Scholar]
- 41.Men M, Xue J, Jiang L, Wang H, Pan Q, Feng Y. Novel multiplex primer extension and denaturing high-performance liquid chromatography for genotyping of the deafness gene mutations. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011;36:1079–84. doi: 10.3969/j.issn.1672-7347.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 42.van Den Bosch BJ, de Coo RF, Scholte HR, Nijland JG, van Den Bogaard R, de Visser M, et al. Mutation analysis of the entire mitochondrial genome using denaturing high performance liquid chromatography. Nucleic Acids Res. 2000;28:E89. doi: 10.1093/nar/28.20.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Yuan YY, Huang DL, Han DY, Dai P. Rapid screening for the mitochondrial DNA C1494T mutation in a deaf population in China using real-time quantitative PCR. Acta Otolaryngol. 2012;132:814–8. doi: 10.3109/00016489.2012.664781. [DOI] [PubMed] [Google Scholar]
- 44.Han B, Dai P, Zhu Q, Liu X, Huang D, Yuan Y, et al. The mitochondrial tRNA (Ala) T5628C variant may have a modifying role in the phenotypic manifestation of the 12S rRNA C1494T mutation in a large Chinese family with hearing loss. Biochem Biophys Res Commun. 2007;357:554–60. doi: 10.1016/j.bbrc.2007.03.199. [DOI] [PubMed] [Google Scholar]
- 45.Guan MX, Fischel-Ghodsian N, Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet. 2001;10:573–80. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- 46.Zhao H, Young WY, Yan Q, Li R, Cao J, Wang Q, et al. Functional characterization of the mitochondrial 12S rRNA C1494T mutation associated with aminoglycoside-induced and non-syndromic hearing loss. Nucleic Acids Res. 2005;33:1132–9. doi: 10.1093/nar/gki262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bykhovskaya Y, Shohat M, Ehrenman K, Johnson D, Hamon M, Cantor RM, et al. Evidence for complex nuclear inheritance in a pedigree with nonsyndromic deafness due to a homoplasmic mitochondrial mutation. Am J Med Genet. 1998;77:421–6. doi: 10.1002/(sici)1096-8628(19980605)77:5<421::aid-ajmg13>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 48.Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum Mutat. 1999;13:261–70. doi: 10.1002/(SICI)1098-1004(1999)13:4<261::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 49.Young WY, Zhao L, Qian Y, Li R, Chen J, Yuan H, et al. Variants in mitochondrial tRNAGlu, tRNAArg, and tRNAThr may influence the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Han Chinese families with hearing loss. Am J Med Genet A. 2006;140:2188–97. doi: 10.1002/ajmg.a.31434. [DOI] [PubMed] [Google Scholar]
- 50.Peng GH, Zheng BJ, Fang F, Wu Y, Liang LZ, Zheng J, et al. Mitochondrial 12S rRNA A1555G mutation associated with nonsyndromic hearing loss in twenty-five Han Chinese pedigrees. Yi Chuan. 2013;35:62–72. doi: 10.3724/sp.j.1005.2013.00062. [DOI] [PubMed] [Google Scholar]
- 51.Qian Y, Guan MX. Interaction of aminoglycosides with human mitochondrial 12S rRNA carrying the deafness-associated mutation. Antimicrob Agents Chemother. 2009;53:4612–8. doi: 10.1128/AAC.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Lu J, Zhu Y, Yang A, Yang L, Li R, et al. Mitochondrial tRNAThr G15927A mutation may modulate the phenotypic manifestation of ototoxic 12S rRNA A1555G mutation in four Chinese families. Pharmacogenet Genomics. 2008;18:1059–70. doi: 10.1097/FPC.0b013e3283131661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forge A, Schacht J. Aminoglycoside antibiotics. Audiol Neurootol. 2000;5:3–22. doi: 10.1159/000013861. [DOI] [PubMed] [Google Scholar]
- 54.Halmagyi GM, Fattore CM, Curthoys IS, Wade S. Gentamicin vestibulotoxicity. Otolaryngol Head Neck Surg. 1994;111:571–4. doi: 10.1177/019459989411100506. [DOI] [PubMed] [Google Scholar]
- 55.Black FO, Gianna-Poulin C, Pesznecker SC. Recovery from vestibular ototoxicity. Otol Neurotol. 2001;22:662–71. doi: 10.1097/00129492-200109000-00018. [DOI] [PubMed] [Google Scholar]
- 56.Nicolau DP, Freeman CD, Belliveau PP, Nightingale CH, Ross JW, Quintiliani R. Experience with a once-daily aminoglycoside program administered to 2,184 adult patients. Antimicrob Agents Chemother. 1995;39:650–5. doi: 10.1128/AAC.39.3.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishiyama G, Ishiyama A, Kerber K, Baloh RW. Gentamicin ototoxicity: Clinical features and the effect on the human vestibulo-ocular reflex. Acta Otolaryngol. 2006;126:1057–61. doi: 10.1080/00016480600606673. [DOI] [PubMed] [Google Scholar]
- 58.Begg EJ, Barclay ML. Aminoglycosides-50 years on. Br J Clin Pharmacol. 1995;39:597–603. [PMC free article] [PubMed] [Google Scholar]
- 59.Black RE, Lau WK, Weinstein RJ, Young LS, Hewitt WL. Ototoxicity of amikacin. Antimicrob Agents Chemother. 1976;9:956–61. doi: 10.1128/aac.9.6.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bravo O, Ballana E, Estivill X. Cochlear alterations in deaf and unaffected subjects carrying the deafness-associated A1555G mutation in the mitochondrial 12S rRNA gene. Biochem Biophys Res Commun. 2006;344:511–6. doi: 10.1016/j.bbrc.2006.03.143. [DOI] [PubMed] [Google Scholar]
- 61.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–77. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]


