Abstract
The half-maximal inhibitory concentration (IC50) is the most widely used and informative measure of a drug’s efficacy. It indicates how much drug is needed to inhibit a biological process by half, thus providing a measure of potency of an antagonist drug in pharmacological research. Most approaches to determine IC50 of a pharmacological compound are based on assays that utilize whole cell systems. While they generally provide outstanding potency information, results can depend on the experimental cell line used and may not differentiate a compound’s ability to inhibit specific interactions. Here we show using the secreted Transforming Growth Factor-β (TGF-β) family ligand BMP-4 and its receptors as example that surface plasmon resonance can be used to accurately determine IC50 values of individual ligand-receptor pairings. The molecular resolution achievable with this approach can help distinguish inhibitors that specifically target individual complexes, or that can inhibit multiple functional interactions at the same time.
Keywords: Cerberus, BMP-4, surface plasmon resonance, SPR, TGF-β, Bone morphogenetic protein, IC50, inhibitor
INTRODUCTION
Cytokines and growth factors play fundamental roles in animal cell physiology and human disease [1, 2]. They help regulate cell differentiation, proliferation, migration, plasticity, and survival, but their dysregulation can lead to inflammation, fibrosis, cancers, and other devastating diseases [3, 4]. Thus, they have elicited much medical interest and a number of therapeutics that target these secreted proteins have been approved for clinical use [5, 6]. The two most common classes of therapeutics used to target cytokines and growth factors are monoclonal antibodies [7] and chimeric receptor-Fc fusion proteins, a.k.a. ligand traps [8]. Both aim to inhibit biological activity by blocking functional interactions with receptors and other binding partners [9].
Possibly the most relevant in vitro measure used to evaluate the therapeutic potential of a monoclonal antibody, ligand trap or other pharmacologic agents is half-maximal inhibitory concentration, a.k.a. IC50 [10–12]. It indicates how much of a specific pharmacologic agent is required to inhibit a given biological activity by half. Methods used to obtain IC50 values generally rely on whole cell systems. They provide essential potency information and are especially useful when evaluating therapeutics that target pathways with few components. However, they can be deficient when evaluating inhibitors of cytokines or growth factors, such as members of the TGF-β, Wnt, FGF, EGF, and interleukin families that have pleiotropic activities and that can interact with many different components. Members of these families can engage multiple receptors for signaling, are also regulated by ‘co-receptors’, which potentiate or suppress biological activity, and even act as regulators of other family members by competing for receptor binding [13–16]. Thus, it is suggested their function is ‘context dependent’ [17–20]. In these cases, IC50 values can vary with experimental cell line used and may not inform well on the efficacy of blocking a specific interaction.
To obtain IC50 values of individual cytokine/growth factor – inhibitor interactions, we implemented an interaction specific surface plasmon resonance (SPR) approach, and turned to the TGF-β family ligand BMP-4 as model cytokine/growth factor and the embryonic Nodal/BMP antagonist Cerberus as model inhibitor. Like all members of the TGF-β family, BMP-4 signals by forming a complex with two ‘type I’ and two ‘type II’ TGF-β receptors, namely the type I receptor ALK3 and the type II receptors ActRIIA or BMPRII [21]. Cerberus is a secreted protein that binds and inhibits several TGF-β family ligands with mid picomolar to low nanomolar affinities, including Activin B, Nodal, BMP-6 and BMP-7 [22, 23].
Here, we show BMP-4 is a new high affinity Cerberus ligand. Using SPR, we demonstrate Cerberus inhibits BMP-4 binding to its type I and type II receptors in a concentration dependent manner, as increasing Cerberus concentrations reduce the BMP-4 dependent SPR signal. Importantly, the Cerberus dependent reduction in the SPR response can be used at any point of the association and dissociation phase to calculate IC50 using standard software such as GraphPad Prism. Similarly, IC50 can be calculated from kinetic parameters obtained by fitting individual concentration curves. We discuss merits of the two different approaches for IC50 determination. In addition, we obtained IC50 using a whole cell system. We discuss differences between the SPR and cell-based approaches in light of the complex composition of the cell surface and the various interactions of BMP-4. Notably, the molecular resolution achievable with the SPR format can help map inhibitor binding sites and thus help guide design of novel agents that selectively block formation of specific biological complexes.
MATERIALS AND METHODS
Proteins expression and purification
Recombinant human BMP-4 (Q53XC5) was obtained from R&D Biosystems (RnD). Synthetic genes consisting of human Cerberus (O95813), ActRIIA (P27037), and ALK3 (P36894) fused to human IgG1-Fc via a linker containing a Tobacco Etch Virus (TEV) protease site were obtained from GeneArt. Two point mutations (R82G and C206A) were introduced in Cerberus by PCR (Cer-Fc) [22]. Human BMPRII (Q13873) was cloned from cDNA and fused to human IgG1-Fc by PCR. NCBI-protein accession numbers are shown in parenthesis. Fc-fusion proteins were expressed using suspension adapted Chinese Hamster Ovary (CHO) cells and purified from condition medium using Protein A capture followed by size exclusion chromatography (SEC). The Fc portion of Cerberus (C206A) was removed with TEV protease, followed by two steps of protein A capture, and SEC. All purified proteins were dialyzed into phosphate-buffered saline, pH 7.5 and stored at −20 °C or −80 °C. The purity of the proteins was checked with SDS-PAGE under reducing and non-reducing conditions.
Surface plasmon resonance
Experiments were performed on a Biacore 2000 at 25 °C using HBS-EPS/BSA as running buffer (0.01 M HEPES, 0.5 M NaCl, 3 mM EDTA, 0.005% (v/v) Tween 20, 0.1 % BSA, pH 7.4). A high flow rate (50 μl/min) and low surface loading were used to minimize mass transport artifacts. Anti-human IgG (Fc) was immobilized onto four channels of a CM5 chip using amine-coupling chemistry. The cross-linked antibody on the experimental flow channels was used to capture receptor-Fc-fusion proteins. Approximately 200–300 RU were loaded on each channel. For binding assays, different concentrations of BMP-4 were injected over experimental and control flow channels loaded with Cer-Fc, ActRIIA-Fc, BMPRII-Fc or ALK3-Fc. As binding curves appear to be bi-phasic [24], data were fitted to a 1:1 Langmuir interaction model with mass transport limitation and to a heterogeneous ligand-binding model using BiaEvaluation software. Both models gave similar kinetic rates for ActRIIA-Fc, BMPRII-Fc and Cerberus-Fc, but not for ALK3 (Table 1). Notably, bi-phasic behavior can be fitted equally well by different models, indicating it is nearly impossible to identify the source of the complex behavior using kinetic data [24]. But the 1:1 Langmuir interaction model with mass transport limitation was selected because it gave the best fit as determined by Chi2 (< 1). Equilibrium dissociation constants (Kd) were determined by calculating the ratio of binding and dissociation rate constants. Results are summarized in Table 1. For inhibition analysis, ActRIIA-Fc, BMPRII-Fc, ALK3-Fc, or Cerberus-Fc were captured on the sensor chip. Between 150 and 200 RUs ActRIIA-Fc, BMPRII-Fc, ALK3-Fc, or Cerberus-Fc were captured to minimize steric hindrance and mass transport artifacts. 60 nM BMP-4 preincubated with different concentrations of Fc-free Cerberus was injected over experimental and control flow channels. IC50 values for SPR inhibition data were determined using GraphPad. Results are summarized in Table 2. After each binding cycle, the antibody surface was regenerated to base line with MgCl2. The reference channel was monitored to account for nonspecific binding, drift, and bulk shifts. Sensograms were analyzed by double referencing. Association rate constants (ka, M−1s−1) of individual curves were obtained by global fitting using BiaEvaluation software. After individual fitting of the curves, local observed rate constants (kobs s−1) values were calculated as ka*Conc+kd using BiaEvaluation software.
Table 1.
BMP-4* binding constants
| Chip: | Model | ka | kd | Kd | Chi2 |
|---|---|---|---|---|---|
| Cerberus | 1:1ms | (1.10 ± 0.12) × 105 | (3.01 ± 0.04) × 10−4 | 2.73 | 0.292 |
| 1:1hl | (8.25 ± 0.92) × 104 | (1.85 ± 0.06) × 10−4 | 2.24 | 0.276 | |
| ActRIIA | 1:1ms | (1.18 ± 0.11) × 105 | (3.37 ± 0.41) × 10−4 | 2.85 | 0.398 |
| 1:1hl | (1.04 ± 0.28) × 105 | (3.39 ± 0.38) × 10−4 | 3.26 | 0.862 | |
| BMPRII | 1:1ms | (7.63 ± 1.4) × 104 | (5.62 ± 0.24) × 10−4 | 7.36 | 0.164 |
| 1:1hl | (3.79 ± 0.69) × 104 | (3.08 ± 0.56) × 10−4 | 8.13 | 0.113 | |
| ALK3 | 1:1ms | (7.78 ± 1.8) × 104 | (1.50 ± 0.74) × 10−5 | 0.193 | 0.177 |
Units are: ka (M−1s−1), kd (s−1), Kd (nM), Chi2 (RU2)
ms: mass transport limited model.
hl: heterogeneous ligand binding model.
Carrier free BMP-4 from RnD Systems
Table 2.
IC50 values
| Interaction: | IC50 (150 s) | IC50 (300 s) | IC50 (750 s) | IC50 (ka) | IC50 (kobs) |
|---|---|---|---|---|---|
| Cerberus | 56.7 ± 3.3 | 57.6 ± 3.3 | 46.6 ± 3.0 | 99.7 ± 21 | 99.7 ± 21 |
| ActRIIA | 266 ± 44 | 305 ± 61 | 239 ± 71 | 56.0 ± 24 | 58.0 ± 25 |
| BMPRII | 166 ± 28 | 153 ± 26 | 132 ± 20 | 138 ± 62 | 153 ± 73 |
| ALK3 | 289 ± 81 | 303 ± 83 | 245 ± 71 | 134 ± 54 | 132 ± 53 |
Reporter assays
HepG2 cells (HB-8065) were obtained from ATCC (American Type Culture Collection). Cells were maintained according to ATCC culture conditions. Freshly thawed cells were passaged at least three times before performing assays. ~ 10,000 HepG2 cells/well in complete medium (Eagle's Minimum Essential Medium supplemented with 10% FBS and 1% P/S) were seeded in a 96-well plate and grown overnight. Each well was transfected with 0.25 μl lipofectamine 2000, 200 ng of the SMAD1/5/8 responsive reporter plasmid pGL3 [luc2P/BRE] pGL3.48 and 2 ng of the [Luc2P/hRluc/TK] vector (control luciferase reporter plasmid, Promega, E6921). Transfection medium was removed the following day, and replaced with assay medium containing BMP-4 (1 nM) ± Cer-Fc (0–600 nM). After adding assay medium, cells were incubated for 16 h and luciferase activity was detected with a homemade dual-glow luciferase assay [25] using a FluoStar Omega plate reader. Relative luciferase units (RLU) were calculated by dividing firefly luciferase units (fLU) with renilla luciferase units (rLU). IC50 values were calculated using GraphPad.
Statistical analysis
Reporter gene assays were repeated at least two times and performed in quadruplicates. One-way analysis of variance (ANOVA) and Dunnett’s post hoc test as implemented in GraphPad Prism 6 was used to calculate statistical significance using all experimental values. Data are expressed as mean of four biological replicates ± standard error S/E.
RESULTS AND DISCUSSION
Establishing the BMP-4 – receptor interaction network
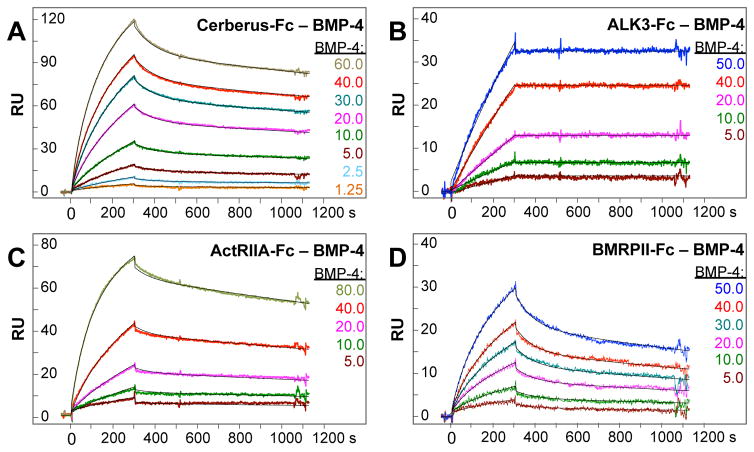
Cerberus is a secreted protein that binds and antagonizes several TGF-β family ligands during embryonic development. To evaluate the inhibitory efficacy of Cerberus, we turned to BMP-4, a newly discovered Cerberus target ligand. We first characterized its interactions with Cerberus and with receptors. Briefly, we captured Cerberus-Fc, or the TGF-β family receptor-Fc fusion proteins ALK3-Fc, ActRIIA-Fc, and BMPRII-Fc on a Biacore sensor chip and injected a concentration series of BMP-4 (Fig. 1, Table 1). BMP-4 bound both Cerberus-Fc and the type I receptor-Fc fusion protein ALK3-Fc with high affinity (Kd = 2.7 nM and 0.2 nM, respectively) (Fig. 1A and B). BMP-4 binding to the type II receptor- Fc fusion protein ActRIIA-Fc was strong, but binding to BMPRII-Fc was slightly weaker (Kd = 2.8 nM, and 7.4 nM, respectively) (Fig. 1C and D). Together, these findings show BMP-4 binds Cerberus, the type I receptor ALK3 and the type II receptors ActRIIA and BMPRII with mid picomolar to low nanomolar affinity.
Figure 1. BMP-4 binding to Cerberus and receptors.
(A) BMP-4 – Cer-Fc interaction. Cer-Fc was captured on the sensor chip and different concentrations of BMP-4 as shown were injected. (B) BMP-4 – ALK3-Fc interaction. ALK3-Fc was captured on the sensor chip and different concentrations of BMP-4 as shown were injected. (C) BMP-4 –ActRIIA-Fc interaction. ActRIIA-Fc was captured on the sensor chip and different concentrations of BMP-4 as shown were injected. (D) BMP-4 –BMPRII-Fc interaction. BMPRII-Fc was captured on the sensor chip and different concentrations of BMP-4 as shown were injected. (A–D) Colors of injection curves are matches with corresponding concentrations. Fitted curves (black lines) are superimposed over all experimental curves. Calculated binding rate constants and equilibrium dissociation rate constants are shown in Table 1.
Obtaining SPR based inhibition data
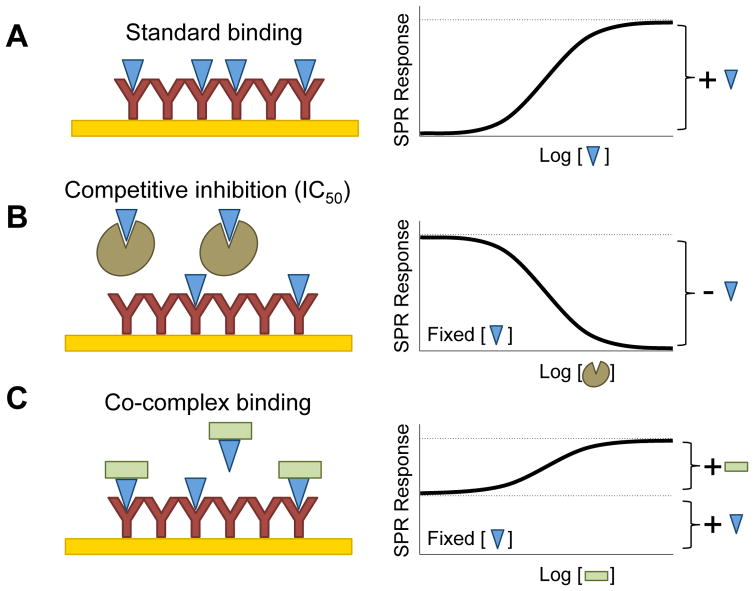
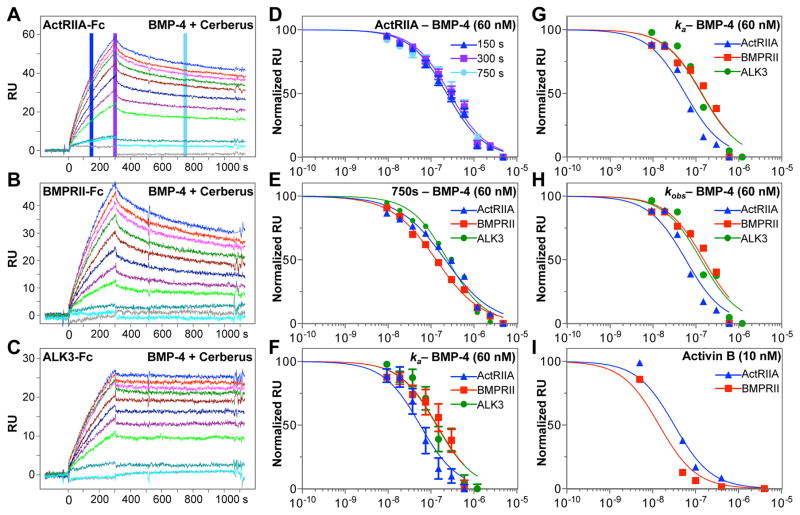
To obtain Cerberus IC50 values for individual BMP-4 – receptor interactions, we established an SPR based approach (see Fig. 2 for experimental design). Briefly, we captured ActRIIA-Fc, BMPRII-Fc or ALK3-Fc on the sensor chip and injected BMP-4 (60 nM) preincubated with different concentrations of Fc free Cerberus (0 nM to 4800 nM) over the captured receptors (Figs. 3 and 4). In this format, Cerberus must be free of Fc to avoid Fc capture by the anti-human Fc antibody that is cross-linked on sensor chip. We hypothesized that the BMP-4 dependent SPR signal will decrease with increasing Cerberus concentrations, if Cerberus inhibits that particular BMP-4 receptor interaction (Figs. 3 and 4). We tested both type I and type II receptors, as each receptor type interacts with a unique BMP-4 epitope [13, 26, 27], offering two independent experimental examples. Cerberus prevented BMP-4 binding to ActRIIA-Fc, BMPRII-Fc, and ALK3-Fc, as seen in the concentration dependent reduction of the SPR signal (Fig. 3A–C). Cerberus alone did not bind receptors or the sensor chip.
Figure 2. Experimental design.
(A) In a typical SPR binding experiment (left panel), the ‘analyte’ (BMP-4, shown as a blue triangle) is injected at increasing concentrations over the captured ‘ligand’ (ActRIIA-Fc, BMPRII-Fc, ALK3-Fc, Cerberus-Fc, etc., shown as a Y shape). When analyte binds to ligand, the SPR response increases with increasing analyte concentration until the surface is saturated by the analyte (see Figure 1). The binding-curve conforms to a Langmuir adsorption isotherm (right panel). (B) In the equilibrium displacement or competitive inhibitor format (left panel), the analyte concentration is fixed at a level that results in ~ 80% saturation. The injected sample is preincubated with increasing concentrations of ‘inhibitor’ (Fc free Cerberus, shown as brown shape). The analyte mediated SPR response decreases with increasing inhibitor concentration. The resulting inhibition curve follows an inversion of the Langmuir adsorption isotherm (right panel). (C) If the putative inhibitor forms a complex with the analyte that can bind the captured ligand (left panel), the starting SPR response corresponds to the free analyte response. The SPR response increases with increasing concentration of the co-binder (shown as green square) until all analyte is captured in a complex. This binding-curve also conforms to a Langmuir adsorption isotherm (right panel).
Figure 3. Inhibition of BMP-4 – receptor binding with Cerberus.
(A) ActRIIA-Fc was captured on the sensor chip. (B) BMPRII-Fc was captured on the sensor chip. (C) ALK3-Fc was captured on the sensor chip. (A–C) In the three experiments, 60 nM BMP-4 was preincubated with 0 nM (blue), 9.375 nM (red), 18.75 nM (magenta), 37.5 nM (green), 75.0 nM (maroon), 150.0 nM (dark blue), 300.0 nM (purple), 600.0 nM (bright green), 1200.0 nM (teal), 2400.0 nM (cyan), and 4800.0 nM (grey) Fc free Cerberus. Cerberus – BMP-4 mixtures were injected over the sensor chip. (D) Analysis of SPR RU ActRIIA-Fc data (2A). Background subtracted raw RU values were taken for each Cerberus concentration at 150 seconds (blue), 300 seconds (purple) and 750 seconds (cyan) following injection of BMP-4 mixtures. RU values for each concentration group were normalized and fitted using the non-linear regression algorithm implemented in GraphPad. Standard errors (SEs) are shown. (E) Analysis of SPR RU data for ActRIIA-Fc (blue, from 2A), BMPRII-Fc (red, from 2B) and ALK3-Fc (green, from 2C). Background subtracted RU values were taken for each Cerberus concentration at 750 seconds of the SPR reaction. RU values for each group were normalized and fitted using non-linear regression in GraphPad. SEs are similar to those observed for ActRIIA-Fc inhibition in D, but are omitted here for clarity. (F) Analysis of ka data for ActRIIA-Fc (blue, from 2A), BMPRII-Fc (red, from 2B) and ALK3-Fc (green, from 2C). ka values were obtained by individually fitting each injection curve. ka values were normalized and fitted using non-linear regression in GraphPad. SEs are shown. (G) Same data as F, but Standard errors are omitted for clarity. (H) Analysis of kobs data for ActRIIA-Fc (blue, from 2A), BMPRII-Fc (red, from 2B) and ALK3-Fc (green, from 2C). kobs values were obtained by individually fitting each injection curve. kobs values were normalized and fitted using non-linear regression in GraphPad. SEs are similar to those observed for ka analysis in F, but are omitted here for clarity. (I) Cerberus prevents Activin B binding to type II receptors. SPR Background subtracted RU values were taken for each Cerberus concentration at 750 seconds of the SPR reaction for ActRIIA-Fc (blue) and BMPRII-Fc (red). RU values for each group were normalized and fitted using non-linear regression in GraphPad. Concentrations in panels D–I are shown in logarithmic scale.
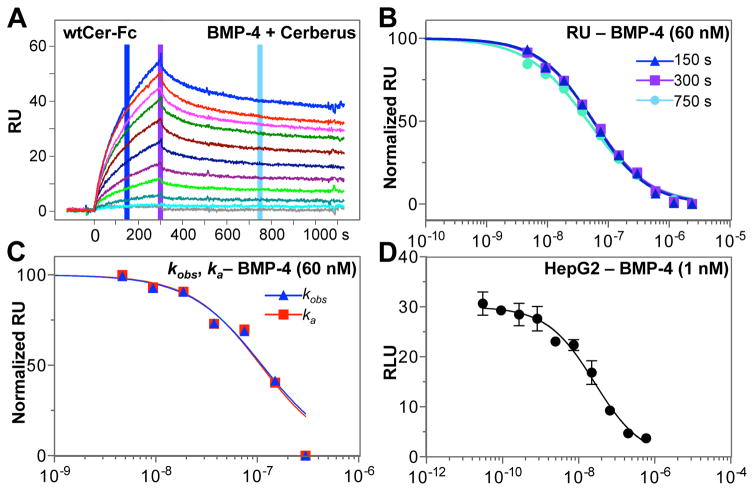
Figure 4. Inhibition of BMP-4 – Cerberus binding with Cerberus.
(A) Cerberus prevents BMP-4 – Cerberus-Fc binding. Cer-Fc was captured on the sensor chip and 60 nM BMP-4 preincubated with 0 nM (blue), 4.6875 nM (red), 9.375 nM (magenta), 18.75 nM (green), 37.5 nM (maroon), 75.0 nM (dark blue), 150.0 nM (purple), 300.0 nM (bright green), 600.0 nM (teal), 1200.0 nM (cyan), and 2400.0 nM (grey), Fc free Cerberus was injected over the sensor chip. (B) Background subtracted raw RU values were taken for each Cerberus concentration at 150 seconds (blue), 300 seconds (purple) and 750 seconds (cyan). RU values for each group were normalized and fitted using non-linear regression in GraphPad. SEs are similar to those observed for ActRIIA-Fc inhibition in 2D, but are omitted here for clarity. (C) Analysis of kobs (blue) and ka (red) data for Cerberus-Fc. kobs and ka values were obtained by individually fitting each injection curve. kobs and ka values were normalized and fitted using non-linear regression in GraphPad. SEs are similar to those observed for ka analysis in 2F, but are omitted here for clarity. (D) BMP-4 mediated Smad1/5/8 signaling and inhibition with Cerberus. HepG2 cells were transfected with SMAD1/5/8 responsive reporter and control plasmids. Cells were treated with 1.0 nM BMP-4, and Cer-Fc at the following concentrations: 0.0 nM, 0.03 nM, 0.09 nM, 0.27 nM, 0.82 nM, 2.47 nM, 7.41 nM, 22.2 nM, 66.6 nM, 200 nM, and 600 nM. Concentrations in panels B–D are shown in logarithmic scale.
Calculating half-maximal inhibitory concentration from SPR data
To quantitate SPR inhibition data and obtain inhibitory potency information, we evaluated different approaches that allowed us to calculate IC50 values. First, we evaluated simple response unit (RU) data taken at different time points of the SPR reaction (Fig. 3D and E). We used the blank-injection adjusted RU values for each Cerberus concentration at a specified time point (Fig. 3A, D and E). To obtain half maximal inhibitory concentration values (IC50), we fitted BMP-4 binding dependent RU values as function of Cerberus concentration using the non-linear regression algorithm for log(concentration) versus normalized response implemented in GraphPad (Table 2). We found IC50 values determined at different time points of the SPR reaction were similar (Fig. 3D, Table 2). With respect to experimental time point selected for the calculations, IC50 values were consistently lower at 750 seconds (middle of the dissociation phase). IC50 values were consistently higher when using RU data at 150 seconds (middle of the association phase) and at 300 seconds (end of the association phase). But at 300 seconds, IC50 values showed the overall greatest trend variability. We speculate data taken in the middle of the association or dissociation phases are more consistent than data taken at the junction between association and dissociation phases (Fig. 3A–C), as signal noise increases during the sample to buffer transition. Regardless, all IC50 values fell within the calculated 95% confidence interval for each of the three time points (Figure 3D, and E), indicating that, in good approximation, SPR RU data can be taken directly at any time point of the SPR reaction to obtain robust information on specific inhibitory potency of a compound. Notably, reproducibility is extremely high once the SPR experiment is properly set up. Errors between measurements are consistently less than 5% (Fig. 3D).
In addition to the RU based approach, we also examined a mathematical approach of IC50 determination. Namely, we obtained IC50 values based on the apparent association rate constant (ka, M−1s−1) (Fig. 3F and G) and the observed rate constant (kobs, s−1) (Fig. 3H) for each Cerberus concentration. Rate constants were calculated individually for each Cerberus concentrations using biaevaluation. Concentration dependent ka and kobs values were fitted using the non-linear regression algorithm implemented in GraphPad. We found both rate constants and fitted curves showed the expected, dose dependent decrease and sigmoidal shape that is characteristic of a competitive inhibitor (Fig. 3G and H). As expected, IC50 values were highly comparable between the ka and kobs determinations given their linear relationship (Table 2). But we found differences when comparing ka and kobs with the simpler RU analysis. IC50 values for ALK3 and ActRIIA were, respectively, 2-fold and 4- to 6-fold lower for IC50 determined using ka or kobs, whereas IC50 values for BMPRII were similar. Fitting errors for IC50 values obtained from kinetic rate constants were significantly larger than fitting errors obtained using RU values (Fig. 3D and F, Table 2), indicating IC50 values obtained from kinetic rate constants are less precise. The decreased precision likely results from estimating ka and kobs values by individual fitting of single concentration curves.
In summary, our data demonstrate Cerberus blocks binding of BMP-4 to the type I receptor ALK3 and the type II receptors ActRIIA and BMPRII with high potency. We describe several approaches for calculating IC50 values for individual BMP-4 receptor interactions. The different approaches are consistent with each other. Importantly, IC50 determination using SPR can be broadly used as demonstrated by analysis of Activin B inhibition with Cerberus (Fig. 3I) [22, 23].
Evaluating RU- and rate constant-based IC50 values
As different SPR based approaches can be used to obtain IC50 values, we asked which approach delivers the most meaningful result. To answer this question we turned to Cerberus inhibition of BMP-4 – Cerberus-Fc binding. Given Cerberus and Cerberus-Fc have nearly identical affinities for BMP-4 (i.e. Ki = Kd), we hypothesized that the most accurate SPR-based approach should produce an IC50 value that is similar to the Kd value from the Cerberus- Fc – BMP-4 binding experiment at the given BMP-4 [S] concentration (i.e., based on the Cheng-Prusoff equation, if IC50 = Ki(1+ [S]/Kd) and Ki = Kd, then IC50 = Kd + [S]) [12]. To test this hypothesis, we obtained equilibrium binding constants for Cerberus-Fc – BMP-4 (Fig. 4A), we generated IC50 values for inhibition of BMP-4 – Cerberus-Fc binding by Cerberus using both RU and rate constant based approaches (Fig. 4B and C), and we compared experimental with calculated IC50 values (Tables 1 and 2). The theoretical IC50 value for the Cerberus – BMP-4 – Cerberus reaction is 62.73 nM ([BMP-4] = 60 nM, Kd for Cerberus – BMP-4 binding = 2.73 nM) [12]. Experimental IC50 values calculated using RU were 56.7 ± 3.3 at 150 seconds, 57.6 ± 3.3 at 300 seconds, and 46.6 ± 3.0 at 750 seconds. Experimental IC50 values calculated using rate constants were 99.7 ± 21 (for ka) and 99.7 ± 21 (for kobs). Comparison of theoretical and experimental IC50 values indicates the most accurate experimental IC50 values are obtained using the simple, RU based approach (Table 2). Moreover, IC50 values obtained using RU data from the association phase match the theoretical value more closely. Together with our findings above (Fig. 3D, and E), these experiments indicate RU values from the middle of the association phase offer the most consistent and accurate results. Nonetheless, all approaches gave IC50 values that were within reasonable range of the theoretical IC50 value (less than 2-fold difference), demonstrating the overall reliability of biosensor- based IC50 determinations.
Comparing SPR based IC50 with whole cell IC50
To evaluate IC50 values obtained with SPR in relation to IC50 values obtained using a whole cell system, we determined the half-maximal inhibitory concentration of Cerberus for BMP-4 using a luciferase reporter gene expression assay (Fig. 4D). Briefly, we transfected HepG2 cells with control plasmid pGL4.74 [hRluc] and the SMAD1/5/8 responsive reporter plasmid pGL3 [luc2P/BRE]. Transfected HepG2 cells were treated with 1 nM BMP-4 and different concentrations of Cerberus-Fc. BMP-4 induced luciferase expression with the SMAD1/5/8 reporter, and Cer-Fc inhibited BMP-4 signaling in a concentration dependent manner (Fig. 4D). Using relative luciferase response unit (RLU) and GraphPad, we obtained an IC50 value of 26 nM for inhibition.
Overall, IC50 values obtained using a whole cell system are in agreement with, but not identical to IC50 values determined by SPR (Figs. 3 and 4, Table 2). There are several reasons for the observed differences, and they relate to the complexity of the BMP-4 signaling pathway. First, the amount of BMP-4 used in the two experimental formats is different, 60 nM for the SPR experiment versus 1 nM for the reporter assay. Thus, in this particular SPR setup a higher concentration of inhibitor is needed. Second, the molar equivalent of captured receptor on the sensor chip is in all likelihood different from the molar equivalent of receptor found on the cell surface. Lastly, IC50 determined with SPR is specific to individual receptors. By contrast, IC50 determined using a whole cell system combines the contributions of several receptors and other co-receptors expressed on the cell surface. Which receptor or co-receptor is dominant depends on expression levels, and on the exact combination of interacting components expressed by the experimental cell line selected for the assay. However resolving these questions is likely not possible for most experimental cell lines. In spite of these differences, our findings demonstrate 1) IC50 values for inhibition of specific interactions can be obtained using SPR, 2) these IC50 values offer inhibitory information that complements inhibition data obtained using whole cell systems, and 3) inhibitory mechanisms and specific potency of inhibitor variants can be investigated using SPR based inhibition analysis, but not using cell lines.
Adapting SPR based IC50 determination to other experimental systems
Here we have demonstrated using the TGF-β family ligand BMP-4, its receptors and the inhibitor Cerberus how to use SPR to elucidate the mechanism of action of a competitive inhibitor and to obtain inhibitory potency information using simple RU responses. Notably, this approach can be used to analyze any system where the goal is to prevent a particular interaction. The inhibitor can be large, such as an antibody or a natural polypeptide antagonist, or it can also be a small molecule. But several criteria must be met for this system to work. As is typical for SPR, both ‘Ligand’-capture and surface-regeneration must be highly reproducible. This point cannot be emphasized enough. More specifically for IC50 determination, the interaction geometry must be such that the inhibitor prevents ‘analyte’ binding to the captured ‘ligand’, and thus causes a reduction in the SPR signal. Importantly, the ‘inhibitor’ must not interact with the biosensor surface. Any example that meets these criteria can be analyzed using this approach.
Conclusions
Our findings show SPR can be used to obtain robust and quantitative functional data for an inhibitor and potential pharmacologic agent. The SPR based approach is especially useful when examining targets with complex biochemistry, such as cytokines or growth factors that interact with multiple receptors and co-receptors. In a best approximation response unit (RU) values from an inhibition reaction can be taken directly to evaluate inhibitory potency. The association phase likely provides the most accurate IC50 values. When established, SPR based IC50 determination allows for a simple and high-throughput comparison of the molecular properties of variants.
Acknowledgments
We thank Jake Reske for assistance with protein purification. This work was funded in part by Michigan State University, the Clinical and Translational Sciences Institute and NIH grants R41AR068804-01 and R43CA203180-01.
ABBREVIATIONS
- SPR
surface plasmon resonance
- SEC
size exclusion chromatography
- IC50
half maximal inhibitory concentration
- Cer-Fc
Cerberus Fc fusion protein
- TGF-β
transforming growth factor-β
- BMP
bone morphogenetic protein
- ActRIIA
Activin receptor 2A
- ActRIIB
Activin receptor 2B
- BMPRII
bone morphogenetic protein receptor 2
- ALK3
Activin-like kinase 3
Footnotes
CONFLICT OF INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
SA and EMH designed experiments. SA performed experiments. SA and EMH interpreted experiments. EMH wrote manuscript. SA and EMH revised manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.LeRoith D, Bondy C. Growth factors and cytokines in health and disease : a multi-volume treatise. JAI Press; Greenwich, Conn: 1996. [Google Scholar]
- 2.Feldmann M. Many cytokines are very useful therapeutic targets in disease. J Clin Invest. 2008;118:3533–3536. doi: 10.1172/JCI37346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vilcek J, Feldmann M. Historical review: Cytokines as therapeutics and targets of therapeutics. Trends in pharmacological sciences. 2004;25:201–209. doi: 10.1016/j.tips.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Oppenheim JJ. Cytokines: past, present, and future. International journal of hematology. 2001;74:3–8. doi: 10.1007/BF02982543. [DOI] [PubMed] [Google Scholar]
- 5.Elvin JG, Couston RG, van der Walle CF. Therapeutic antibodies: market considerations, disease targets and bioprocessing. International journal of pharmaceutics. 2013;440:83–98. doi: 10.1016/j.ijpharm.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 6.Kinch MS. An overview of FDA-approved biologics medicines. Drug discovery today. 2015;20:393–398. doi: 10.1016/j.drudis.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. British journal of pharmacology. 2009;157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Botran R, Crespo FA, Sun X. Soluble cytokine receptors in biological therapy. Expert opinion on biological therapy. 2002;2:585–605. doi: 10.1517/14712598.2.6.585. [DOI] [PubMed] [Google Scholar]
- 9.Schreiber G, Walter MR. Cytokine-receptor interactions as drug targets. Current opinion in chemical biology. 2010;14:511–519. doi: 10.1016/j.cbpa.2010.06.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck B, Chen YF, Dere W, Devanarayan V, Eastwood BJ, Farmen MW, Iturria SJ, Iversen PW, Kahl SD, Moore RA, Sawyer BD, Weidner J. Assay Operations for SAR Support. In: Sittampalam GS, Coussens NP, Nelson H, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Iversen PW, Li Z, McGee J, McManus O, Minor L, Napper A, Peltier JM, Riss T, Trask OJ Jr, Weidner J, editors. Assay Guidance Manual. Bethesda (MD): 2004. [Google Scholar]
- 11.Cortes A, Cascante M, Cardenas ML, Cornish-Bowden A. Relationships between inhibition constants, inhibitor concentrations for 50% inhibition and types of inhibition: new ways of analysing data. Biochem J. 2001;357:263–268. doi: 10.1042/0264-6021:3570263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical pharmacology. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 13.Hinck AP. Structural studies of the TGF-betas and their receptors - insights into evolution of the TGF-beta superfamily. FEBS Lett. 2012;586:1860–1870. doi: 10.1016/j.febslet.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 14.Ivashkiv LB. Cross-regulation of signaling by ITAM-associated receptors. Nature immunology. 2009;10:340–347. doi: 10.1038/ni.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aykul S, Martinez-Hackert E. Transforming Growth Factor-beta Family Ligands Can Function as Antagonists by Competing for Type II Receptor Binding. J Biol Chem. 2016;291:10792–10804. doi: 10.1074/jbc.M115.713487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oshimori N, Fuchs E. The harmonies played by TGF-beta in stem cell biology. Cell Stem Cell. 2012;11:751–764. doi: 10.1016/j.stem.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massague J. TGFbeta signalling in context. Nature reviews. Molecular cell biology. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwanbeck R, Martini S, Bernoth K, Just U. The Notch signaling pathway: molecular basis of cell context dependency. European journal of cell biology. 2011;90:572–581. doi: 10.1016/j.ejcb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 21.Massague J. TGF-beta signal transduction. Annual review of biochemistry. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 22.Aykul S, Martinez-Hackert E. New Ligand Binding Function of Human Cerberus and Role of Proteolytic Processing in Regulating Ligand-Receptor Interactions and Antagonist Activity. J Mol Biol. 2016;428:590–602. doi: 10.1016/j.jmb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aykul S, Ni W, Mutatu W, Martinez-Hackert E. Human Cerberus Prevents Nodal-Receptor Binding, Inhibits Nodal Signaling, and Suppresses Nodal-Mediated Phenotypes. PLoS One. 2015;10:e0114954. doi: 10.1371/journal.pone.0114954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich RL, Myszka DG. Survey of the year 2007 commercial optical biosensor literature. Journal of molecular recognition : JMR. 2008;21:355–400. doi: 10.1002/jmr.928. [DOI] [PubMed] [Google Scholar]
- 25.Baker JM, Boyce FM. High-throughput functional screening using a homemade dual-glow luciferase assay. Journal of visualized experiments : JoVE. 2014 doi: 10.3791/50282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci U S A. 2006;103:7643–7648. doi: 10.1073/pnas.0602558103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klammert U, Mueller TD, Hellmann TV, Wuerzler KK, Kotzsch A, Schliermann A, Schmitz W, Kuebler AC, Sebald W, Nickel J. GDF-5 can act as a context-dependent BMP-2 antagonist. BMC biology. 2015;13:77. doi: 10.1186/s12915-015-0183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]