Abstract
Oncogenic Ras proteins are a driving force in a significant set of human cancers and wild-type, unmutated Ras proteins likely contribute to the malignant phenotype of many more. The overall challenge of targeting activated Ras proteins has great promise to treat cancer, but this goal has yet to be achieved. Significant efforts and resources have been committed to inhibiting Ras, but these energies have so far made little impact in the clinic. Direct attempts to target activated Ras proteins have faced many obstacles, including the fundamental nature of the gain-of-function oncogenic activity being produced by a loss-of-function at the biochemical level. Nevertheless, there has been very promising recent pre-clinical progress. The major strategy that has so far reached the clinic aimed to inhibit activated Ras indirectly through blocking its post-translational modification and inducing its mislocalization. While these efforts to indirectly target Ras through inhibition of farnesyl transferase (FTase) were rationally designed, this strategy suffered from insufficient attention to the distinctions between the isoforms of Ras. This led to subsequent failures in large-scale clinical trials targeting K-Ras driven lung, colon, and pancreatic cancers. Despite these setbacks, efforts to indirectly target activated Ras through inducing its mislocalization have persisted. It is plausible that FTase inhibitors may still have some utility in the clinic, perhaps in combination with statins or other agents. Alternative approaches for inducing mislocalization of Ras through disruption of its palmitoylation cycle or interaction with chaperone proteins are in early stages of development.
Keywords: H-Ras, K-Ras, lovastatin, N-Ras, prenylation inhibitors, statins, subcellular localization
1. INTRODUCTION
Mutated Ras proteins play critical roles in both the development [1] and maintenance of tumors [2, 3]. In contrast to the increasingly successful therapeutic targeting of other driving oncogenes to treat human cancer [4], Ras has yet to be effectively blocked in the clinic [5]. This void is particularly serious because Ras mutations are the most prevalent oncogenic events in human cancer. Although previous estimates of 30% incidence of Ras mutations in human cancers are likely high due to selection bias for certain tumor types with particularly high rates, an average pan-Ras mutation incidence of 16% points to the potential that would result if effective targeting could be achieved [6]. Positive Ras mutation status is not only a major driver of disease, it also correlates with poor prognosis and resistance to therapy [7], for example in colorectal carcinoma and lung adenocarcinoma [8, 9].
Ras proteins are the products of genes that were the first identified human oncogenes [10]. There are three human Ras genes that code for four distinct proteins: H-Ras; N-Ras; and two variants of K-Ras produced by alternative splicing, K-Ras4A and K-Ras4B. We are still discovering fundamental new information about these isoforms. For example, K-Ras4A used to be viewed as a minor variant and was less studied, recent application of isoform-specific PCR and antibodies has shown expression at significant levels in all human cancer cell lines and tissue specimens tested [11]. This result is particularly notable in light of previous evidence that K-Ras4A may have an important role in lung carcinoma [12]. While the Ras proteins all have identical effector domains, studies have shown that these isoforms have some distinct functions [13–15]. For example, K-Ras is the only form that binds to calmodulin [16], that can confer stem-like properties to certain cell types [7], and that has essential functions in mouse embryogenesis [17, 18]. Insertion of H-Ras at the K-Ras locus allows embryonic development, but then reveals a late-onset cardiomyopathy [19].
In regard to the pathologic expression of each isoform, K-Ras is mutated most often, particularly in pancreatic, intestinal, cholangio, and lung carcinomas, while N-Ras is mutated more in certain skin and hematopoietic cancers. H-Ras mutations are less common, but occur more in salivary gland and urinary tract cancers [6]. Further examination of the mouse model that has H-Ras inserted at the K-Ras locus shows that carcinogen-induced pulmonary tumorigenesis is maintained at a similar rate, with the lung cancers now driven by oncogenic H-Ras [12]. This spectrum of results suggests that the pattern, timing and level of expression of the Ras protein, rather than the specific isoform, may be more crucial for some aspects of development and tumorigenesis. Nevertheless, there are also some isoform-specific functions. Such discrepancies between isoforms are generally ascribed to differences in their short hypervariable regions (which are located just before the C-termini) and associated with their distinct subcellular locales [20–22].
In addition to the driving roles played by oncogenically-mutated Ras, it is likely that unmutated Ras proteins may also contribute to human cancers. For example, oncogenic K-Ras activity may require functional H-Ras or N-Ras to drive its effects [23, 24]. Further, activated wild-type Ras proteins may also promote cancers that do not harbor mutated Ras. For example, in breast cancer Ras mutations are rare, but there is strong evidence that increased growth factor and HER2 signaling induce over-activated Ras proteins to produce the transformed phenotype [25–27]. In type 1 neurofibromatosis (NF1), the loss of expression of neurofibromin, which normally functions to deactivate Ras, provides a route for over-activated Ras proteins to drive tumor formation [28–30]. Sporadic loss of neurofibromin expression likely drives Ras activation in other cancers, such as melanoma and lung adenocarcinoma, where it has also been linked to treatment resistance [31, 32].
Ras proteins drive pathways that can regulate perhaps all of the characterized hallmarks of cancer [33]. Thus, effective blockade of activated Ras could lead to beneficial outcomes if it could be achieved with acceptable levels of toxicity. The current status of several potential approaches are discussed below, with reference to the complexity of the Ras isoforms, and the degree to which there is a need to consider both the mutated and wild-type proteins. The focus will be on the status of attempts to directly target the protein, and to indirectly target it through induced mislocalization. The ongoing efforts to target pathways downstream of activated Ras, and that are beginning to prove useful in cancer treatment [34], have been reviewed elsewhere [5, 35].
1.1. Direct Targeting of Ras
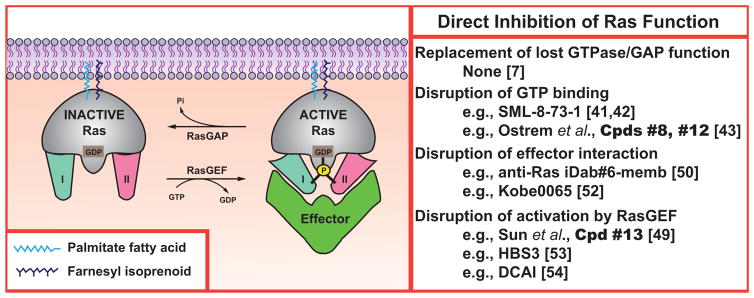
There are several barriers to direct therapeutic targeting of activated Ras. The first problem is that Ras is a small GTPase that is regulated through a cycle of GTP binding for activation (which is stimulated by guanine nucleotide exchange factors or GEFs) and then GTP hydrolysis to GDP for deactivation (which is stimulated by GTPase-activating proteins, or GAPs) [36], see (Fig. 1). Thus the conventional understanding of direct inhibition of an enzyme, i.e., to block its enzymatic activity, would actually produce an increase in the GTP-bound, activated fraction of Ras and so be the opposite of the mechanistic goal. Structural analyses of Ras indicated that it was unlikely that a small molecule could be designed to restore to oncogenic variants the lost GTPase activity and sensitivity to GAPs [37]. Furthermore, screening efforts to discover such compounds failed [7]. A related problem underlies the challenge of targeting Ras for treatment of NF1 [38]. The Ras in this case is wild-type and so maintains its endogenous GTPase activity, with the over-activation caused by loss of expression of the GAP, neurofibromin [28, 29]. There is no clear route to design a small molecule that could replace the lost expression and activity of neurofibromin.
Fig. (1). Direct inhibition of Ras Function.
The Ras activation/deactivation cycle and interaction with downstream effectors provides several potential therapeutic targets. Mature Ras proteins are anchored at the membrane and achieve an active conformation that interacts with effectors following the binding of GTP. This active conformation is lost upon GTP hydrolysis.
The second problem is that Ras binds GTP with picomolar affinity, and so it has not generally been feasible to design a small molecule that can displace the activating nucleotide [39]. This situation differs significantly from protein kinases, where ATP binding typically occurs with micromolar affinity. As a result, small molecule nucleotide analogues (typically with nanomolar affinities) effectively block ATP binding to kinases, but are unable to disrupt GTP binding to Ras [40]. Recent publications involving the GDP analog named SML-8-73-1 by Gray, Westover and colleagues provide the first example of success in this area [41, 42]. These groups, and that of Shokat [43], have addressed an interesting premise: why not target the activating, mis-sense substitution itself? Such an approach would have elegant selectivity for the pathological driver. Their efforts have produced small molecules that target K-Ras with the G12C mutation, which commonly occurs in tobacco-induced lung cancer [44]. The feasibility of such covalent targeting is supported by recent studies with ibrutinib, which binds a cysteine in the active site to inhibit Bruton’s tyrosine kinase. Although the compounds targeting Ras have only been shown to act in vitro so far, the approval of ibrutinib for treatment of relapsed mantle cell lymphoma provides a paradigm for this approach [45]. Shokat and colleagues developed a set of small molecules that could irreversibly bind to K-Ras G12C and prevent mutant protein—but not wild-type—from entering the GTP-bound state [43]. In parallel efforts, Gray and Westover and colleagues identified a GDP analog (SML-8-73-1) and a prodrug derivative (SML-10-70-1) that had the ability to covalently bind and specifically inactivate K-Ras G12C by leaving it in an open conformation that cannot interact productively with effectors [41, 42]. Although the compounds will require significant further pre-clinical optimization [46], these developments have rejuvenated interest in directly targeting Ras.
The third problem is that the function of activated Ras-GTP is transmitted through its formation of complexes with effectors [47], and small molecule inhibition of such protein:protein contacts has often proved difficult [48]. The structure of Ras does not have any clearly exploitable pockets to target, and allosteric regulation sites have not been revealed [43, 49]. A proof-of-principle study used expression of a blocking antibody fragment to demonstrate that oncogenic function of mutated K-Ras could be inhibited in a mouse model [50]. These results are a successor to earlier studies in which micro-injection of Ras antibodies into fibroblasts demonstrated the essential role of proto-oncogenic Ras function in serum stimulation of G1-to-S phase progression [51]. Recently, Kataoka and colleagues demonstrated that binding of H-Ras.GTP to c-Raf1 could be inhibited by small molecules both in vitro and in vivo. These inhibitors were also capable of down-regulating a number of Ras-driven pathways, and were orally active against a K-Ras driven colon cancer xenograft [52].
Another potential mechanism to inhibit Ras activation would be to block its activation by a GEF. Progress has recently been made in this area by disruption of Ras interaction with Son of Sevenless (Sos) [49, 53, 54]. On the other hand, the ultimate utility of targeting the GEF step has not yet been determined. Given that Ras mutations favor constitutive signaling, it seems likely that wild-type proteins would be significantly more dependent on GEF interaction, i.e., there is probably differential reliance on basal vs. stimulated nucleotide exchange [24, 54, 55]. Thus it is plausible that disrupting Sos activation of normal Ras could lead to unwanted toxicity if oncogenic Ras is the desired target.
The degree to which inhibitors of Sos/Ras interaction will also block Ras activation by other GEFs has generally not been defined. Sos is likely the principal activating pathway for Ras as a driver of cellular proliferation [56] and thus the logical target for these efforts. If compounds also block other GEFs that activate Ras in different contexts, then adverse events could occur. For example, Ras-GRP GEFs play critical roles in regulation of Ras in lymphocytes to control immune functions [57]. In T cells, subtle regulation of Ras determines development and selection, and small perturbations in Ras activation could lead to auto-immune dysfunction [57–59]. Another important example is provided by Ras-GRF GEFs that regulate neuronal Ras, particularly the H-Ras isoform [60], to control synaptic plasticity [61, 62]. Ras-GRF control of Ras is likely to integrate multiple neurotransmitter and neurotrophic pathways [63–67]. Disruption of Ras activation in the brain is strongly linked to disordered cognition and mental retardation [68]. It is clear that the accurate regulation of Ras is vital for normal functioning.
1.2. Indirect Targeting of Activated Ras Through Inhibition of Prenylation to Cause Mislocalization
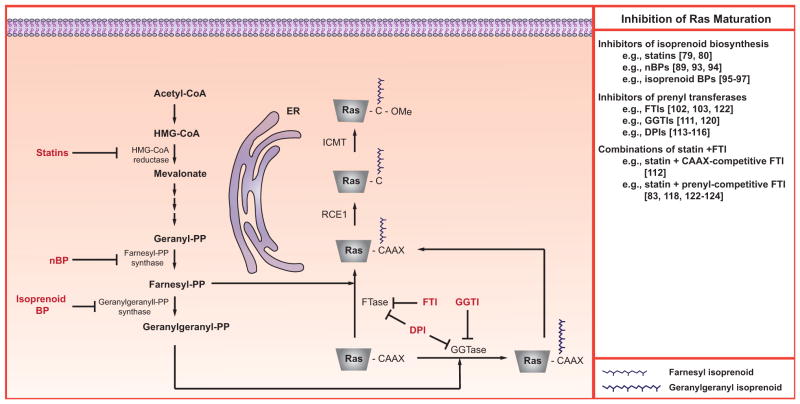
Ras proteins require a sequence of post-translational modifications to appropriately localize to membranes and become functionally active [69–71], see (Fig. 2). The demonstration that correct localization was essential for Ras’s transformative activity was the basis for a strategy to block Ras-driven cancers by inhibiting these modifications [72, 73]. In addition, selective toxicity might occur if mislocalized, constitutively-active Ras brought related signaling molecules to an abnormal environment [72, 74]. The initial step in the series that leads to membrane association and activity is the post-translational prenylation of the cysteine residue in the CaaX sequence (where C is cysteine, “a” is any residue (typically aliphatic), and X is glutamine, methionine, serine or leucine) found at the carboxyl terminus of Ras. Prenylation entails the covalent attachment of farnesyl or geranylgeranyl moieties to the cysteine catalyzed by farnesyl transferase (FTase) or geranylgeranyl transferase-I (GGTase-I), respectively [75]. Which moiety is attached depends on the CaaX sequence, and if a farnesyltransferase inhibitor (FTI) is present. All Ras molecules preferably undergo farnesylation. However, if an FTI is present, then K- and N-Ras, because of a leucine at the “X” position, can undergo geranylgeranylation by GGTase-I [76–78].
Fig. (2). Inhibition of Ras maturation.
Newly-synthesized Ras polypeptides undergo complex, multi-step maturation pathways. Many of the enzymes shown have been considered as potential therapeutic targets. In addition to those detailed in this figure, both Rce1 and Icmt have also been investigated [see 5 for review].
1.2.1. Inhibition of Prenylation by Suppressing Isoprenoid Synthesis
A popular strategy for disrupting Ras localization targeted prenylation. One general approach targets isoprenoid biosynthesis. Farnesyl pyrophosphate (FPP) and geranylgeranyl pyrophosphate (GGPP), which are co-substrates in FTase and GGTase-I reactions respectively, are intermediates in the cholesterol biosynthetic pathway [73], see (Fig. 2). Presumably, limiting the supply of precursors needed for FPP and GGPP synthesis should reduce prenylation. Early studies focused on the use of statins (e.g., particularly lovastatin) to inhibit HMG-CoA reductase. Preclinical studies with lovastatin indicated that it could inhibit prenylation in cultured cells. Furthermore, it showed promising therapeutic effects in murine tumor xenograft studies and, in some cases, the therapeutic effect correlated with reductions in the tumors’ prenylated Ras content [79, 80]. However, the concentrations needed to affect Ras prenylation in these preclinical studies are unattainable in humans because of dose limiting toxicity [81]. Specifically, although a lovastatin plasma level of 3.9 μM was achieved in a phase I clinical trial, it could not be maintained because of toxicity [82]. Treatment of cultured vascular smooth muscle cells with 3 μM lovastatin was sufficient to greatly impair their proliferation and reduced prenylation of RhoB, but had negligible effect on Ras [83]. The upper range of tolerable lovastatin dosing in humans results in plasma levels of ~225 nM and does not affect Ras processing in peripheral blood cells [84]. Irrespective of this limitation, statins continue to be widely used in preclinical studies, especially in combinational protocols, at concentrations ≥ 10 μM. Although sufficient to modulate Ras prenylation, such concentrations are physiologically irrelevant since they cannot be achieved in humans. There have also been recent clinical trials with statins with the goal of improving cognitive functions in children and adults with NF1. Despite the previous results in humans, the rationale for these studies is that there will be an inhibition of Ras [85–87]. The largest trial, with 84 children and a placebo-controlled, double-blind design, failed to demonstrate any beneficial effects of simvastatin treatment [87].
Nitrogen-containing bisphosphonates (nBPs) are synthetic analogues of inorganic pyrophosphate. They readily bind Ca2+ and have a very high affinity for areas of bone undergoing osteoclastic resorption [88–90]. Because of this affinity, and their cytotoxicity towards osteoclasts, nBPs have been used in the treatment of osteoporosis, Paget’s disease, and metastatic bone disease [90, 91]. Like statins, nBPs suppress isoprenoid synthesis, but do so by inhibiting farnesyl pyrophosphate synthase (FPP synthase), the enzyme immediately responsible for the generation of FPP, which is the precursor to GGPP [89, 92], see (Fig. 2). In vitro studies, with a variety of cell types, demonstrated that nBPs suppress the conversion of [14C]mevalonate into [14C]FPP and [14C]GGPP [89, 93], reduce the prenylation of Ras [93] and Rap1A [89], and cause a loss of membrane-associated Ras [94]. In addition to their proven effectiveness in the treatment of a variety of osteoclast-mediated bone conditions, mouse xenograft studies suggest that nBPs may be useful in the treatment of some non-bone-related cancers [89, 95]. At issue is whether these latter in vivo anti-cancer effects are mediated by protein deprenylation.
An alternative approach for modifying production of isoprenoids entails the targeted inactivation of geranylgeranyl diphosphate synthase (GGDPS), a cytosolic enzyme responsible for the conversion of FPP to GGPP [95]. A variety of isoprenoid bisphosphonates have been synthesized that selectively inhibit the in vitro activity of purified GGDPS with high nM to low micromolar potency [95–97], see (Fig. 2). Cell culture studies confirmed that the more potent of these also suppressed the prenylation of Rap1A (a GGTase-I substrate) and Rab6 (a GGTase-II substrate) to a level comparable to 10 μM lovastatin [96, 97]. However, unlike lovastatin, the GGDPS inhibitors did not affect the prenylation of Ras [96, 97]. Furthermore, it has been reported that cotreatment of cultured K562 leukemia cells with lovastatin and the GGDPS inhibitor digeranyl bisphosphonate resulted in a synergistic suppression of both Rap1a and Rab6 prenylation, but an antagonism of lovastatin’s inhibitory effects on Ras prenylation [96]. This is not surprising since inhibition of GGDPS activity would lead to a build up of FPP, and thus favor the farnesylation of Ras. Interestingly, concentrations of the GGDPS inhibitor digeranyl bisphosphonate sufficient to inhibit prenylation in cultured K562 cells also suppressed cell growth and induced apoptosis [96]. Furthermore, the anti-proliferative and pro-apoptotic activities of digeranyl bisphosphonate were synergistically enhanced by co-treatment with lovastatin [96]. These latter findings suggest that prenylated proteins other than Ras may be the targets and basis for the anti-proliferative/pro-apoptotic activities of some prenylation inhibitors.
1.2.2. Inhibitors of Prenylation Enzymes
A second general approach for modulating Ras prenylation involves suppression of FTase or GGTase-I catalytic activities. This entails the use of agents that suppress prenylation by competing with FPP, GGPP, or CaaX containing protein substrates for binding to FTase or GGTase-I, see (Fig. 2). Early attempts focused on the development of FTIs, and an extensive preclinical literature accumulated indicating that FTIs were effective at reversing H-Ras-mediated transformation, and suppressing H-Ras driven tumor xenografts in mice [98–100]. This strategy became the dominant one for targeting oncogenic Ras [101]. Ultimately, six different FTIs entered clinical trials [i.e., tipifarnib (aka R115777 or Zarnestra), lonafarnib (aka SCH66336 or Sarasar), BMS-214662, L778123, L744832 and FTI-277]. Of the six, the CaaX box competitive inhibitor tipifarnib advanced the farthest and was tested in stage III trials involving colorectal and pancreatic cancers [102, 103]. The results of these stage III tipifarnib studies were very disappointing. No significant anti-tumor activities were observed. In retrospect, an obvious explanation for the failure of these trials was the focus on malignancies driven by activated K-Ras, which are able to undergo alternative geranylgeranylation in the presence of an FTI [76–78].
Although a logical explanation exists for the negative outcomes of the tipifarnib phase III trials, the lack of efficacy branded FTIs as being failed drugs for the treatment of human cancer. Unfortunately that perception has persisted in spite of more recent studies indicating that FTIs may be useful in the treatment of astrocytomas, gliomas, and a subset of patients with hematological cancers [104–106]. It is not clear whether inhibition of Ras prenylation underlies any of these responses, however. Lonafarnib, which also advanced as far as phase II/III cancer trials, has more recently shown great promise as a novel therapy for progeria [107]. In this case the rationale is to block farnesylation of a variant of lamin-A, not Ras. It is, however, still plausible that FTIs may be effective against cancers that are driven by oncogenic H-Ras, which would not be able to escape inhibition through undergoing alternative prenylation [see [5] for review]. Mutated H-Ras is present in approximately 3% of human cancers [35]. Tumor profiling is already identifying patients with oncogenic H-Ras [108]. Perhaps by supporting such results with bioinformatic analysis to delineate driving pathways [109], it might be possible to select a patient population who may be primed to respond in a FTI clinical trial.
Both pharmacological and molecular approaches have documented the contributions of GGTase-I to Ras prenylation, so it is reasonable to ask whether it would be possible to achieve effective inhibition of K-Ras prenylation and localization by the combined inhibition of both FTase and GGTase-I. For example, K-Ras prenylation is maintained in cells/tissues derived from mice deficient in either FTase or GGTase-I due to a conditional knock out of the b-subunit of either enzyme [110]. However, dual deficiency in FTase and GGTase-1 reduces K-Ras prenylation in both cell lines and tissues derived from knockout mice [110]. Similarly, cotreatment with FTI and GGTase-I inhibitors (GGTIs) facilitates the deprenylation of both N- and K-Ras in cultured cells and murine xenograft models [99, 111, 112]. However, doses of GGTIs sufficient to block K-Ras prenylation in vivo were observed to be toxic, and lethal if chronically administered [111]. Furthermore, the anti-tumor activity of GGTIs towards models driven by activated K-Ras has been reported to be independent of K-Ras prenylation status [99, 113].
In addition to combinations of molecules that selectively target FTase or GGTase-I, considerable effort has gone into the synthesis of dual prenyltransferase inhibitors (DPIs), see (Fig. 2). As nicely documented in a paper by Tucker et al., the ratio of FTI to GGTI activity can vary markedly from DPI to DPI [114]. Of the many DPIs synthesized, L-778,123 is probably the most extensively examined. Low micromolar concentrations of L-778,123 were reported to suppress the prenylation of H-, N- and K-Ras in HL-60 leukemia cells [115]. However, doses of L-778,123 sufficient to inhibit the prenylation of Rab6 and HDJ2 in the peripheral blood mononuclear cells (PBMCs) of treated dogs had no effect on canine K-Ras prenylation [113]. Similarly, two phase I pre-clinical studies with L-778,123 [113, 116] reported that doses and scheduling protocols having no, or acceptable side effects, partially suppressed patient PBMC Rab6 and HDJ2 prenylation. However, no deprenylation of K-Ras occurred, even at doses that had to be discontinued because of adverse side effects. In the clinical studies the level of suppression of HDJ2 and Rab6 deprenylation correlated with plasma L-778,123 concentrations.
1.2.3. Combinations of Statins and Inhibitors of Prenylation Enzymes
A third general approach for modulating Ras prenylation entails treatment with a statin plus FTI or GGTI. The statin dosage in such protocols is insufficient by itself to inhibit prenylation, but sufficient to decrease the isoprenoid pool available for FPP and GGPP synthesis. Several studies document the synergistic anti-proliferative/cytotoxic effects of statin and FTI cotreatment on H-, K- and N-Ras driven tumors. For example, Ding et al. reported that a combination of atorvastatin and tipifarnib inhibited the growth of K-Ras mutated pancreatic cancer cells and xenografts in excess of the additive inhibitory effects of the two agents [117]. Unfortunately, there was no determination of whether the observed effect correlated with the deprenylation of Ras. Similarly, Morgan et al. reported that lovastatin synergized with the FTI L-744,832 to inhibit the in vitro growth of primary myeloma cells isolated from bone marrow aspirates of patients with multiple myeloma. In this study, the concentration of lovastatin (20 μM) used for analyses of Ras prenylation status was by itself sufficient to cause deprenylation of both N- and K-Ras [112]. In contrast, Yonemoto et al. reported that cotreatment of H-Ras transfected NIH3T3 fibroblasts with the FTI J-104,871 and 2.5 μM lovastatin shifted the IC50 for the FTI from 3.1 to 0.5 μM, and that this change was accompanied by a parallel deprenylation of H-Ras [118].
The strongest rationale for combinations of a statin with inhibitors of FTase or GGTase likely applies to FTIs or GGTIs that compete with the prenyl co-substrate of the enzyme rather than with the CaaX peptide substrate. In such situations, the limitation of the prenyl substrate pools imposed by the statin would most likely potentiate the action of the FTI or GGTI [83, 119–121]. For example, combinations of lovastatin and farnesyl pyrophosphate prodrug FTIs dramatically synergize with one another in their killing of NF1 malignant peripheral nerve sheath tumors [122–124]. Prenylation analyses in these studies indicated that the combinational treatment decreased the prenylation of not only Ras, but also of Rap1A and Rab5, proteins normally geranylgeranylated by GGTase-I and GGTase-II, respectively. Importantly, these cells express little or no H-Ras and the predominant activated isoform is N-Ras [125]. Further, the synergy occurred with concentrations of lovastatin as low as 100–250 nM, i.e., levels that are achievable and tolerated in humans.
1.2.4. Summary of Prenylation as a Target for Inhibition of Activated Ras
Studies of Ras prenylation and inhibitors have yielded several key insights. First, statins and prenylation inhibitors often exhibit therapeutic effectiveness against tumors that are either not Ras-driven, or, if they are Ras-driven, there is no change in Ras prenylation status following inhibitor treatment [126–128]. Hence, in some cases, the anti-cancer activities of statins or FTIs and GGTIs appear to be independent of effects on Ras prenylation.
Second, the effects of prenylation inhibitors and high dose statin treatment are not limited to Ras. Hundreds of proteins are predicted to be prenylated [129]. It is likely that the deprenylation of some of these non-Ras proteins (e.g., RhoB, Rheb) contributes to the cytostatic/cytotoxic activities of statins and FTIs and GGTIs. It is also plausible, as discussed above, that endogenous H-Ras might be a critical target for FTI action [5].
Third, individual prenylated proteins differ in their sensitivities to FTIs and GGTIs. For example, K-Ras is less sensitive to deprenylation than is N-Ras [130]. Similarly, in a study in which K-Ras farnesylation could be investigated independent of any confounding geranylgeranylation, comparable deprenylation of K-Ras and NDJ2 (both of which are normally farnesylated) required a 10-fold higher concentration of the FTI tipifarnib to block K-Ras [111]. Such findings emphasize the limitations of using the deprenylation of marker proteins, such as Rap1A and NDJ2, as surrogates for Ras prenylation status. There is also the general problem of studying easily biopsied tissues in clinical studies, such as peripheral blood mononuclear cells, rather than the more difficult-to-biopsy tumor itself. Thus, it is often difficult to know in clinical studies if deprenylating agents actually affect Ras in the target tissue.
Fourth, the prenylation enzymes and the effects of many prenylation inhibitors need to be considered in terms of relative selectivity rather than specificity. For example, analyses of 23 CaaX box competitive FTIs indicated that 17 were also potent inhibitors of GGTase-II [131]. In the case of GGTase-I, it is well established that it can farnesylate prenyl acceptors that have a C-terminal leucine in their CaaX box motif [132, 133]. Similarly, in vitro studies indicate that FPP binds to GGTase-II, and that GGTase-II can farnesylate Rab7 if the ratio of FPP to GGPP is high [134]. This ability of GGTase-I and -II to catalyze farnesylation may provide an explanation as to why FTIs designed to compete with FPP might also inhibit the prenylation of proteins that are normally GGTase-I and -II substrates.
1.3. Alternative Approaches to Target Activated Ras by Preventing Ras Maturation and Localization
Ras prenylation is the first step in its process of maturation and localization. The sequence continues with proteolytic cleavage after the prenylated cysteine by Ras converting enzyme (Rce1), and subsequent methylation by isoprenylcysteine carboxyl methyltransferase (Icmt), see (Fig. 2). Both of these steps have been considered as potential therapeutic targets, though such work does not seem particularly promising. Likely problems include lack of specificity for Ras proteins over the many other substrates of these enzymes and consequent toxicity [see [5] for review].
Ubiquitination has also been implicated in controlling the cellular localization of Ras. In a series of papers, de la Vega and colleagues described the dynamic relationship between the de-ubiquitinase USP17 and Rce1 to control H- and N-Ras trafficking, but leaving K-Ras4B unaffected [135]. Briefly, ubiquitination of an Rce1 isoform localizes it to the endoplasmic reticulum (ER). In the presence of active USP17, deubiquitination occurs, causing this isoform to leave the ER and be degraded. As a result of this degradation, CaaX modification is blocked [136]. Research in this area could lead to another potential alternative for targeting Ras.
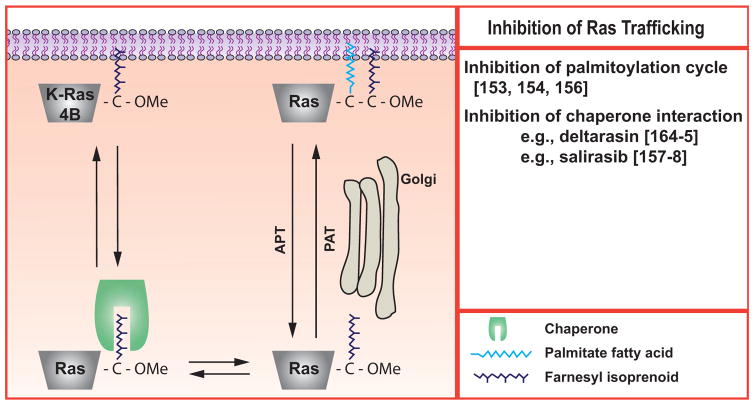
Subsequent to the prenylation, proteolysis and methylation reactions, an additional palmitoylation step is required for membrane association of H-, N-, and K-Ras4A [137], see (Fig. 3). K-Ras4B and 4A have stretches of lysine residues near their C-termini that serve as polybasic signals for membrane association [71]; K-Ras4A achieves membrane association due to the effects of both its polybasic residues and palmitoylation [11, 138]. In addition to the isoform-specific differences, another distinction at this step is that palmitoylation is clearly reversible, with specific enzymes catalyzing both the addition and removal of the palmitoyl moiety, and that this reversibility governs the subcellular localization and function of H- and N-Ras [139–143]. There is increasing interest in whether these reactions may provide therapeutic targets to block activated Ras (but not K-Ras4B) function [144–147]. The work is at a comparatively early stage and the complexity of the system makes it hard to study [148], suggesting that there may be a number of unanticipated obstacles. Yet the complexity also gives several reasons for optimism. As we learn more about the palmitoylation/depalmitoylation cycle of Ras, regulatory mechanisms will be revealed [149]. It is plausible that such steps may ultimately be exploited as targets for intervention to block Ras function. One reason why effective therapeutic targeting of the prenylation, proteolytic and methylation steps has proven to be difficult is that there is generally a single enzyme for each reaction that has many substrates in addition to Ras. In contrast, the enzymology of palmitoylation is extremely complex with many identified genes [150, 151]. Early results suggested that H- and N-Ras palmitoylation was under control of specific enzymes [152]. For example, DHHC9, which can be inhibited by microRNA-134, may be predominantly responsible for palmitoylation of H-Ras in cortical neurons [153]. If a specific subset of palmitoylation/depalmitoylation enzymes with limited redundancy controls Ras, then some selective targeting might be achieved [154]. On the other hand, dynamic palmitoylation is required for synaptic remodeling [155], which, together with the evidence that H-Ras function is critical here [see [5] for review], could suggest that blocking these reactions may lead to cognitive problems.
Fig. (3). Inhibition of Ras trafficking.
The prenylated Ras proteins require additional steps to achieve the membrane localization that is required for their activity. H-Ras, K-Ras4A, and N-Ras are reversibly palmitoylated. K-Ras4B requires interactions with chaperone proteins such as galectins or PDEδ. This figure is adapted and re-drawn from [156].
The functions of the lipid modifications of Ras proteins are expressed through induced interactions with both the lipid bilayer and also targeting or chaperone proteins [156]. These interactions have also been developed as potential therapeutic targets for activated Ras, see (Fig. 3). One that has made it to the clinic is the compound salirasib, which is an analog of the farnesyl isoprenoid [157], and that may interfere with the chaperoning activity of galectins toward farnesylated Ras proteins to disrupt their localization and activity [158]. It is likely that interactions with other, non-Ras farnesylated proteins will also be disrupted. A related compound that has been tested in patients is TLN-4601 [159]. Neither drug has been demonstrated to affect K-Ras function in patients [159–161], although a decrease in total K-Ras protein was reported in paired (pre- and post-treatment) tumor biopsies in two patients taking salirasib [161]. In a recent trial on hematological malignancies, salirasib efficacy was modest and did not correlate with Ras mutation status [162]. Although the diseases under study would presumably have made biochemical examination of peripheral blood mononuclear cells relevant, results from such studies were not reported.
Another chaperone for farnesylated proteins, including Ras, is phosphodiesterase-delta (PDEδ) [163]. Recently Zimmerman and colleagues have described small molecules that disrupt the interaction of K-Ras and PDEδ. One of these, termed deltarasin, decreased K-Ras function in pancreatic cells and inhibited growth of xenografts [164, 165]. The degree to which there is a selective effect on Ras as opposed to other farnesylated proteins and whether the inhibition will be effective in vivo and exerted through block of K-Ras function remain to be defined.
CONCLUSION
When it was realized that oncogenic Ras mutants were a driving force in many human cancers, great efforts and resources were committed to targeting these proteins. Unfortunately, these energies have so far produced little impact in the clinic. To an extent, directly targeting activated Ras parallels the trials and tribulations faced by scientists trying to therapeutically exploit tumor suppressors like p53, in that the biochemical lesion in oncogenic Ras is actually a loss of GTPase function. While the efforts to indirectly target Ras through FTIs were rationally designed, this strategy suffered from insufficient attention to the distinctions between the isoforms and lack of consideration of the fundamental biology of Ras prenylation. This led to their subsequent failure in large-scale clinical trials targeting K-Ras driven lung, colon, and pancreatic cancers. Despite these setbacks, efforts to indirectly target activated Ras through inducing its mislocalization have persisted and progress continues.
Acknowledgments
The authors acknowledge their longstanding collaboration with the late Prof. R.A. Gibbs for the joint development of several of the ideas presented here.
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest. E.B. has been supported by the Wayne State University School of Medicine, the cancer biology graduate program, and the training program in the biology of cancer [T32-CA009531]. Relevant work in J.J.R.’s laboratory has been supported by Department of Army [W81XWH-13-1-0097], and the Developmental Therapeutics Program, Karmanos Cancer Institute. Relevant work in R.R.M.’s laboratory has been supported by the National Cancer Institute [R01 CA81150 and CA131990], the Department of the Army [NF99035, NF020054, NF043107, and NF093146], by NTAP (Neurofibromatosis Therapeutic Acceleration Program), and by philanthropic support from Dan and Jennifer Gilbert. The content of the information does not necessarily reflect the position or policy of the U.S. government, and no official endorsement should be inferred.
References
- 1.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Jiang G, Yang F, Wang J. Knockdown of mutant K-ras expression by adenovirus-mediated siRNA inhibits the in vitro and in vivo growth of lung cancer cells. Cancer Biol Ther. 2006;5:1481–1486. doi: 10.4161/cbt.5.11.3297. [DOI] [PubMed] [Google Scholar]
- 3.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Adjei AA. Targeting oncogenic drivers. Prog Tumor Res. 2014;41:1–14. doi: 10.1159/000355895. [DOI] [PubMed] [Google Scholar]
- 5.Mattingly RR. Activated Ras as a Therapeutic Target: Constraints on Directly Targeting Ras Isoforms and Wild-Type versus Mutated Proteins. ISRN Oncol. 2013:536529. doi: 10.1155/2013/536529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prior IA, Lewis PD, Mattos C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Starmans MH, Pintilie M, Chan-Seng-Yue M, Moon NC, Haider S, Nguyen F, Lau SK, Liu N, Kasprzyk A, Wouters BG, et al. Integrating RAS Status into Prognostic Signatures for Adenocarcinomas of the Lung. Clin Cancer Res. 2015;21:1477–1486. doi: 10.1158/1078-0432.CCR-14-1749. [DOI] [PubMed] [Google Scholar]
- 9.Chang YY, Lin JK, Lin TC, Chen WS, Jeng KJ, Yang SH, Wang HS, Lan YT, Lin CC, Liang WY, et al. Impact of KRAS mutation on outcome of patients with metastatic colorectal cancer. Hepatogastroenterology. 2014;61:1946–1953. [PubMed] [Google Scholar]
- 10.Barbacid M. Ras genes. Ann Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 11.Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, Gierut JJ, Cox AD, Haigis KM, Philips MR. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci USA. 2015;112:779–784. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.To MD, Wong CE, Karnezis AN, Del Rosario R, Di Lauro R, Balmain A. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat Genet. 2008;40:1240–1244. doi: 10.1038/ng.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotiadou PP, Takahashi C, Rajabi HN, Ewen ME. Wild-Type NRas and KRas Perform Distinct Functions during Transformation. Mol Cell Biol. 2007;27:6742–6755. doi: 10.1128/MCB.00234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellano E, Santos E. Functional Specificity of Ras Isoforms: So Similar but So Different. Genes Cancer. 2011;2:216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton M, Wolfman A. Ha-ras and N-ras regulate MAPK activity by distinct mechanisms in vivo. Oncogene. 1998;16:1417–1428. doi: 10.1038/sj.onc.1201653. [DOI] [PubMed] [Google Scholar]
- 16.Villalonga P, Lopez-Alcala C, Bosch M, Chiloeches A, Rocamora N, Gil J, Marais R, Marshall CJ, Bachs O, Agell N. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol Cell Biol. 2007;21:7345–7354. doi: 10.1128/MCB.21.21.7345-7354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteban LM, Vicario-Abejón C, Fernández-Salguero P, Fernández-Medarde A, Swaminathan N, Yienger K, Lopez E, Malumbres M, McKay R, Ward JM, et al. Targeted Genomic Disruption of H-ras and N-ras, Individually or in Combination, Reveals the Dispensability of Both Loci for Mouse Growth and Development. Mol Cell Biol. 2011;21:1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potenza N, Vecchione C, Notte A, De Rienzo A, Rosica A, Bauer L, Affuso A, De Felice M, Russo T, Poulet R, et al. Replacement of K-Ras with H-Ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep. 2005;6:432–437. doi: 10.1038/sj.embor.7400397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ten Klooster JP, Hordijk PL. Targeting and localized signalling by small GTPases. Biol Cell. 2007;99:1–12. doi: 10.1042/BC20060071. [DOI] [PubMed] [Google Scholar]
- 21.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 23.Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentley C, Jurinka SS, Kljavin NM, Vartanian S, Ramani SR, Gonzalez LC, Yu K, Modrusan Z, Du P, Bourgon R, et al. A requirement for wild-type Ras isoforms in mutant KRas-driven signalling and transformation. Biochem J. 2013;452:313–320. doi: 10.1042/BJ20121578. [DOI] [PubMed] [Google Scholar]
- 25.Eckert LB, Repasky GA, Ulku AS, McFall A, Zhou H, Sartor CI, Der CJ. Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res. 2004;64:4585–4592. doi: 10.1158/0008-5472.CAN-04-0396. [DOI] [PubMed] [Google Scholar]
- 26.Clark GJ, Der CJ. Aberrant function of the Ras signal transduction pathway in human breast cancer. Breast cancer Res Treat. 1992;35:133–144. doi: 10.1007/BF00694753. [DOI] [PubMed] [Google Scholar]
- 27.Mittal S, Sharma A, Balaji SA, Gowda MC, Dighe RR, Kumar RV, Rangarajan A. Coordinate hyperactivation of Notch1 and Ras/MAPK pathways correlates with poor patient survival: novel therapeutic strategy for aggressive breast cancers. Mol Cancer Ther. 2014;13:3198–3209. doi: 10.1158/1535-7163.MCT-14-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosiss. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 29.Feldkamp MM, Angelov L, Guha A. Neurofibromatosis type 1 peripheral nerve tumors: aberrant activation of the Ras pathway. Surg Neurol. 1999;51:211–218. doi: 10.1016/s0090-3019(97)00356-x. [DOI] [PubMed] [Google Scholar]
- 30.Kraniak JM, Sun D, Mattingly RR, Reiners JJ, Jr, Tainsky MA. The role of neurofibromin in N-Ras mediated AP-1 regulation in malignant peripheral nerve sheath tumors. Mol Cell Biochem. 2014;344:267–276. doi: 10.1007/s11010-010-0551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nissan MH, Pratilas CA, Jones AM, Ramirez R, Won H, Liu C, Tiwari S, Kong L, Hanrahan AJ, Yao Z, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bruin EC, Cowell C, Warne PH, Jiang M, Saunders RE, Melnick MA, Gettinger S, Walther Z, Wurtz A, Heynen GJ, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014;4:606–619. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Wright CJ, McCormack PL. Trametinib: first global approval. Drugs. 2013;73:1245–1254. doi: 10.1007/s40265-013-0096-1. [DOI] [PubMed] [Google Scholar]
- 35.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Fut Med Chem. 2011;3:1787–1808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macara IG. The ras superfamily of molecular switches. Cell Signal. 1991;3:179–187. doi: 10.1016/0898-6568(91)90043-t. [DOI] [PubMed] [Google Scholar]
- 37.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 38.Dilworth JT, Kraniak JM, Wojtkowiak JW, Gibbs RA, Borch RF, Tainsky MA, Reiners JJ, Jr, Mattingly RR. Molecular targets for emerging anti-tumor therapies for neurofibromatosis type 1. Biochem Pharmacol. 2006;72:1485–1492. doi: 10.1016/j.bcp.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 39.John J, Rensland H, Schlichting I, Vetter I, Borasio GD, Goody RS, Wittinghofer A. Kinetic and structural analysis of the Mg(2+)-binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem. 1993;268:923–929. [PubMed] [Google Scholar]
- 40.Goekjian PG. Protein kinase C in the treatment of disease: signal transduction pathways, inhibitors, and agents in development. Curr Med Chem. 1999;6:877–903. [PubMed] [Google Scholar]
- 41.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, Carrasco M, Hunter J, Kim ND, Xie T, et al. Therapeutic Targeting of Oncogenic K-Ras by a Covalent Catalytic Site Inhibitor. Angewandte Chemie Intl Ed. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter JC, Gurbani D, Ficarro SB, Carrasco MA, Lim SM, Choi HG, Xie T, Marto JA, Chen Z, Gray NS, et al. In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc Nat Acad Sci USA. 2014;111:8895–8900. doi: 10.1073/pnas.1404639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dogan S, Shen R, Ang DC, Johnson ML, D’Angelo SP, Paik PK, Brzostowski EB, Riely GJ, Kris MG, Zakowski MF, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrera AF, Jacobsen ED. Ibrutinib for the treatment of mantle cell lymphoma. Clin Cancer Res. 2014;20:5365–5371. doi: 10.1158/1078-0432.CCR-14-0010. [DOI] [PubMed] [Google Scholar]
- 46.Ledford H. Cancer: The Ras renaissance. Nature. 2015;520:278–280. doi: 10.1038/520278a. [DOI] [PubMed] [Google Scholar]
- 47.Moodie SA, Willumsen BM, Weber MJ, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 48.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of alpha-helix-mediated protein-protein interactions using designed molecules. Nat Chem. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 49.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka T, Rabbitts TH. Interfering with RAS-effector protein interactions prevent RAS-dependent tumour initiation and causes stop-start control of cancer growth. Oncogene. 2010;29:6064–6070. doi: 10.1038/onc.2010.346. [DOI] [PubMed] [Google Scholar]
- 51.Mulcahy LS, Smith MR, Stacey DW. Requirement for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells. Nature. 1985;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- 52.Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, et al. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras–effector interaction. Proc Nat Acad Sci USA. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. An orthosteric inhibitor of the Ras-Sos interaction. Nat chem Boil. 2011;7:585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Nat Acad Sci USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864; a novel small molecule inhibitor of Rac family small GTPases. J Boil Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- 56.Nimnual A, Bar-Sagi D. The two hats of SOS. Science’s STKE : signal transduction knowledge environment. 2002;2002:pe36. doi: 10.1126/stke.2002.145.pe36. [DOI] [PubMed] [Google Scholar]
- 57.Jun JE, Rubio I, Roose JP. Regulation of ras exchange factors and cellular localization of ras activation by lipid messengers in T cells. Front Immunol. 2013;4:239. doi: 10.3389/fimmu.2013.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattingly RR, Felczak A, Chen CC, McCabe MJ, Jr, Rosenspire AJ. Low concentrations of inorganic mercury inhibit Ras activation during T cell receptor-mediated signal transduction. Toxicol Appl Pharmacol. 2001;176:162–168. doi: 10.1006/taap.2001.9272. [DOI] [PubMed] [Google Scholar]
- 59.Ziemba SE, Mattingly RR, McCabe MJ, Jr, Rosenspire AJ. Inorganic mercury inhibits the activation of LAT in T-cell receptor-mediated signal transduction. Toxicol Sci. 2006;89:145–153. doi: 10.1093/toxsci/kfj029. [DOI] [PubMed] [Google Scholar]
- 60.Yang H, Mattingly RR. The Ras-GRF1 exchange factor coordinates activation of H-Ras and Rac1 to control neuronal morphology. Mol Boil Cell. 2006;17:2177–2189. doi: 10.1091/mbc.E05-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez-Medarde A, Santos E. The RasGrf family of mammalian guanine nucleotide exchange factors. Biochim Biophys Acta. 2011;1815:170–188. doi: 10.1016/j.bbcan.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Sturani E, Abbondio A, Branduardi P, Ferrari C, Zippel R, Martegani E, Vanoni M, Denis-Donini S. The Ras Guanine nucleotide Exchange Factor CDC25Mm is present at the synaptic junction. Exp Cell Res. 1997;235:117–123. doi: 10.1006/excr.1997.3660. [DOI] [PubMed] [Google Scholar]
- 63.Yang H, Cooley D, Legakis JE, Ge Q, Andrade R, Mattingly RR. Phosphorylation of the Ras-GRF1 exchange factor at Ser916/898 reveals activation of Ras signaling in the cerebral cortex. J Biol Chem. 2003;278:13278–13285. doi: 10.1074/jbc.M209805200. [DOI] [PubMed] [Google Scholar]
- 64.Mattingly RR. Phosphorylation of serine 916 of Ras-GRF1 contributes to the activation of exchange factor activity by muscarinic receptors. J Boil Chem. 1999;274:37379–37384. doi: 10.1074/jbc.274.52.37379. [DOI] [PubMed] [Google Scholar]
- 65.Mattingly RR, Saini V, Macara IG. Activation of the Ras-GRF/CDC25Mm exchange factor by lysophosphatidic acid. Cell Signal. 1999;11:603–610. doi: 10.1016/s0898-6568(99)00034-0. [DOI] [PubMed] [Google Scholar]
- 66.Mattingly RR, Macara IG. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- 67.Krapivinsky G, Krapivinsky L, Manasian Y, Ivanov A, Tyzio R, Pellegrino C, Ben-Ari Y, Clapham DE, Medina I. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron. 2003;40:775–784. doi: 10.1016/s0896-6273(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 68.Weeber EJ, Sweatt JD. Molecular neurobiology of human cognition. Neuron. 2002;33:845–848. doi: 10.1016/s0896-6273(02)00634-7. [DOI] [PubMed] [Google Scholar]
- 69.Willumsen BM, Christensen A, Hubbert NL, Papageorge AG, Lowy DR. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984;310:583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- 70.Jackson JH, Cochrane CG, Bourne JR, Solski PA, Buss JE, Der CJ. Farnesol modification of Kirsten-ras exon 4B protein is essential for transformation. Proc Nat Acad Sci USA. 1990;87:3042–3046. doi: 10.1073/pnas.87.8.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 72.Gibbs JB, Oliff A, Kohl NE. Farnesyltransferase inhibitors: Ras research yields a potential cancer therapeutic. Cell. 1994;77:175–178. doi: 10.1016/0092-8674(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 73.Kim R, Rine J, Kim SH. Prenylation of mammalian Ras protein in Xenopus oocytes. Mol Cell Biol. 1990;10:5945–5949. doi: 10.1128/mcb.10.11.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lerner EC, Qian Y, Blaskovich MA, Fossum RD, Vogt A, Sun J, Cox AD, Der CJ, Hamilton AD, Sebti SM. Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic Ras signaling by inducing cytoplasmic accumulation of inactive Ras-Raf complexes. J Biol Chem. 1995;270:26802–26806. doi: 10.1074/jbc.270.45.26802. [DOI] [PubMed] [Google Scholar]
- 75.Fu HW, Casey PJ. Enzymology and biology of CaaX protein prenylation. Rec Prog Hormone Res. 1999;54:315–342. discussion 342–313. [PubMed] [Google Scholar]
- 76.Zhang FL, Kirschmeier P, Carr D, James L, Bond RW, Wang L, Patton R, Windsor WT, Syto R, Zhang R, et al. Characterization of Ha-ras, N-ras, Ki-Ras4A, and Ki-Ras4B as in vitro substrates for farnesyl protein transferase and geranylgeranyl protein transferase type I. J Biol Chem. 1997;272:10232–10239. doi: 10.1074/jbc.272.15.10232. [DOI] [PubMed] [Google Scholar]
- 77.Rowell CA, Kowalczyk JJ, Lewis MD, Garcia AM. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J Biol Chem. 1997;272:14093–14097. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- 78.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 79.Khan SG, Saxena R, Bickers DR, Mukhtar H, Agarwal R. Inhibition of ras p21 membrane localization and modulation of protein kinase C isozyme expression during regression of chemical carcinogen-induced murine skin tumors by lovastatin. Mol Carcinog. 1995;12:205–212. doi: 10.1002/mc.2940120405. [DOI] [PubMed] [Google Scholar]
- 80.Sebti SM, Tkalcevic GT, Jani JP. Lovastatin, a cholesterol biosynthesis inhibitor, inhibits the growth of human H-ras oncogene transformed cells in nude mice. Cancer commun. 1991;3:141–147. doi: 10.3727/095535491820873371. [DOI] [PubMed] [Google Scholar]
- 81.Knox JJ, Siu LL, Chen E, Dimitroulakos J, Kamel-Reid S, Moore MJ, Chin S, Irish J, LaFramboise S, Oza AM. A Phase I trial of prolonged administration of lovastatin in patients with recurrent or metastatic squamous cell carcinoma of the head and neck or of the cervix. Eur J Cancer. 2005;41:523–530. doi: 10.1016/j.ejca.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 82.Thibault A, Samid D, Tompkins AC, Figg WD, Cooper MR, Hohl RJ, Trepel J, Liang B, Patronas N, Venzon DJ, et al. Phase I study of lovastatin, an inhibitor of the mevalonate pathway, in patients with cancer. Clin Cancer Res. 1996;2:483–491. [PubMed] [Google Scholar]
- 83.Mattingly RR, Gibbs RA, Menard RE, Reiners JJ., Jr Potent suppression of proliferation of a10 vascular smooth muscle cells by combined treatment with lovastatin and 3-allylfarnesol, an inhibitor of protein farnesyltransferase. J pharmacol Exper Ther. 2002;303:74–81. doi: 10.1124/jpet.102.036061. [DOI] [PubMed] [Google Scholar]
- 84.Lewis KA, Holstein SA, Hohl RJ. Lovastatin alters the isoprenoid biosynthetic pathway in acute myelogenous leukemia cells in vivo. Leuk Res. 2005;29:527–533. doi: 10.1016/j.leukres.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 85.Chabernaud C, Mennes M, Kardel PG, Gaillard WD, Kalbfleisch ML, Vanmeter JW, Packer RJ, Milham MP, Castellanos FX, Acosta MT. Lovastatin regulates brain spontaneous low-frequency brain activity in neurofibromatosis type 1. Neurosci Lett. 2012;515:28–33. doi: 10.1016/j.neulet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mainberger F, Jung NH, Zenker M, Wahllander U, Freudenberg L, Langer S, Berweck S, Winkler T, Straube A, Heinen F, et al. Lovastatin improves impaired synaptic plasticity and phasic alertness in patients with neurofibromatosis type 1. BMC Neurol. 2013;13:131. doi: 10.1186/1471-2377-13-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van der Vaart T, Plasschaert E, Rietman AB, Renard M, Oostenbrink R, Vogels A, de Wit MC, Descheemaeker MJ, Vergouwe Y, Catsman-Berrevoets CE, et al. Simvastatin for cognitive deficits and behavioural problems in patients with neurofibromatosis type 1 (NF1-SIMCODA): a randomised, placebo-controlled trial. Lancet Neurol. 2013;12:1076–1083. doi: 10.1016/S1474-4422(13)70227-8. [DOI] [PubMed] [Google Scholar]
- 88.Reszka AA, Rodan GA. Nitrogen-containing bisphosphonate mechanism of action. Mini-Rev Med Chem. 2004;4:711–719. [PubMed] [Google Scholar]
- 89.Roelofs AJ, Thompson K, Gordon S, Rogers MJ. Molecular mechanisms of action of bisphosphonates: current status. Clin Cancer Res. 2006;12:6222s–6230s. doi: 10.1158/1078-0432.CCR-06-0843. [DOI] [PubMed] [Google Scholar]
- 90.Kimmel DB. Mechanism of action, pharmacokinetic and pharmacodynamic profile, and clinical applications of nitrogen-containing bisphosphonates. J Dent Res. 2007;86:1022–1033. doi: 10.1177/154405910708601102. [DOI] [PubMed] [Google Scholar]
- 91.Delmas PD, Meunier PJ. The management of Paget’s disease of bone. N Engl J Med. 1997;336:558–566. doi: 10.1056/NEJM199702203360807. [DOI] [PubMed] [Google Scholar]
- 92.van Beek E, Pieterman E, Cohen L, Lowik C, Papapoulos S. Nitrogen-containing bisphosphonates inhibit isopentenyl pyrophosphate isomerase/farnesyl pyrophosphate synthase activity with relative potencies corresponding to their antiresorptive potencies in vitro and in vivo. Biochem Biophys Res Commun. 1999;255:491–494. doi: 10.1006/bbrc.1999.0224. [DOI] [PubMed] [Google Scholar]
- 93.Luckman SP, Hughes DE, Coxon FP, Graham R, Russell G, Rogers MJ. Nitrogen-containing bisphosphonates inhibit the mevalonate pathway and prevent post-translational prenylation of GTP-binding proteins, including Ras. J Bone Mineral Res: the Official J Amer Soc Bone Mineral Res. 1998;13:581–589. doi: 10.1359/jbmr.1998.13.4.581. [DOI] [PubMed] [Google Scholar]
- 94.Oades GM, Senaratne SG, Clarke IA, Kirby RS, Colston KW. Nitrogen containing bisphosphonates induce apoptosis and inhibit the mevalonate pathway, impairing Ras membrane localization in prostate cancer cells. J Urol. 2003;170:246–252. doi: 10.1097/01.ju.0000070685.34760.5f. [DOI] [PubMed] [Google Scholar]
- 95.Wiemer AJ, Wiemer DF, Hohl RJ. Geranylgeranyl diphosphate synthase: an emerging therapeutic target. Clin Pharmacol Ther. 2011;90:804–812. doi: 10.1038/clpt.2011.215. [DOI] [PubMed] [Google Scholar]
- 96.Dudakovic A, Wiemer AJ, Lamb KM, Vonnahme LA, Dietz SE, Hohl RJ. Inhibition of geranylgeranyl diphosphate synthase induces apoptosis through multiple mechanisms and displays synergy with inhibition of other isoprenoid biosynthetic enzymes. J Pharmacol Exp Ther. 2008;324:1028–1036. doi: 10.1124/jpet.107.132217. [DOI] [PubMed] [Google Scholar]
- 97.Wiemer AJ, Yu JS, Lamb KM, Hohl RJ, Wiemer DF. Mono- and dialkyl isoprenoid bisphosphonates as geranylgeranyl diphosphate synthase inhibitors. Bioorg Med Chem. 2008;16:390–399. doi: 10.1016/j.bmc.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 98.Nagasu T, Yoshimatsu K, Rowell C, Lewis MD, Garcia AM. Inhibition of human tumor xenograft growth by treatment with the farnesyl transferase inhibitor B956. Cancer Res. 1995;55:5310–5314. [PubMed] [Google Scholar]
- 99.Sun J, Qian Y, Hamilton AD, Sebti SM. Both farnesyltransferase and geranylgeranyltransferase I inhibitors are required for inhibition of oncogenic K-Ras prenylation but each alone is sufficient to suppress human tumor growth in nude mouse xenografts. Oncogene. 1998;16:1467–1473. doi: 10.1038/sj.onc.1201656. [DOI] [PubMed] [Google Scholar]
- 100.Brunner TB, Hahn SM, Gupta AK, Muschel RJ, McKenna WG, Bernhard EJ. Farnesyltransferase inhibitors: an overview of the results of preclinical and clinical investigations. Cancer Res. 2003;63:5656–5668. [PubMed] [Google Scholar]
- 101.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Nat Cancer Ins. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 102.Rao S, Cunningham D, de Gramont A, Scheithauer W, Smakal M, Humblet Y, Kourteva G, Iveson T, Andre T, Dostalova J, et al. Phase III double-blind placebo-controlled study of farnesyl transferase inhibitor R115777 in patients with refractory advanced colorectal cancer. J Clin Oncol. 2004;22:3950–3957. doi: 10.1200/JCO.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 103.Van Cutsem E, van de Velde H, Karasek P, Oettle H, Vervenne WL, Szawlowski A, Schoffski P, Post S, Verslype C, Neumann H, et al. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol. 2004;22:1430–1438. doi: 10.1200/JCO.2004.10.112. [DOI] [PubMed] [Google Scholar]
- 104.Haas-Kogan DA, Banerjee A, Poussaint TY, Kocak M, Prados MD, Geyer JR, Fouladi M, Broniscer A, Minturn JE, Pollack IF, et al. Phase II trial of tipifarnib and radiation in children with newly diagnosed diffuse intrinsic pontine gliomas. Neuro Oncol. 2011;13:298–306. doi: 10.1093/neuonc/noq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Desjardins A, Reardon DA, Peters KB, Threatt S, Coan AD, Herndon JE, 2nd, Friedman AH, Friedman HS, Vredenburgh JJ. A phase I trial of the farnesyl transferase inhibitor SCH 66336; with temozolomide for patients with malignant glioma. J Neurooncol. 2011;105:601–606. doi: 10.1007/s11060-011-0627-0. [DOI] [PubMed] [Google Scholar]
- 106.Witzig TE, Tang H, Micallef IN, Ansell SM, Link BK, Inwards DJ, Porrata LF, Johnston PB, Colgan JP, Markovic SN, et al. Multi-institutional phase 2 study of the farnesyltransferase inhibitor tipifarnib (R115777) in patients with relapsed and refractory lymphomas. Blood. 2011;118:4882–4889. doi: 10.1182/blood-2011-02-334904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Nat Acad Sci USA. 2012;109:16666–16671. doi: 10.1073/pnas.1202529109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beadling C, Heinrich MC, Warrick A, Forbes EM, Nelson D, Justusson E, Levine J, Neff TL, Patterson J, Presnell A, et al. Multiplex mutation screening by mass spectrometry evaluation of 820 cases from a personalized cancer medicine registry. J Mol Diagnost: JMD. 2011;13:504–513. doi: 10.1016/j.jmoldx.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaur H, Mao S, Shah S, Gorski DH, Krawetz SA, Sloane BF, Mattingly RR. Next-generation sequencing: a powerful tool for the discovery of molecular markers in breast ductal carcinoma in situ. Exp Rev Mol Diagn. 2013;13:151–165. doi: 10.1586/erm.13.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu M, Sjogren AK, Karlsson C, Ibrahim MX, Andersson KM, Olofsson FJ, Wahlstrom AM, Dalin M, Yu H, Chen Z, et al. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc Nat Acad Sci USA. 2010;107:6471–6476. doi: 10.1073/pnas.0908396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lobell RB, Omer CA, Abrams MT, Bhimnathwala HG, Brucker MJ, Buser CA, Davide JP, de Solms SJ, Dinsmore CJ, Ellis-Hutchings MS, et al. Evaluation of farnesyl:protein transferase and geranylgeranyl:protein transferase inhibitor combinations in preclinical models. Cancer Res. 2001;61:8758–8768. [PubMed] [Google Scholar]
- 112.Morgan MA, Sebil T, Aydilek E, Peest D, Ganser A, Reuter CW. Combining prenylation inhibitors causes synergistic cytotoxicity, apoptosis and disruption of RAS-to-MAP kinase signalling in multiple myeloma cells. Br J Haematol. 2005;130:912–925. doi: 10.1111/j.1365-2141.2005.05696.x. [DOI] [PubMed] [Google Scholar]
- 113.Lobell RB, Liu D, Buser CA, Davide JP, DePuy E, Hamilton K, Koblan KS, Lee Y, Mosser S, Motzel SL, et al. Preclinical and clinical pharmacodynamic assessment of L-778, 123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol Cancer Ther. 2002;1:747–758. [PubMed] [Google Scholar]
- 114.Tucker TJ, Abrams MT, Buser CA, Davide JP, Ellis-Hutchings M, Fernandes C, Gibbs JB, Graham SL, Hartman GD, Huber HE, et al. The synthesis and biological evaluation of a series of potent dual inhibitors of farnesyl and geranyl-Geranyl protein transferases. Bioorg Med Chem Lett. 2002;12:2027–2030. doi: 10.1016/s0960-894x(02)00308-6. [DOI] [PubMed] [Google Scholar]
- 115.Morgan MA, Onono FO, Spielmann HP, Subramanian T, Scherr M, Venturini L, Dallmann I, Ganser A, Reuter CW. Modulation of anthracycline-induced cytotoxicity by targeting the prenylated proteome in myeloid leukemia cells. J Mol Med (Berl) 2012;90:149–161. doi: 10.1007/s00109-011-0814-7. [DOI] [PubMed] [Google Scholar]
- 116.Britten CD, Rowinsky EK, Soignet S, Patnaik A, Yao SL, Deutsch P, Lee Y, Lobell RB, Mazina KE, McCreery H, et al. A phase I and pharmacological study of the farnesyl protein transferase inhibitor L-778,123 in patients with solid malignancies. Clin Cancer Res. 2001;7:3894–3903. [PubMed] [Google Scholar]
- 117.Ding N, Cui XX, Gao Z, Huang H, Wei X, Du Z, Lin Y, Shih WJ, Rabson AB, Conney AH, et al. A triple combination of atorvastatin, celecoxib and tipifarnib strongly inhibits pancreatic cancer cells and xenograft pancreatic tumors. Intl J Oncol. 2014;44:2139–2145. doi: 10.3892/ijo.2014.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yonemoto M, Satoh T, Arakawa H, Suzuki-Takahashi I, Monden Y, Kodera T, Tanaka K, Aoyama T, Iwasawa Y, Kamei T, et al. J-104,871, a novel farnesyltransferase inhibitor, blocks Ras farnesylation in vivo in a farnesyl pyrophosphate-competitive manner. Mol Pharmacol. 1998;54:1–7. doi: 10.1124/mol.54.1.1. [DOI] [PubMed] [Google Scholar]
- 119.Wojtkowiak JW, Gibbs RA, Mattingly RR. Working together: Farnesyl transferase inhibitors and statins block protein prenylation. Mol Cell Pharmacol. 2009;1:1–6. doi: 10.4255/mcpharmacol.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sane KM, Mynderse M, Lalonde DT, Dean IS, Wojtkowiak JW, Fouad F, Borch RF, Reiners JJ, Jr, Gibbs RA, Mattingly RR. A novel geranylgeranyl transferase inhibitor in combination with lovastatin inhibits proliferation and induces autophagy in STS-26T MPNST cells. J pharmacol Exp Ther. 2010;333:23–33. doi: 10.1124/jpet.109.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gibbs BS, Zahn TJ, Mu Y, Sebolt-Leopold JS, Gibbs RA. Novel farnesol and geranylgeraniol analogues: A potential new class of anticancer agents directed against protein prenylation. J Med Chem. 1999;42:3800–3808. doi: 10.1021/jm9902786. [DOI] [PubMed] [Google Scholar]
- 122.Clark MK, Scott SA, Wojtkowiak J, Chirco R, Mathieu P, Reiners JJ, Jr, Mattingly RR, Borch RF, Gibbs RA. Synthesis, biochemical, and cellular evaluation of farnesyl monophosphate prodrugs as farnesyltransferase inhibitors. J Med Chem. 2007;50:3274–3282. doi: 10.1021/jm0701829. [DOI] [PubMed] [Google Scholar]
- 123.Wojtkowiak JW, Fouad F, LaLonde DT, Kleinman MD, Gibbs RA, Reiners JJ, Jr, Borch RF, Mattingly RR. Induction of apoptosis in neurofibromatosis type 1 malignant peripheral nerve sheath tumor cell lines by a combination of novel farnesyl transferase inhibitors and lovastatin. J pharmacol Exp Ther. 2008;326:1–11. doi: 10.1124/jpet.107.135830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wojtkowiak JW, Sane KM, Kleinman M, Sloane BF, Reiners JJ, Jr, Mattingly RR. Aborted autophagy and nonapoptotic death induced by farnesyl transferase inhibitor and lovastatin. J pharmacol Exp Ther. 2011;337:65–74. doi: 10.1124/jpet.110.174573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mattingly RR, Kraniak JM, Dilworth JT, Mathieu P, Bealmear B, Nowak JE, Benjamins JA, Tainsky MA, Reiners JJ., Jr The mitogen-activated protein kinase/extracellular signal-regulated kinase kinase inhibitor PD184352 (CI-1040) selectively induces apoptosis in malignant schwannoma cell lines. J pharmacol Exp Ther. 2006;316:456–465. doi: 10.1124/jpet.105.091454. [DOI] [PubMed] [Google Scholar]
- 126.Rose WC, Lee FY, Fairchild CR, Lynch M, Monticello T, Kramer RA, Manne V. Preclinical antitumor activity of BMS-214662; a highly apoptotic and novel farnesyltransferase inhibitor. Cancer Res. 2001;61:7507–7517. [PubMed] [Google Scholar]
- 127.Chan KK, Oza AM, Siu LL. The statins as anticancer agents. Clin Cancer Res. 2003;9:10–19. [PubMed] [Google Scholar]
- 128.Appels NM, Beijnen JH, Schellens JH. Development of farnesyl transferase inhibitors: a review. Oncologist. 2005;10:565–578. doi: 10.1634/theoncologist.10-8-565. [DOI] [PubMed] [Google Scholar]
- 129.Maurer-Stroh S, Koranda M, Benetka W, Schneider G, Sirota FL, Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.End DW, Smets G, Todd AV, Applegate TL, Fuery CJ, Angibaud P, Venet M, Sanz G, Poignet H, Skrzat S, et al. Characterization of the antitumor effects of the selective farnesyl protein transferase inhibitor R115777 in vivo and. in vitro Cancer Res. 2001;61:131–137. [PubMed] [Google Scholar]
- 131.Lackner MR, Kindt RM, Carroll PM, Brown K, Cancilla MR, Chen C, de Silva H, Franke Y, Guan B, Heuer T, et al. Chemical genetics identifies Rab geranylgeranyl transferase as an apoptotic target of farnesyl transferase inhibitors. Cancer Cell. 2005;7:325–336. doi: 10.1016/j.ccr.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 132.Armstrong SA, Hannah VC, Goldstein JL, Brown MS. CAAX geranylgeranyl transferase transfers farnesyl as efficiently as geranylgeranyl to RhoB. J Biol Chem. 1995;270:7864–7868. doi: 10.1074/jbc.270.14.7864. [DOI] [PubMed] [Google Scholar]
- 133.Yokoyama K, McGeady P, Gelb MH. Mammalian protein geranylgeranyltransferase-I: substrate specificity, kinetic mechanism, metal requirements, and affinity labeling. Biochemistry. 1995;34:1344–1354. doi: 10.1021/bi00004a029. [DOI] [PubMed] [Google Scholar]
- 134.Thoma NH, Iakovenko A, Owen D, Scheidig AS, Waldmann H, Goody RS, Alexandrov K. Phosphoisoprenoid binding specificity of geranylgeranyltransferase type II. Biochemistry. 2000;39:12043–12052. doi: 10.1021/bi000835m. [DOI] [PubMed] [Google Scholar]
- 135.de la Vega M, Burrows JF, McFarlane C, Govender U, Scott CJ, Johnston JA. The deubiquitinating enzyme USP17 blocks N-Ras membrane trafficking and activation but leaves K-Ras unaffected. J Biol Chem. 2010;285:12028–12036. doi: 10.1074/jbc.M109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jaworski J, Govender U, McFarlane C, de la Vega M, Greene MK, Rawlings ND, Johnston JA, Scott CJ, Burrows JF. A novel RCE1 isoform is required for H-Ras plasma membrane localization and is regulated by USP17. Biochem J. 2014;457:289–300. doi: 10.1042/BJ20131213. [DOI] [PubMed] [Google Scholar]
- 137.Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 138.Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;121:421–427. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- 139.Agudo-Ibanez L, Herrero A, Barbacid M, Crespo P. H-Ras distribution and signaling in plasma-membrane microdomains are regulated by acylation and deacylation events. Mol Cell Biol. 2015;35(11):1898–914. doi: 10.1128/MCB.01398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lynch SJ, Snitkin H, Gumper I, Philips MR, Sabatini D, Pellicer A. The differential palmitoylation states of N-Ras and H-Ras determine their distinct Golgi subcompartment localizations. J Cell Physiol. 2015;230:610–619. doi: 10.1002/jcp.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Song SP, Hennig A, Schubert K, Markwart R, Schmidt P, Prior IA, Bohmer FD, Rubio I. Ras palmitoylation is necessary for N-Ras activation and signal propagation in growth factor signaling. Biochem J. 2013;454:323–332. doi: 10.1042/BJ20121799. [DOI] [PubMed] [Google Scholar]
- 142.Cuiffo B, Ren R. Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood. 2010;115:3598–3605. doi: 10.1182/blood-2009-03-213876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Coats SG, Booden MA, Buss JE. Transient palmitoylation supports H-Ras membrane binding but only partial biological activity. Biochemistry. 1999;38:12926–12934. doi: 10.1021/bi9909290. [DOI] [PubMed] [Google Scholar]
- 144.Hernandez JL, Majmudar JD, Martin BR. Profiling and inhibiting reversible palmitoylation. Curr Opin Chem Boil. 2013;17:20–26. doi: 10.1016/j.cbpa.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu J, Hedberg C, Dekker FJ, Li Q, Haigis KM, Hwang E, Waldmann H, Shannon K. Inhibiting the palmitoylation/depalmitoylation cycle selectively reduces the growth of hematopoietic cells expressing oncogenic Nras. Blood. 2012;119:1032–1035. doi: 10.1182/blood-2011-06-358960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dekker FJ, Hedberg C. Small molecule inhibition of protein depalmitoylation as a new approach towards downregulation of oncogenic Ras signaling. Bioorg Med Chem. 2011;19:1376–1380. doi: 10.1016/j.bmc.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 147.Ducker CE, Griffel LK, Smith RA, Keller SN, Zhuang Y, Xia Z, Diller JD, Smith CD. Discovery and characterization of inhibitors of human palmitoyl acyltransferases. Mol Cancer Ther. 2006;5:1647–1659. doi: 10.1158/1535-7163.MCT-06-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Peng T, Hang HC. Bifunctional fatty acid chemical reporter for analyzing S-palmitoylated membrane protein-protein interactions in mammalian cells. J Am Chem Soc. 2007;137:556–559. doi: 10.1021/ja502109n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, Fehrenbacher N, Linder ME, Philips MR. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol Cell. 2011;41:173–185. doi: 10.1016/j.molcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 151.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2015;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 153.Chai S, Cambronne XA, Eichhorn SW, Goodman RH. MicroRNA-134 activity in somatostatin interneurons regulates H-Ras localization by repressing the palmitoylation enzyme DHHC9. Proc Natl Acad Sci USA. 2013;110:17898–17903. doi: 10.1073/pnas.1317528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chavda B, Arnott JA, Planey SL. Targeting protein palmitoylation: selective inhibitors and implications in disease. Exp Opin Drug Discov. 2013;9:1005–1019. doi: 10.1517/17460441.2014.933802. [DOI] [PubMed] [Google Scholar]
- 155.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cox AD, Der CJ, Philips MR. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin Cancer Res. 2015;21:1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marciano D, Ben-Baruch G, Marom M, Egozi Y, Haklai R, Kloog Y. Farnesyl derivatives of rigid carboxylic acids-inhibitors of ras-dependent cell growth. J Med Chem. 1995;38:1267–1272. doi: 10.1021/jm00008a004. [DOI] [PubMed] [Google Scholar]
- 158.Rotblat B, Niv H, Andre S, Kaltner H, Gabius HJ, Kloog Y. Galectin-1(L11A) predicted from a computed galectin-1 farnesyl-binding pocket selectively inhibits Ras-GTP. Cancer Res. 2004;64:3112–3118. doi: 10.1158/0008-5472.can-04-0026. [DOI] [PubMed] [Google Scholar]
- 159.Boufaied N, Wioland MA, Falardeau P, Gourdeau H. TLN-4601; a novel anticancer agent, inhibits Ras signaling post Ras prenylation and before MEK activation. Anti-cancer Drugs. 2010;21:543–552. doi: 10.1097/CAD.0b013e328337f373. [DOI] [PubMed] [Google Scholar]
- 160.Riely GJ, Johnson ML, Medina C, Rizvi NA, Miller VA, Kris MG, Pietanza MC, Azzoli CG, Krug LM, Pao W, et al. A phase II trial of Salirasib in patients with lung adenocarcinomas with KRAS mutations. J Thorac Oncol. 2011;6:1435–1437. doi: 10.1097/JTO.0b013e318223c099. [DOI] [PubMed] [Google Scholar]
- 161.Laheru D, Shah P, Rajeshkumar NV, McAllister F, Taylor G, Goldsweig H, Le DT, Donehower R, Jimeno A, Linden S, et al. Integrated preclinical and clinical development of S-trans, trans-Farnesylthiosalicylic Acid (FTS, Salirasib) in pancreatic cancer. Invest New Drugs. 2012;30:2391–2399. doi: 10.1007/s10637-012-9818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Badar T, Cortes JE, Ravandi F, O’Brien S, Verstovsek S, Garcia-Manero G, Kantarjian H, Borthakur G. Phase I Study of S-Trans Trans-Farnesylthiosalicylic Acid (Salirasib), a Novel Oral RAS Inhibitor in Patients With Refractory Hematologic Malignancies. Clin Lymph Myeloma Leukemia. 2015 doi: 10.1016/j.clml.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang H, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W. Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J Biol Chem. 2014;279:407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 164.Zimmermann G, Schultz-Fademrecht C, Kuchler P, Murarka S, Ismail S, Triola G, Nussbaumer P, Wittinghofer A, Waldmann H. Structure guided design and kinetic analysis of highly potent benzimidazole inhibitors targeting the PDEdelta prenyl binding site. J Med Chem. 2014;57:5435–5448. doi: 10.1021/jm500632s. [DOI] [PubMed] [Google Scholar]
- 165.Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signaling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]