Abstract
BACKGROUND
Recent gains in reducing the global burden of malaria are threatened by the emergence of Plasmodium falciparum resistance to artemisinins. The discovery that mutations in portions of a P. falciparum gene encoding kelch (K13)–propeller domains are the major determinant of resistance has provided opportunities for monitoring such resistance on a global scale.
METHODS
We analyzed the K13-propeller sequence polymorphism in 14,037 samples collected in 59 countries in which malaria is endemic. Most of the samples (84.5%) were obtained from patients who were treated at sentinel sites used for nationwide surveillance of antimalarial resistance. We evaluated the emergence and dissemination of mutations by haplotyping neighboring loci.
RESULTS
We identified 108 nonsynonymous K13 mutations, which showed marked geographic disparity in their frequency and distribution. In Asia, 36.5% of the K13 mutations were distributed within two areas — one in Cambodia, Vietnam, and Laos and the other in western Thailand, Myanmar, and China — with no overlap. In Africa, we observed a broad array of rare nonsynonymous mutations that were not associated with delayed parasite clearance. The gene-edited Dd2 transgenic line with the A578S mutation, which expresses the most frequently observed African allele, was found to be susceptible to artemisinin in vitro on a ring-stage survival assay.
CONCLUSIONS
No evidence of artemisinin resistance was found outside Southeast Asia and China, where resistance-associated K13 mutations were confined. The common African A578S allele was not associated with clinical or in vitro resistance to artemisinin, and many African mutations appear to be neutral.
Increased efforts have substantially reduced the global burden of malaria caused by Plasmodium falciparum,1,2 but the recent gains are threatened by emerging resistance to artemisinins, the cornerstone of current first-line combination treatment.1,3,4 Artemisinins are active against a large range of intraerythrocytic developmental stages, but their usefulness is curtailed by ring-stage resistance.5,6 Clinical artemisinin resistance, which was first documented in western Cambodia,7–10 is now prevalent across Southeast Asia and South China.11–17 Widespread artemisinin resistance would have dramatic consequences, since replacement therapies are limited and threatened by resistance.18–22
Therapeutic efficacy studies are the standard method for determining the efficacy of antimalarial drugs,23 but resource constraints restrict the numbers of sites and patients studied each year. The recent discovery of mutations in portions of a P. falciparum gene encoding kelch (K13)–propeller domains as the primary determinant of artemisinin resistance provided unprecedented opportunities for improving resistance monitoring.24,25 To date, 13 independent K13 mutations have been shown to be associated with clinical resistance,3,12,14,26–28 with evidence of independent emergence of the same mutation in different geographic areas.28,29 Four Asian mutations (C580Y, R539T, I543T, and Y493H) have been validated in vitro.24–27 In regions with no documented clinical artemisinin resistance, scattered studies have indicated that K13 mutations are rare.30–41 The few examples of reduced susceptibility to artemisinin-based combination therapy that have been reported in Africa were unrelated to K13 polymorphism, apart from three severe pediatric cases.42–44 No data are available for large regions of Africa, South America, Oceania, Central and East Asia, Indonesia, and the Philippines. The risk of the emergence or dissemination of resistance has spurred increased efforts to track artemisinin resistance in all geographic areas in which malaria is endemic. The use of a molecular indicator of artemisinin resistance is particularly timely, since rapid, large-scale screening is needed to provide an early warning about emergence or invasion events.45,46
To assess the global distribution of K13 polymorphisms, we launched in 2014 the K13 Artemisinin Resistance Multicenter Assessment (KARMA) study, in which we analyzed parasites that were collected from regions in which malaria is endemic, using a dedicated molecular toolbox and validation procedures for sequence data. Sequencing of the K13-propeller domain was combined with an analysis of flanking haplotypes to ascertain the origin and dissemination of specific mutations. We performed genome editing to confirm the phenotypic effect of the most frequent African A578S mutation. This worldwide survey was designed to provide critical information for drug policymakers and to outline methods for future surveillance activities.
METHODS
STUDY DESIGN, SAMPLING, AND OVERSIGHT
Investigators from Institut Pasteur in Paris and in Cambodia designed the study, which was coordinated by the Cambodian branch. According to the study design, the investigators planned to analyze 200 blood samples positive for P. falciparum that had been collected since 2012 and to contribute blood samples, K13 products obtained on polymerase-chain-reaction (PCR) assay, or K13 sequence data (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Sites that had few available samples or samples that were collected before 2012 were included if they were located in regions in which K13 diversity had not been documented. The study was approved by a national or institutional ethics committee or other appropriate authority at each site, and all investigators signed a declaration of ethical clearance.
Samples were obtained from patients seeking treatment at sites involved in national surveys of antimalarial drug resistance, from patients enrolled in therapeutic efficacy studies,23 from asymptomatic participants who were enrolled in surveillance programs, and from travelers returning to Europe with malaria. Investigators at Institut Pasteur in Cambodia collected all biologic material and performed K13 sequence and haplotype analysis; they also conducted quality-control analyses, and they vouch for the accuracy and completeness of the molecular data. Investigators who conducted therapeutic efficacy studies vouch for the accuracy and completeness of the clinical data. The sponsors had no role in the study design or in the collection or analysis of the data. There was no confidentiality agreement between the sponsors and the investigators.
GENOTYPING
The Institut Pasteur in Cambodia provided reagents, blinded quality-control samples, and standard operating procedures as a K13 toolbox (Table S1 in the Supplementary Appendix). DNA extraction and amplification of the K13-propeller domain (codons 440–680, 720 bp)24 were performed accordingly. PCR products were sequenced by Macrogen. We analyzed electropherograms on both strands, using PF3D7_1343700 as the reference sequence. We performed external quality assessment that included proficiency testing (in which 6 blinded quality-control samples were tested in each 96-well sequencing plate by each partner) and external quality control (in which 359 blood samples [2.6%] were independently retested) (Fig. S2 in the Supplementary Appendix). Isolates with mixed alleles were considered to be mutated for the purposes of mutation-frequency estimation.
The PF3D7_1337500 (K13_151) and PF3D7_1339700 (K13_159) loci were amplified (Table S2 in the Supplementary Appendix) and DNA sequences were analyzed as indicated above. Individual alleles were identified for each locus and haplotypes generated.
A578S GENE EDITING
We performed zinc-finger nuclease engineering, plasmid construction, and gene editing of the Dd2 line (which was collected in Indochina) with the A578S mutation.25 We assessed the in vitro susceptibility to artemisinin of the Dd2 A578S mutation, the Dd2 parent, and the Dd2 C580Y mutation (as a positive control), using the ring-stage survival assay (RSA) performed in red cells that had been infected with the malaria parasite in its ring stage (the developmental stage that is associated with resistance to artemisinin) for an estimated 0 to 3 hours. The cells were then exposed to dihydroartemisinin (the main metabolite of all artemisinin derivatives), and drug susceptibility was measured at 72 hours.5,25
GEOGRAPHICAL MAPPING
We calculated the proportion of parasites with a 3D7, wild-type allele (which is associated with artemisinin susceptibility) in each country and recorded the geospatial coordinates. To generate graphical maps, we interpolated data using ordinary kriging methods (or Gaussian process regression) or inverse distance weighting. (Details about the methods are provided in the Supplementary Appendix.)
STATISTICAL ANALYSIS
We expressed quantitative data as medians and interquartile ranges or as means and 95% confidence intervals. We used the Mann–Whitney test or t-tests of independent samples to compare continuous variables and the chi-square test or Fisher’s exact test to compare categorical variables. We obtained estimates of nucleotide diversity, 47 haplotype diversity,48 and Tajima’s D test49 using DnaSP software, version 5.50 All reported P values are two-sided, and a P value of less than 0.05 was considered to indicate statistical significance. Data were analyzed with the use of Microsoft Excel and MedCalc software, version 12.
RESULTS
SAMPLE COLLECTION
From May through December 2014, we gathered 14,037 samples from 163 sites in 59 countries. The samples included 11,854 (84.4%) from resident malaria patients in 40 countries, 1232 (8.8%) from residents with asymptomatic infections in 11 countries, and 951 (6.8%) from travelers returning with falciparum malaria from 40 countries (Fig. S3 in the Supplementary Appendix). The samples included those obtained from 2450 patients with malaria (2367 residents and 83 travelers) for whom follow-up data on day 3 were available (Table S3 in the Supplementary Appendix). We collected 4156 samples in 2012, 6440 samples in 2013, and 1671 samples in 2014 (>87% of all samples). Data regarding sample size, sex ratio, age, and parasitemia ranges according to study site are provided in Table S4 in the Supplementary Appendix.
K13-PROPELLER SEQUENCE POLYMORPHISMS
Sequences were generated for 13,157 (93.7%) samples; 880 samples from 35 sites had sequences that could not be interpreted because of poor quality or an insufficient quantity of DNA. Nearly all the samples (99.2%) contained a single K13 allele; 108 of 111 polyclonal infections were identified in samples obtained in Africa. The minor allele in a mixed sample was detected when its proportion was more than 20% (Fig. S4 in the Supplementary Appendix).
Overall, 1250 samples (9.5%) had 1295 K13 mutations, including 1097 (84.7%) with nonsynonymous mutations and 198 (15.3%) with synonymous mutations; 186 alleles were identified (Table S5 and Fig. S5 in the Supplementary Appendix). Among these alleles, there were 108 nonsynonymous mutations, of which 70 were newly identified and 38 were among 103 that had been reported previously.12,14,17,24,28,31–35,37–40,51–57 Only 9 nonsynonymous mutations (C580Y, F446I, R539T, A578S, Y493H, P574L, P553L, N458Y, and R561H) had a frequency of more than 1%; 72 alleles were observed only once (Tables S6 and S7 in the Supplementary Appendix).
There was marked geographic disparity in the proportion and distribution of K13 polymorphisms. We measured nucleotide diversity, haplotype diversity, and Tajima’s D ratio to highlight the continent-specific frequency of mutant alleles (Fig. S6 in the Supplementary Appendix). Of the 1097 nonsynonymous mutations, 957 (87.2%) were observed in samples obtained in Asia, and 169 of the 198 synonymous mutations (85.4%) were observed in samples obtained in Africa.
CONTINENTAL DISTRIBUTION AND PROPORTION OF K13-PROPELLER MUTATIONS
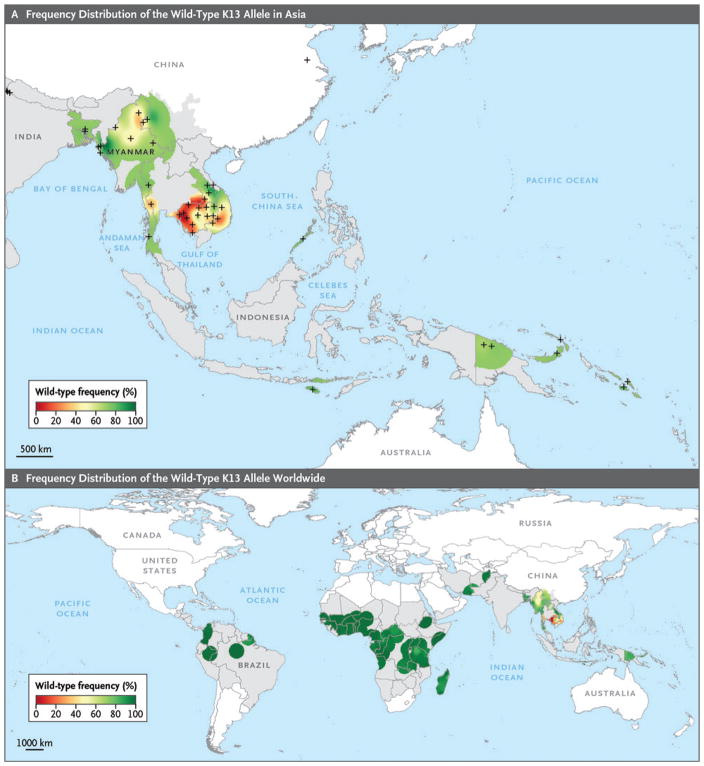
The proportion of K13 nonsynonymous mutations was heterogeneous in Asia, ranging from fixed (>95%) to very high (80 to 94%) in western Cambodia (Fig. S7 in the Supplementary Appendix), to intermediate (40 to 50%) in Myanmar and Vietnam, to moderate (10 to 20%) in eastern Cambodia, Thailand, China, and Laos, to low (<5%) elsewhere (Fig. 1). In South America, Oceania, and Africa, K13 nonsynonymous mutations were uncommon, except for a few African countries (range, 3.0 to 8.3% in Gambia, Central African Republic, Zambia, Comoros, Guinea, Kenya, and Chad). K13 nonsynonymous mutations were not detected in 27 countries from which samples were obtained (19 in Africa, 2 in Asia, 1 in Oceania, and 5 in South America), as shown on maps of wild-type allele distribution (Fig. 2).
Figure 1. K13 Nonsynonymous Mutations, According to Country and Continent.
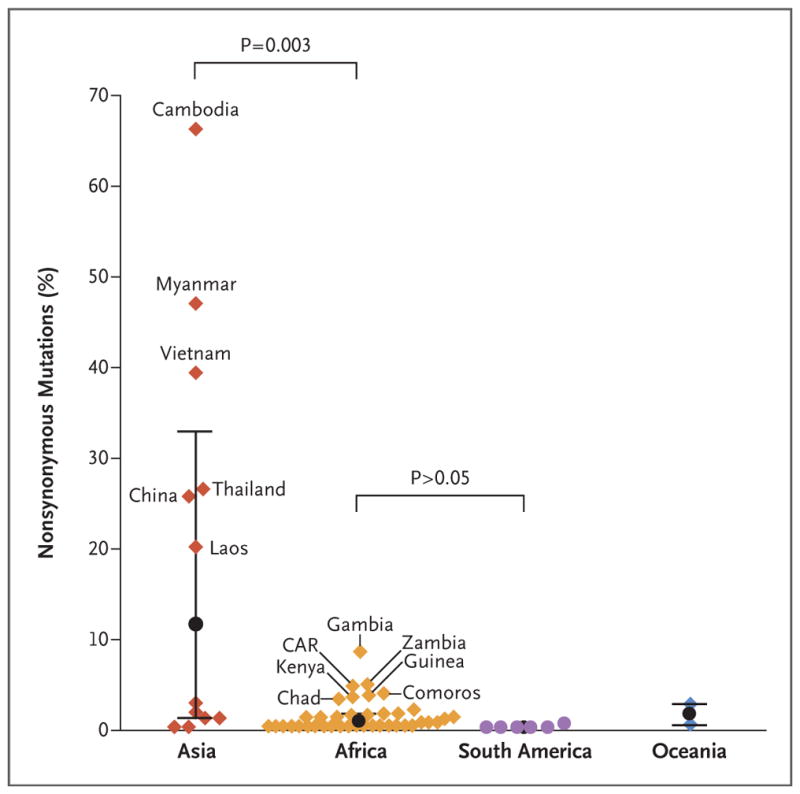
Shown are the percentages of nonsynonymous mutations that have been identified in the portion of the Plasmodium falciparum K13 gene encoding the kelchpropeller domain in Asia, Africa, South America, and Oceania. Synonymous mutations that do not modify the protein sequence are not indicated. At present, all the mutations that have been associated with resistance to artemisinin derivatives have resulted in nonsynonymous amino acid changes. The black circles indicate medians and the I bars interquartile ranges for each continent. K13 nonsynonymous mutations were not detected in 27 countries from which samples were obtained (19 in Africa, 2 in Asia, 1 in Oceania, and 5 in South America). Names are not shown (owing to a lack of space) for the following countries: in Asia: Afghanistan, Iran, Bangladesh, Nepal, Indonesia, and Philippines; in Africa: Cameroon, Congo, Democratic Republic of Congo, Equatorial Guinea, Gabon, Burundi, Ethiopia, Rwanda, Sudan, South Sudan, Somalia, Tanzania, Uganda, Madagascar, Angola, Malawi, Mozambique, South Africa, Zimbabwe, Benin, Burkina Faso, Ghana, Guinea Bissau, Ivory Coast, Liberia, Mali, Mauritania, Niger, Nigeria, Senegal, Sierra Leone, and Togo; in South America: Brazil, Colombia, Ecuador, French Guiana, Peru, and Venezuela; and in Oceania: Papua New Guinea and Solomon Islands. The percentages for Chad and Gambia were derived from a sample size of less than 50. Details regarding sampling and K13 diversity according to country are provided in Tables S4, S5, and S7 and Fig. S3 in the Supplementary Appendix. CAR denotes Central African Republic.
Figure 2. Frequency Distribution of the Wild-Type K13 Allele.
Shown are the distributions of the wild-type K13 allele in Asia (Panel A) and around the world (Panel B). Areas in which malaria is endemic are shaded in gray, and areas that are considered to be malaria-free are shown in white. The mean frequency of the wild-type allele is indicated by the color code. In Panel A, the individual sites of sample collection are indicated with a cross. In Panel B, a 100-km radius was used for the area centered on each sampling site or on the capital city of the country if no specific site was used for sampling. Data regarding sampling methods and K13 diversity according to country are provided in Tables S4, S5, and S7 and Fig. S3 in the Supplementary Appendix.
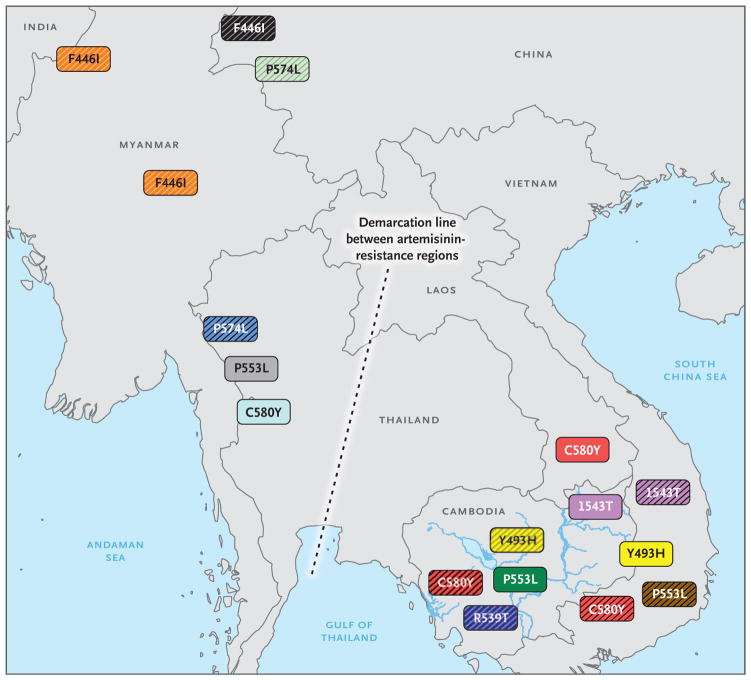
Individual K13 nonsynonymous mutations showed restricted geographic localization. In Asia, two distinct areas were identified, with different frequencies of individual mutations: one area that includes Cambodia, Vietnam, and Laos, where C580Y, R539T, Y493H, and I543T mutations were frequent or specific, and a second area that includes western Thailand, Myanmar, and China, where F446I, N458Y, P574L, and R561H mutations were specific (Fig. S8 in the Supplementary Appendix). The P553L allele was distributed in the two areas (Fig. 3).
Figure 3. Overview of the Distribution of the Flanking Haplotypes of the C580Y, Y493H, R539T, I543T, P553L, P574L, and F446I Nonsynonymous Mutations in K13 in Two Regions in Asia.
Shown are two areas in Asia in which individual K13 nonsynonymous mutations (which are labeled in the colored boxes) were frequently identified: one area that includes Cambodia, Vietnam, and Laos and a second area that includes western Thailand, Myanmar, and China. The haplotype groups of C580Y, F446I, P574L, and P553L are shown in distinct colors. Similarly colored boxes indicate shared haplotypes, and hatched boxes indicate the presence of additional countryspecific haplotypes. For example, C580Y haplotypes that are shared between Cambodia, Vietnam, and Laos (H29, H33, or H43) are shown in red, whereas the C580Y haplotype found in Thailand (H42) is shown in light blue. Full details about the distribution of haplotypes are provided in Table S9 and Fig. S9 in the Supplementary Appendix.
In Africa, no Asian artemisinin-resistance allele was observed among 9542 sequences, but 150 distinct alleles were identified, 92% of which were found in only one or two samples. Apart from A578S, V589I, S522C, V534A, F583L, and G665C, most alleles were Africa-specific and localized. A578S, which ranked fourth in abundance among mutant isolates, was observed in 1 sample from Thailand and 41 samples from Africa (Tables S6 and S7 in the Supplementary Appendix).
NEW EMERGENCE OR DISSEMINATION?
We investigated the genetic relatedness of isolates harboring the same frequent K13 mutation by assessing the polymorphism of two neighboring loci. We observed 10 alleles for K13_151 and 42 alleles for K13_159, which resulted in 80 flanking haplotypes (Tables S8 and S9 in the Supplementary Appendix). This assessment identified numerous emergence events alongside spreading of a small group of mutations for artemisinin resistance (Fig. 3). Of 17 distinct C580Y haplotypes, 3 common haplotypes were distributed across Cambodia, Vietnam, and Laos and 14 in specific areas of Cambodia, Thailand, and Vietnam. Similar observations were made for the Y493H, R539T, and I543T haplotypes, with probable west-to-east dissemination for the Y493H and R539T mutations and movement from Vietnam to eastern Cambodia for I543T mutations. Three prevalent F446I haplotypes were distributed across Myanmar, and 8 haplotypes were localized within China or Myanmar. Site-specific localization was observed for the 5 P553L haplotypes, the 6 P574L haplotypes, and the 2 E605K and R561H haplotypes (Fig. S9 in the Supplementary Appendix). For the common African mutation A578S, we identified multiple independent events of emergence (31 different haplotypes among 35 isolates) (Fig. S10 in the Supplementary Appendix).
TESTING OF THE A578S ALLELE FOR ARTEMISININ RESISTANCE
We wanted to evaluate whether the A578S mutation had a selective advantage against artemisinin, since there have been reports about a conflicting phenotype of the allele,12,31,36,44 which has been found at multiple sites in Africa and Asia12,31,32,34–40,57 (Table S6 and Fig. S10 in the Supplementary Appendix). We therefore introduced the A578S mutation in the Dd2 line that had been used to investigate the effect of several Asian K13 mutations.25 The Dd2 line with the A578S mutation was susceptible to artemisinin on RSA. The survival rate (mean [±SE], 0.29±0.11%) was equivalent to that in the parental line (mean, 0.14±0.03%), which was unlike the survival rate (mean, 4.01±0.16%) in the positive control Dd2 line with the C580Y mutation (Fig. S11 in the Supplementary Appendix).
K13 MUTATIONS AND POSITIVITY RATE
Our sampling included 2450 patients in whom the presence of parasites was assessed on day 3 after 7 days of artesunate monotherapy or a standard 3-day course of artemisinin-based combination therapy. Of these patients, 121 had positive parasite results on day 3 (Table S3 in the Supplementary Appendix). Of 40 nonsynonymous mutations that were detected in isolates obtained from these patients, only 8 were associated with positivity on day 3 (F446I, N458Y, N537D, R539T, I543T, P553L, P574L, and C580Y); all these alleles were observed only in Southeast Asia and China (Fig. S12 in the Supplementary Appendix). No circulating blood-stage parasites were found on day 3 in samples obtained from 1533 African patients in 25 countries, except for 9 patients who were each carrying a wild-type allele. The 9 African patients who were carrying an A578S allele on day 0 were parasite-free on day 3.
DISCUSSION
In this study, we have attempted to profile the global distribution of K13 polymorphisms. The breadth of this survey, with 163 sites that were sampled for molecular mapping and 36 countries with data regarding therapeutic efficacy, led us to conclude that artemisinin resistance is confined to Southeast Asia and China, where resistance-associated K13 mutations have reached intermediate frequency to fixation. This status contrasts with the situation in South America, Oceania, the Philippines, and Central and South Asia, which are essentially free of nonsynonymous K13 mutations. We observed many highly diverse, low-frequency nonsynonymous mutations in Africa, although none of these mutations were associated with clinical artemisinin resistance, as assessed by the presence of parasites on day 3 after artesunate monotherapy or 3-day treatment with artemisinin-based combination therapy. We saw no evidence of invasion of Africa by Asian resistance-conferring alleles, a finding that was consistent with the results of previous smaller studies.35–38,40,41,44,58,59 Finally, no positive selection was observed aside from that in Asia, which suggests no immediate threat to artemisinin efficacy in most countries in which malaria is endemic.
In the two independent foci of resistance that have been identified in the region that includes Cambodia, Vietnam, and Laos and the one that includes western Thailand, Myanmar, and China, we observed many more examples of emergence of resistance-associated mutations than had been reported previously.28,29,55 This finding indicates that contemporary artemisinin resistance is a result of numerous independent emergence events involving the same mutations together with the dissemination of a small group of endemic mutations (Fig. 3). The endemic mutations are probably the oldest ones, as suggested by an analysis of our archived samples, which showed that all four common contemporary C580Y haplotypes that were observed in Cambodia were already quite frequent 10 years earlier in Pailin province, the epicenter of the emergence of artemisinin-resistant parasites (Table S9 in the Supplementary Appendix).9,10 We observed no overlap between the sets of mutations and haplotypes in the two main resistance areas in Asia, which suggests the presence of different selective pressures in the two areas. Such differences may stem from the use of different artemisinin-based combination therapies as first-line treatment, different types of founder populations,29,59 or different epidemiologic ecosystems.
To date, 13 nonsynonymous mutations have been associated with slow parasite clearance, 3,12,24,25,28,32 and 4 mutations have been validated as conferring an increased rate of ringstage survival in drug-resistant field isolates in vitro24 or in gene-edited parasite lines.45 In the KARMA study, 8 nonsynonymous mutations that were observed in Southeast Asia and China — F446I, N458Y, N537D, R539T, I543T, P553L, P574L, and C580Y — were associated with positive results on day 3, findings that were consistent with data from previous studies that used parasite-clearance half-lives to identify artemisinin resistance12,14,24,26–28 and that provide further evidence of the positivity rate on day 3 as a sensitive indicator of clinical resistance to artemisinins.
The A578S mutation that has been commonly observed in Africa12,35–40,44 and detected in India,31 Bangladesh,34 and Thailand was sensitive on in vitro RSAs; this finding was unrelated to positivity on day 3. Numerous events of independent emergence were observed with no evidence of dissemination. These findings are consistent with the sensitive phenotype of one A578S Ugandan parasite as seen on RSA performed ex vivo (i.e., in samples obtained directly from patients)36 or the lack of association with slow-clearing infections in earlier studies.12,31 Thus, except for one recent study involving a few severely ill children, 44 the available data suggest that A578S is not an artemisinin-resistance mutation.
Aside from nine patients who carried wild-type K13 alleles, there was no positivity on day 3 among African patients. This result is consistent with the finding that a large number of rare K13 alleles are neutral with respect to artemisinin susceptibility and with the lack of evidence of artemisinin-driven selection on the K13 locus in Africa (Table S5 in the Supplementary Appendix). However, clinical detection of artemisinin resistance in Africa is complicated by the contribution of acquired immunity and maintained efficacy of the partner drugs (amodiaquine and lumefantrine) to parasite clearance.60,61 In vitro phenotyping with the use of RSA5 and allele-specific genome-editing studies25 can provide important information about the potential phenotypic effect of nonsynonymous mutations with respect to artemisinin susceptibility, as we provided for A578S in this study. Similar approaches could be used to assess the potential effect of frequently observed K13 nonsynonymous mutations associated with clinical artemisinin resistance (e.g., F446I, N458Y, P574L, and P553L). Of 108 nonsynonymous mutations that were detected, we observed 72 only once, which was consistent with the results of earlier studies.35,37–41 The criteria for prioritizing further laboratory studies include the following: the frequent observation of a new allele with a nonsynonymous mutation, evidence of dissemination (since the absence of dissemination probably indicates that the mutation was lost to genetic drift or intrapopulation competition), and preliminary association with clinical data whenever possible.
Some clues about the phenotypic effect of newly discovered mutations might also be obtained from exploring specific metabolic changes (e.g., in phosphatidylinositol 3-phosphate levels62) in parasites with K13 mutations or modeling the consequences of the mutations on K13 protein structure. The recently solved BTB–POZ (broad complex–tramtrack–bric-a-brac–poxvirus and zinc-finger) propeller structure of K13 protein domains (UniProtKB accession number, PDB AYY8) showed that F446, Y493, R539, and C580 are located in a beta sheet of their respective kelch domains (Fig. S13 in the Supplementary Appendix); these mutations should alter the overall structure (Fig. S14 in the Supplementary Appendix). We predict that A578S, located in the flexible junction between blade 3 and blade 4 of the beta sheet, has a limited effect on the propeller structure.
Our study has several weaknesses. First, the isolates that we evaluated represent only a convenience sample of those in the total population areas that we surveyed. Second, we lacked information about regions in which malaria is endemic such as India, where we needed to clarify the western boundary of resistance distribution. Third, in some countries, the numbers of available samples were small. The numbers of sites surveyed in East Africa, the Philippines, Indonesia, and South America need to be increased in future studies, although we could still discern clear geographic patterns in the distribution of haplotypes. Investigators should be able to implement the K13 toolbox in the field. We expect that future surveillance will fill in the gaps of the existing map.
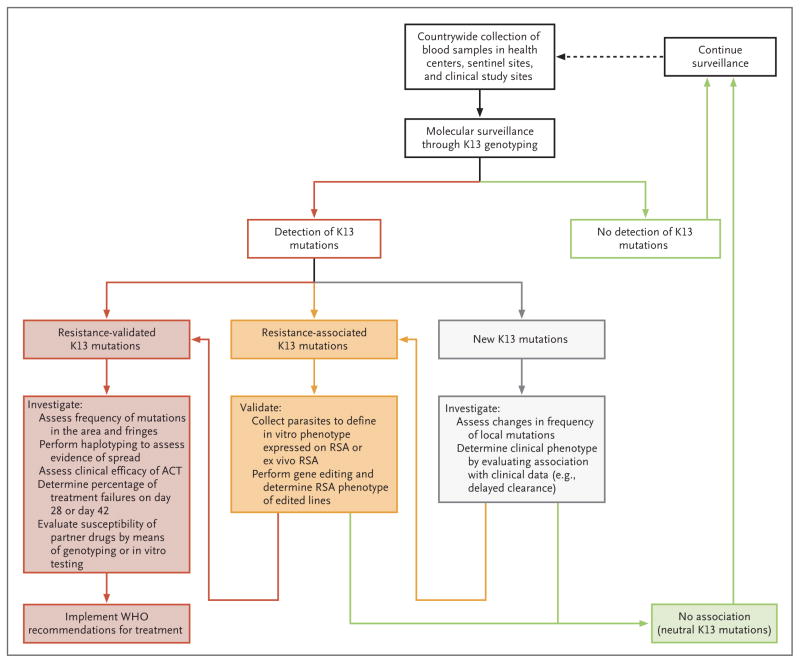
In conclusion, our study clarifies how K13 monitoring can assist future surveillance of artemisinin resistance (Fig. 4). In regions where resistance is established, K13 sequencing can map its geographic distribution and invasion of the fringes. The dissemination of K13 mutations can be conveniently assessed by determining the identity of flanking haplotypes, as we did here, which provides a more accessible approach than whole-genome sequencing29 or microsatellite typing. 55 In resistance-free areas, molecular surveillance will need to detect possible invasion by known alleles associated with K13 resistance and track any temporal increase in the proportion and dissemination of newly identified nonsynonymous mutations. This strategy should identify foci to be targeted for further in vitro phenotyping and therapeutic efficacy studies as well as for surveillance of other candidate artemisinin-susceptibility loci.42,43 This effort can contribute information to characterize these areas with respect to potential artemisinin resistance and inform treatment guidelines before large-scale dissemination has occurred.
Figure 4. Algorithm for Surveillance of Artemisinin Resistance on the Basis of K13 Mutations.
Shown is a step-by-step procedure for acquiring information to help develop policies regarding the use of artemisinin-based combination therapy (ACT) and strategies to eliminate malaria. K13 genotyping is integrated into surveillance activities by combining clinical efficacy studies with in vitro susceptibility testing and is supported by gene-editing studies. The validated or confirmed K13 mutations that are associated with resistance to artemisinins are Y493H, R539T, I543T, and C580Y, and the associated K13 mutations are P441L, F446I, G449A, N458Y, P553L, R561H, V568G, P574L, and A675V.3 The validation of a K13 mutation as a resistance marker (i.e., confirmed K13 mutation) is based on the following criteria: a significant association with delayed clearance of Plasmodium falciparum and a reduced drug sensitivity (survival rate, >1%) on ex vivo ring-stage survival assay (RSA) or in vitro RSA performed on field isolates, culture-adapted parasites, or gene-edited parasite lines, as compared with the K13 wild-type parent control. A K13 mutation is deemed to be associated with artemisinin resistance if it has a significant association with delayed clearance. A K13 mutation is said to be neutral if it has no significant association with delayed clearance or reduced drug sensitivity. A new K13 mutation is one that has been never observed and thus does not appear in mutation databases. Recommendations regarding efficacy trials and treatment policies are provided by the World Health Organization (WHO).4
Supplementary Material
Acknowledgments
Funded by Institut Pasteur Paris and others.
Supported by the Institut Pasteur Paris, Institut Pasteur International Division, Institut Pasteur Cambodia, and the World Health Organization; by a grant (ANR-10-LABX-62-IBEID) from the French Government Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases”; a grant from Natixis Banques; a grant (R01 AI109023, to Dr. Fidock) from the National Institutes of Health; grants from the Fiocruz Fundação Oswaldo Cruz, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro, Fundação de Amparo à Pesquisa do Estado do Amazonas, the Brazilian National Council for Scientific and Technological Development, the Agence Nationale de la Recherche (13-BSV3-0018-01 and 11-BSV7-009-01), the Austrian Federal Ministry of Science, Research, and Economy, the Calgary Laboratory Services, the Centre International de Recherches Médicales de Franceville, the European and Developing Countries Clinical Trials Partnership (CT-2004-31070-001), the Drugs for Neglected Diseases Initiative, the Else Kroener Fresenius Stiftung, the Holger Poehlmann Stiftung, the European Community African–European Research Initiative “IDEA” (HEALTH-F3-2009-241642), the Fonds Wetenschappelijk Onderzoek, the Vlaamse Interuniversitaire Raad–Universitaire Ontwikkelingssamenwerking, the Belgian Technical Cooperation in Democratic Republic of Congo, the European Community Seventh Framework Program (FP7/2007-2013, 242095, and 223601), the European Commission (REGPOT-CT-2011-285837-STRONGER), the Ministère de la Santé Publique du Niger (Laboratoire National de Référence Résistance aux Antipaludiques), the Foundation of National Science and Technology Major Program (2012ZX10004-220), the French Ministry of Health (Institut National de Veille Sanitaire), the Global Fund to Fight AIDS, Tuberculosis and Malaria, the 5% Initiative program (French Ministry of Foreign Affairs, France Expertise Internationale, 12INI109), the Institut Pasteur de Madagascar, the Government of the Philippines, the Institut de Recherche pour le Développement, the Foundation des Treilles, the Délégation Générale pour l’Armement (PDH-2-NRBC-4-B1-402), the Institut Pasteur de Bangui, the International Society for Health Research and Training, the Malaria Research Initiative Bandarban, Vienna, International Centre for Diarrhoeal Disease Research, Bangladesh, the Médecins sans Frontières (Centre Opérationnel Paris, France), Medicines for Malaria Venture, the National Research Council of Thailand, the Thammasat University, the National Natural Science Foundation of China (81271870, 81361120405, and 81271863), the Natural Science Foundation of Jiangsu Province (BK20130114 and BK20150001), the Jiangsu Science and Technology Department (BM2015024), the National Institutes of Health (R01 AI11646601, AI109023, and ICEMR U19AI089702, U19AI089672), the Pasteur Institute of Iran, the Malaria Division of the Iranian Center for Diseases Management and Control, Public Health England (Malaria Reference Service Contract), the Government of Rwanda, the U.S. Department of Defense Armed Forces Health Surveillance Center, Global Emerging Infections Surveillance and Response System (P0463-14-N6), the Fogarty International Center of the National Institutes of Health training (D43 TW007393), the Mahidol-Oxford Research Unit, the Government of Japan (Science and Technology Agency, Agency for Medical Research and Development, Japan International Cooperation Agency, and Science and Technology Research Partnership for Sustainable Development), and the President’s Malaria Initiative of the U.S. Agency for International Development.
We thank all the patients, health workers, and research teams involved in the worldwide sample collection; Prof. C. Brechot and M. Jouan of Institut Pasteur for their support and advice; the shipping department team of the Institut Pasteur, Paris, for help in shipping the K13 toolbox; and Dr. Raymond Hui for providing information on the crystal structure of the K13 protein.
APPENDIX
The authors’ full names and academic degrees are as follows: Didier Ménard, Ph.D., Nimol Khim, Ph.D., Johann Beghain, M.Sc., Ayola A. Adegnika, M.D., Ph.D., Mohammad Shafiul-Alam, Ph.D., Olukemi Amodu, Ph.D., Ghulam Rahim-Awab, Ph.D., Céline Barnadas, Ph.D., Antoine Berry, M.D., Ph.D., Yap Boum, Ph.D., Maria D. Bustos, M.D., Ph.D., Jun Cao, Ph.D., Jun-Hu Chen, Ph.D., Louis Collet, M.D., Liwang Cui, Ph.D., Garib-Das Thakur, M.D., Alioune Dieye, Pharm.D., Ph.D., Djibrine Djallé, M.Sc., Monique A. Dorkenoo, M.D., Carole E. Eboumbou-Moukoko, Ph.D., Fe-Esperanza-Caridad J. Espino, M.D., Ph.D., Thierry Fandeur, Ph.D., Maria-Fatima Ferreira-da-Cruz, Ph.D., Abebe A. Fola, M.Sc., Hans-Peter Fuehrer, Ph.D., Abdillahi M. Hassan, B.Sc., Socrates Herrera, M.D., Bouasy Hongvanthong, M.D., Sandrine Houzé, M.D., Ph.D., Maman L. Ibrahim, M.V.D., Ph.D., Mohammad Jahirul-Karim, M.B., B.S., Lubin Jiang, Ph.D., Shigeyuki Kano, M.D., Ph.D., Wasif Ali-Khan, M.B., B.S., Maniphone Khanthavong, M.D., Peter G. Kremsner, M.D., Marcus Lacerda, M.D., Ph.D., Rithea Leang, M.D., Mindy Leelawong, Ph.D., Mei Li, Ph.D., Khin Lin, M.D., Jean-Baptiste Mazarati, Ph.D., Sandie Ménard, M.Sc., Isabelle Morlais, Ph.D., Hypolite Muhindo-Mavoko, M.D., Lise Musset, Pharm.D., Ph.D., Kesara Na-Bangchang, Ph.D., Michael Nambozi, M.P.H., Karamoko Niaré, Pharm.D., Harald Noedl, M.D., Ph.D., Jean-Bosco Ouédraogo, M.D., Ph.D., Dylan R. Pillai, M.D., Ph.D., Bruno Pradines, Pharm.D., Ph.D., Bui Quang-Phuc, M.D., Michael Ramharter, M.D., D.T.M.H., Milijaona Randrianarivelojosia, Ph.D., Jetsumon Sattabongkot, Ph.D., Abdiqani Sheikh-Omar, M.D., Kigbafori D. Silué, Ph.D., Sodiomon B. Sirima, M.D., Ph.D., Colin Sutherland, Ph.D., M.P.H., Din Syafruddin, M.D., Ph.D., Rachida Tahar, Ph.D., Lin-Hua Tang, M.D., Ph.D., Offianan A. Touré, Ph.D., Patrick Tshibangu-wa-Tshibangu, M.D., Inès Vigan-Womas, Ph.D., Marian Warsame, M.D., Ph.D., Lyndes Wini, M.B., B.S., Sedigheh Zakeri, Ph.D., Saorin Kim, B.S., Rotha Eam, B.S., Laura Berne, M.Sc., Chanra Khean, B.S., Sophy Chy, B.S., Malen Ken, B.S., Kaknika Loch, B.S., Lydie Canier, M.Sc., Valentine Duru, M.Sc., Eric Legrand, Ph.D., Jean-Christophe Barale, Ph.D., Barbara Stokes, B.Sc., Judith Straimer, Ph.D., Benoit Witkowski, Ph.D., David A. Fidock, Ph.D., Christophe Rogier, M.D., Ph.D., Pascal Ringwald, M.D., Frederic Ariey, M.D., Ph.D., and Odile Mercereau-Puijalon, Ph.D.
Footnotes
The authors’ full names and academic degrees are listed in the Appendix. The authors’ affiliations are listed in the Supplementary Appendix, available at NEJM.org.
The views expressed in this article are those of the authors and do not necessarily reflect the official policies of the World Health Organization, the Department of the Navy, the Department of Defense, or the U.S. Government.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.World malaria report 2014. Geneva: World Health Organization; 2014. ( http://www.who.int/malaria/publications/world_malaria_report_2014/en/) [Google Scholar]
- 2.Bhatt S, Weiss DJ, Cameron E, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–11. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Status report on artemisinin and ACT resistance. Geneva: World Health Organization; Sep, 2015. ( http://www.who.int/malaria/publications/atoz/status-repartemisinin-resistance-sept2015.pdf) [Google Scholar]
- 4.Guidelines for the treatment of malaria. 3. Geneva: World Health Organization; Apr, 2015. ( http://www.who.int/malaria/publications/atoz/9789241549127/en/) [PubMed] [Google Scholar]
- 5.Witkowski B, Amaratunga C, Khim N, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–9. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witkowski B, Khim N, Chim P, et al. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother. 2013;57:914–23. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denis MB, Tsuyuoka R, Lim P, et al. Efficacy of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in northwest Cambodia. Trop Med Int Health. 2006;11:1800–7. doi: 10.1111/j.1365-3156.2006.01739.x. [DOI] [PubMed] [Google Scholar]
- 8.Denis MB, Tsuyuoka R, Poravuth Y, et al. Surveillance of the efficacy of artesunate and mefloquine combination for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2006;11:1360–6. doi: 10.1111/j.1365-3156.2006.01690.x. [DOI] [PubMed] [Google Scholar]
- 9.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 11.Amaratunga C, Sreng S, Suon S, et al. Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: a parasite clearance rate study. Lancet Infect Dis. 2012;12:851–8. doi: 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashley EA, Dhorda M, Fairhurst RM, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–23. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hien TT, Thuy-Nhien NT, Phu NH, et al. In vivo susceptibility of Plasmodium falciparum to artesunate in Binh Phuoc Province, Vietnam. Malar J. 2012;11:355. doi: 10.1186/1475-2875-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang F, Takala-Harrison S, Jacob CG, et al. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis. 2015;212:1629–35. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyaw MP, Nyunt MH, Chit K, et al. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One. 2013;8(3):e57689. doi: 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phyo AP, Nkhoma S, Stepniewska K, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–6. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thriemer K, Hong NV, Rosanas-Urgell A, et al. Delayed parasite clearance after treatment with dihydroartemisinin-piperaquine in Plasmodium falciparum malaria patients in central Vietnam. Antimicrob Agents Chemother. 2014;58:7049–55. doi: 10.1128/AAC.02746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 19.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–18. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 20.Trape JF. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg. 2001;64(Suppl):12–7. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- 21.Trape JF, Pison G, Spiegel A, Enel C, Rogier C. Combating malaria in Africa. Trends Parasitol. 2002;18:224–30. doi: 10.1016/s1471-4922(02)02249-3. [DOI] [PubMed] [Google Scholar]
- 22.Vinayak S, Alam MT, Mixson-Hayden T, et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 2010;6(3):e1000830. doi: 10.1371/journal.ppat.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Methods for surveillance of antimalarial drug efficacy. Geneva: World Health Organization; 2009. ( http://apps.who.int/iris/bitstream/10665/44048/1/9789241597531_eng.pdf) [Google Scholar]
- 24.Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straimer J, Gnädig NF, Witkowski B, et al. Drug resistance: K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–31. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amaratunga C, Witkowski B, Dek D, et al. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob Agents Chemother. 2014;58:4935–7. doi: 10.1128/AAC.03055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaratunga C, Witkowski B, Khim N, Menard D, Fairhurst RM. Artemisinin resistance in Plasmodium falciparum. Lancet Infect Dis. 2014;14:449–50. doi: 10.1016/S1473-3099(14)70777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takala-Harrison S, Jacob CG, Arze C, et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis. 2015;211:670–9. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miotto O, Amato R, Ashley EA, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–34. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter TE, Boulter A, Existe A, et al. Artemisinin resistance-associated polymorphisms at the K13-propeller locus are absent in Plasmodium falciparum isolates from Haiti. Am J Trop Med Hyg. 2015;92:552–4. doi: 10.4269/ajtmh.14-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra N, Prajapati SK, Kaitholia K, et al. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob Agents Chemother. 2015;59:2548–53. doi: 10.1128/AAC.04632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang F, Tang L, Yang H, et al. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J. 2012;11:243. doi: 10.1186/1475-2875-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng J, Zhou D, Lin Y, Xiao H, Yan H, Xia Z. Amplification of pfmdr1, pfcrt, pvmdr1, and K13 propeller polymorphisms associated with Plasmodium falciparum and Plasmodium vivax isolates from the China-Myanmar border. Antimicrob Agents Chemother. 2015;59:2554–9. doi: 10.1128/AAC.04843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohon AN, Alam MS, Bayih AG, et al. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013) Malar J. 2014;13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conrad MD, Bigira V, Kapisi J, et al. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One. 2014;9(8):e105690. doi: 10.1371/journal.pone.0105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper RA, Conrad MD, Watson QD, et al. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother. 2015;59:5061–4. doi: 10.1128/AAC.00921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isozumi R, Uemura H, Kimata I, et al. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis. 2015;21:490–2. doi: 10.3201/eid2103.140898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamau E, Campino S, Amenga-Etego L, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2015;211:1352–5. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouattara A, Kone A, Adams M, et al. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg. 2015;92:1202–6. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor SM, Parobek CM, DeConti DK, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:680–8. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torrentino-Madamet M, Fall B, Benoit N, et al. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J. 2014;13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borrmann S, Straimer J, Mwai L, et al. Genome-wide screen identifies new candidate genes associated with artemisinin susceptibility in Plasmodium falciparum in Kenya. Sci Rep. 2013;3:3318. doi: 10.1038/srep03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henriques G, Hallett RL, Beshir KB, et al. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis. 2014;210:2001–8. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkes M, Conroy AL, Opoka RO, et al. Slow clearance of Plasmodium falciparum in severe pediatric malaria, Uganda, 2011–2013. Emerg Infect Dis. 2015;21:1237–9. doi: 10.3201/eid2107.150213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menard D, Ariey F. Towards real-time monitoring of artemisinin resistance. Lancet Infect Dis. 2015;15:367–8. doi: 10.1016/S1473-3099(15)70046-0. [DOI] [PubMed] [Google Scholar]
- 46.Roper C, Alifrangis M, Ariey F, et al. Molecular surveillance for artemisinin resistance in Africa. Lancet Infect Dis. 2014;14:668–70. doi: 10.1016/S1473-3099(14)70826-6. [DOI] [PubMed] [Google Scholar]
- 47.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76:5269–73. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 49.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 50.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 51.Cheeseman IH, McDew-White M, Phyo AP, Sriprawat K, Nosten F, Anderson TJ. Pooled sequencing and rare variant association tests for identifying the determinants of emerging drug resistance in malaria parasites. Mol Biol Evol. 2015;32:1080–90. doi: 10.1093/molbev/msu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nyunt MH, Hlaing T, Oo HW, et al. Molecular assessment of artemisinin resistance markers, polymorphisms in the k13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis. 2015;60:1208–15. doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, Shrestha S, Li X, et al. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007–2012. Malar J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tun KM, Imwong M, Lwin KM, et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–21. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talundzic E, Okoth SA, Congpuong K, et al. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11(4):e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng J, Li J, Yan H, Feng X, Xia Z. Evaluation of antimalarial resistance marker polymorphism in returned migrant workers in China. Antimicrob Agents Chemother. 2015;59:326–30. doi: 10.1128/AAC.04144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MalariaGen. Oxford, United Kingdom: MalariaGen; 2015. P. falciparum Community Project data (beta release) ( https://www.malariagen.net/projects/parasite/pf) [Google Scholar]
- 58.Escobar C, Pateira S, Lobo E, et al. Polymorphisms in Plasmodium falciparum K13-propeller in Angola and Mozambique after the introduction of the ACTs. PLoS One. 2015;10(3):e0119215. doi: 10.1371/journal.pone.0119215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miotto O, Almagro-Garcia J, Manske M, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45:648–55. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopera-Mesa TM, Doumbia S, Chiang S, et al. Plasmodium falciparum clearance rates in response to artesunate in Malian children with malaria: effect of acquired immunity. J Infect Dis. 2013;207:1655–63. doi: 10.1093/infdis/jit082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.WWARN Artemisinin based Combination Therapy (ACT) Africa Baseline Study Group. Clinical determinants of early parasitological response to ACTs in African patients with uncomplicated falciparum malaria: a literature review and meta-analysis of individual patient data. BMC Med. 2015;13:212. doi: 10.1186/s12916-015-0445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mbengue A, Bhattacharjee S, Pandharkar T, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–7. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.