Abstract
In a decade with over half a billion dollars of investment, more than 300 chemical probes have been identified to have biological activity through NIH funded screening efforts. We have collected the evaluations of an experienced medicinal chemist on the likely chemistry quality of these probes based on a number of criteria including literature related to the probe and potential chemical reactivity. Over 20% of these probes were found to be undesirable. Analysis of the molecular properties of these compounds scored as desirable suggested higher pKa, molecular weight, heavy atom count and rotatable bond number. We were particularly interested whether the human evaluation aspect of medicinal chemistry due diligence could be computationally predicted. We used a process of sequential Bayesian model building and iterative testing as we included additional probes. Following external validation of these methods and comparing different machine learning methods we identified Bayesian models with accuracy comparable to other measures of drug-likeness and filtering rules created to date.
Keywords: Bayesian models, Chemical probes, Function class fingerprints
INTRODUCTION
In the past decade the National Institutes of Health (NIH) has funded extensive high throughput screening (HTS) efforts in both intra-mural and academic centers to identify small molecule chemical probes or tool compounds via the Molecular Libraries Screening Center Network (MLSCN) and the Molecular Library Probe Production Center Network (MLPCN). By 2009 it was estimated to have cost $385 million1 and by 2010 $576.6 million2 but since then funding was dramatically scaled back3. Various definitions for compounds to become probes based on a combination of potency, selectivity, solubility, and availability, have been used1. To date the NIH-funded academic screening centers have discovered just over 300 chemical probes (at the time of writing) and some of the groups have demonstrated sophisticated drug discovery capabilities4. The NIH has subsequently made publically accessible an extensive compilation of chemical and biology data on these probes. From medicinal and computational chemistry and biology perspectives, we thought this an opportune time to perform a subjective analysis in order to learn from this massive effort. We posit that medicinal chemistry and computational considerations are relevant and complementary to biological activity considerations. Presenting our probe analysis in the context of medicinal chemistry due diligence is timely given the emerging understanding of the relationship of chemistry ligand structure to biology target topology5–7. Moreover we used our analysis to test whether it was possible to computationally predict the evaluations and eventual decisions of an experienced medicinal chemist.
In the same period that these probes were generated, there have been extensive assessments of medicinal chemists' appraisal of drug- or lead-likeness. For example, Lajiness et al., have evaluated the ability of medicinal chemists to assess the drug- or lead-likeness of molecules8. Thirteen medicinal chemists assessed approximately 22,000 compounds, broken into 11 lists of approximately 2000 compounds each. It was found that they were not very consistent in the compounds they rejected as being undesirable8. Cheshire described how modern medicinal chemists are often “over-productive” synthesizing many more compounds than are required to achieve the objectives of the project9. In contrast Hack et al., used the wisdom of crowds to fill holes in a screening library by leveraging 145 global medicinal chemists at J&J10. A recent study by Kutchukian et al., investigated medicinal chemists behavior using surveys in which they selected chemical fragments for development into a lead compound from a set of ~4,000 available fragments11. Computational Bayesian Classifiers were also built for each chemist to model their selection strategy. These models were not used to prospectively predict compounds. The results suggested the chemists greatly simplified the problem, using only 1–2 of many possible parameters. Overall there was a lack of consensus in compound selections. Cumming et al., have proposed that the `quality' of small-molecule drug candidates, are under the control of chemists during the identification and optimization of lead compounds12. The fusion of all of these studies suggests that decision making by the medicinal chemist while variable, is still widely regarded and critical to drug discovery. However, since the development of the Rule of 513 there has been an apparent focus on similar rules to filter undesirable compounds and influence synthetic decisions.
Previously others have suggested that marketed drugs contain a high percentage of such undesirable groups (277 out of 1070 compounds in one study)14. Filters or rules are widely used by pharmaceutical companies to flag molecules that may be false positives and frequent hitters from HTS screening libraries as well as select compounds from commercial vendors15. Some examples of widely used substructure filters include REOS from Vertex16, as well as filters from GlaxoSmithKline17, BristolMyersSquibb18 and Abbott19–21. These pick up a range of chemical substructures such as thiol traps and redox-active compounds, epoxides, anhydrides, and Michael acceptors. Another group has developed a series of over 400 substructural features for removal of Pan Assay INterference compoundS (PAINS) from screening libraries that is likely considered a definitive rule set22. Bruns and Watson have also described a set of 275 rules they developed at Eli Lilly over an 18-year period, that were used to identify compounds that may interfere with biological assays23. The structural queries were profiled for frequency of occurrence in drug-like and non-drug-like compound sets and were extensively reviewed by a panel of experienced medicinal chemists. Drug-likeness itself has been suggested to not reflect adequately compound quality and rules may lead to undesirable molecular property inflation. Bickerton et al., proposed a measure of drug-likeness based on the concept of desirability called the quantitative estimate of drug-likeness (QED)24. A recently developed computational method termed Bioactivity Data Associative Promiscuity pattern Learning Engine (BadApple) uses public data to predict promiscuity based on comparison of scaffolds25, 26. Finally, a retrospective analysis has shown that recently approved oral drugs are highly optimized for ligand efficiency, a metric that integrates binding affinity with molecular properties27.
The only critical evaluation to date of the NIH probes, was a study in 2009 that took a crowdsourcing approach to evaluation of 64 NIH chemical probes1. Eleven experts subjectively scored the probes and found 48 of 64 (75%) probes were of medium or high confidence. We proposed we could use this type of expert-derived classification data to learn what had been classed as desirable and then using this prospectively to help score additional molecules computationally. One of the chemists involved in the 2009 crowdsourcing study was enlisted to test the approach of whether an algorithm could learn from his selections. We have taken the approach of using several machine learning methods and have focused on Naïve Bayesian classification which has been used for modeling in vitro and in vivo data28 by us and many others29. In addition we have compared the results of this effort with PAINS22, QED24, BadApple25 and ligand efficiency27, 30.
Experimental
Dataset
With just a few exceptions NIH probe compounds were identified from NIH's Pubchem web based book25 summarizing five years of probe discovery efforts. Probes are identified by ML number and by PubChem CID number. NIH probe compounds were compiled using the NIH PubChem Compound Identifier (CID) as the defining field for associating chemical structure. For chiral compounds, two dimensional depictions were searched in CAS SciFinder™ (CAS, Columbus OH) and associated references were used to define the intended structure.
A medicinal chemist,with more than 40 years of experience (C.A.L.) followed a consistent protocol for determining if compounds should be considered undesirable or desirable. The number of biological literature references associated with each compound was determined. Probes with more than 150 references to biological activity were considered unlikely to be selective despite any PubChem HTS assay data. Alternatively, probes with zero references were considered to have uncertain biological quality if the probe was not of recent vintage. The idea is that if a probe report is at least several years old and if neither the probe originator nor anyone else has published on the probe then one might conclude there was some sort of problem. The idea that the presence of a given probe in a US patent application31 across many probes might be a pointer to promiscuous activity was shown on detailed examination to be incorrect. CAS SciFinder™ was used to determine the CAS RegNo. Probes with no CAS RegNo were considered problematic if associated with chemistry generally unexplored in drugs. Finally, probes with predicted chemical reactivity were considered to lead to uncertainty about the cause of the biological effect. Judgment was used for this criterion. Only the most flagrant offenders were flagged and then only with caution often in the face of considerable HTS experimental data. The reactivity criterion is likely the “softest” since this is a criteria where different experts clearly often disagree. Probes that met any of these criteria were considered undesirable and given a score of 0. A summary of the percentage of undesirable compounds that fell within each criterion is shown in Fig. 1. All other probes were given a score of 1 for desirable. These are binary choices with no degree of gradation and thus carry the biases inherent of any binary yes no methodology32 The data and molecular structures have been made publically available in the CDD Public database (Collaborative Drug Discovery Inc. Burlingame, CA)33.
Figure 1.
Relative contribution of each criteria for considering compounds as undersirable.
Molecular properties and filtering methods
Salts were removed from molecules prior to calculation of molecular properties and computational modeling. One compound that was a complex, ML134, was also removed from analysis. Several descriptors were calculated in the CDD database using the Marvin suite (ChemAxon, Budapest, Hungary) namely: molecular weight, logP, H bond donors, H bond acceptors, Lipinski score, pKa, heavy atom count, polar surface area, rotatable bond number. The JChem suite (ChemAxon) was also used to generate two values for pKa namely first the average charge at pH 7.4, where negative values are acidic, positive basic, and close to zero neutral, and second the distribution of the major microspecies.
Similarly descriptors were calculated including AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area from input SD files using Discovery Studio 3.5 (Biovia, San Diego, CA).
BadApple learns from “frequent hitters” found in the Molecular Libraries Screening program, and flags promiscuous compounds. This method was used to score the compounds in this study at the website25,26.
PAINS is a set of filters determined by identifying compounds that were frequent hitters in numerous high throughput screens. PAINS filters were determined using the FAFDrugs2 program34, 35.
The desirability of the NIH chemical probes was also compared with QED24 which was calculated using open source software from SilicosIt (Schilde, Belgium).
For determination of ligand efficiency, the IC50, AC50, and EC50 values associated with each chemical probe were accessed from PubChem using a java script. The calculations function in CDD Vault software was used to determine ligand efficiency with the formula LE= 1.4*−log(EC50 or IC50 or AC50)/(Heavy atom count).30, 36
Analysis of the 307 compounds was performed with Principal Component Analysis (PCA) using Discovery Studio was generated with the interpretable descriptors chosen previously (AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area). The desirable and undesirable compounds were also treated as unique “libraries” that were also compared through the “compare libraries” protocol in Discovery Studio via the use of assemblies (Murcko Assemblies)37.
Machine learning models
We have previously described the generation and validation of the Laplacian-corrected Bayesian classifier models developed for various datasets using Discovery Studio 3.528. This approach was utilized with the literature probe data from PubChem. A set of simple molecular descriptors were used: molecular function class fingerprints of maximum diameter 6 (FCFP_6)38, AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area were calculated from input SD files. Models were validated using leave-one-out cross-validation in which each sample was left out one at a time, a model was built using the remaining samples, and that model utilized to predict the left-out sample. Each of the models was internally validated, receiver operator (ROC) plots were generated, and the cross validated (XV) ROC area under the curve (AUC) calculated. The Bayesian model was additionally evaluated by performing 5 fold cross validation in which 20% of the data set is left out 5 times. Additionally leaving out 50% of the data and rebuilding the model 100 times using a custom protocol was used for validation, to generate the ROC AUC, concordance, specificity and selectivity as described previously39, 40. The Internal ROC value represents the training set value while the external ROC represents the test set molecules left out. For the largest model created we also compared the Bayesian model with SVM and RP Forest and single tree models built with the same molecular descriptors. For SVM models we calculated interpretable descriptors in Discovery Studio then used Pipeline Pilot to generate the FCFP_6 descriptors followed by integration with R41. RP Forest and RP Single Tree models used the standard protocol in Discovery Studio. In the case of RP Forest models ten trees were created with bagging. RP Single Trees had a minimum of ten samples per node and a maximum tree depth of 20. In all cases, 5-fold cross validation (leave out 20% of the database 5 times) was used to calculate the ROC for the models generated and for comparison.
Model predictions for additional compounds identified after model building
After each model was built additional compounds were identified and these were used as an external test set for prospective analysis. These compounds were then later combined to enable model rebuilding. This approach was repeated 3 times. Finally a further 15 compounds initially not scored were identified and these were used as a test set for all models created. For each molecule, the closest distance to the training set was also calculated (a value of zero represents a molecule in the training set).
Statistical analysis
The mean calculated molecular property values for compounds were compared using two tailed t-test with JMP v. 8.0.1 (SAS Institute, Cary, NC). For external test set prediction evaluation, the number of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) were determined and used to generate ROC plots and determine accuracy ((TP+TN)/total), specificity (TN/(TN+FP)), sensitivity (TP/(TP+FN), and precision (TP/(TP+FP) using the standard indicated formula. Mean ligand efficiency, was assessed using a student's t-test, and PAINS and BadApple predicted scores were assessed using Fisher's exact test.
RESULTS
Determining NIH chemical probe quality
For a medicinal chemist to accurately assess the quality of probes identified through HTS, it is necessary to assess previous findings in the literature and in patent applications. Unfortunately, this type of due diligence is not easily performed and requires labor intensive mixing of publically available data and the use of commercial software. In the process of this study we have noted a number of areas where optimizing integration of this information would benefit all participants in the drug discovery community and these will be detailed elsewhere. Eventually, it should be possible to integrate and automate this type of due diligence analysis. With just a few exceptions (ML032, ML049 and ML287) NIH probe compounds were identified from NIH's PubChem web based book. ML032, ML049, ML287 are identified in a spreadsheet available on the MLP website42. ML049 is associated with a probe report “Fluorescent Cross-Reactive FPR/FPRL1 Hexapeptide Ligand”. Neither ML032 and ML287 is associated with probe reports as listed on the MLP website42.
We have noticed that in terms of selectivity, the probes fall into two main categories: 1) the probe is selective in a drug discovery sense, i.e. one or two orders of magnitude more selective for the target versus all salient anti-targets 2) the probe is 3 to 10 times more selective for the target than for any single anti-target, is novel for the target and the breadth of activity against all anti-targets is experimentally very well characterized. Probes in this second category are in the majority.
Molecular properties
The 307 molecules in the complete dataset (excluding the final 15 molecule test set) used for the largest model consisted of 240 scored as desirable and 67 as undesirable. Using 9 descriptors from the CDD Vault software, the molecular weight, rotatable bond number and heavy atom count were all statistically significantly larger in the desirable compound set (Table 1). When we focused on just the acidic or basic compounds we found a statistically significant higher pKa value for acidic compounds only (Table 1). Using 8 partially overlapping descriptors calculated in Discovery Studio the number of molecular weight and rotatable bonds was also greater in desirable compounds while the molecular fractional polar surface area was lower (Table 2). Other descriptors showed no statistically significant difference between groups. Diversity analysis of the desirable and undesirable compounds using FCFP_6 fingerprints, Murcko assemblies, Tanimoto similarity, and default properties indicated few differences apart from undesirable molecules displaying a higher diversity of Fingerprint features (Table S1). Library analysis using Murcko Assemblies showed little similarity (Table S2).
Table 1.
Mean ± SD molecular properties calculated in CDD Vault using ChemAxon software for the 307 molecules used in model building,
| Molecular weight | LogP | H-bond donors | H-bond acceptors | Lipinski score | pKa (6 cpds missing) | Heavy atom count | Polar Surface Area | Rotatable bond number | |
|---|---|---|---|---|---|---|---|---|---|
| Undesirable | 359.87 ± 83.26 | 3.43 ± 1.27 | 1.07 ± 1.13 | 4.14 ± 1.52 | 0.18 ± 0.39 | 5.14 ± 5.13 | 24.70 ± 5.69 | 73.03 ± 29.3 | 4.20 ± 2.25 |
| Desirable | 383.53 ± 87.17* | 3.43 ± 1.36 | 1.23 ± 1.01 | 4.11 ± 1.61 | 0.21 ± 0.49 | 6.52 ± 4.94 | 26.92 ± 6.15* | 72.61 ± 28.10 | 4.92 ± 2.40* |
statistically significant p <0.05 two sided t-test.
For acidic compounds (n = 142) the mean pKa was 8.12 ± 3.95 for undesirable and 9.71 ± 3.84 for desirable compounds (p <0.05 two sided t-test). For basic compounds (n = 160) the mean pKa was 2.25 ± 4.47 for undesirable and 3.75 ± 4.04 for desirable compounds,
Table 2.
Mean ± SD molecular properties calculated in with Discovery Studio 3.5 software. ± SD for the 307 molecules used in model building,
| Molecular weight | ALogP | Number of Rings | Number Aromatic rings | Number H-bonds donors | Number H-bond acceptors | Molecular fractional polar surface area | Number rotatable bonds | |
|---|---|---|---|---|---|---|---|---|
| Undesirable | 359.87 ± 83.26 | 3.55 ± 1.31 | 3.29 ± 1.00 | 2.35± 1.06 | 1.07 ± 1.13 | 4.77 ± 1.75 | 0.28 ± 0.11 | 4.30 ± 2.31 |
| Desirable | 383.53 ± 87.16* | 3.50 ± 1.33 | 3.47 ± 1.02 | 2.55 ± 0.98 | 1.23 ± 1.00 | 4.36 ± 1.63 | 0.23 ± 0.08* | 5.13 ± 2.41 * |
statistically significant p <0.05 two sided t-test.
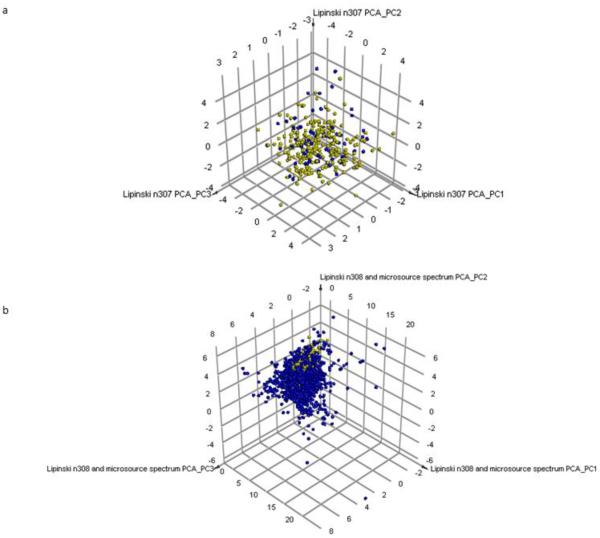
PCA analysis showed the desirable and non-desirable molecules overlapped based on these molecular properties (Fig. 2A) and that these were within the more diverse chemical property space of the Microsource spectrum commercial library of drugs and natural products (Fig. 2B). Library analysis using Murcko Assemblies showed higher similarity to the Microsource library than between the NIH undesirable and undesirable compounds. In contrast the use of a global fingerprint suggested the NIH probes and Microsource libraries were less similar than between the NIH desirable and undesirable compounds (Table S2).
Figure 2. Visualizing chemical property space of desirable and undesirable probes.
A. Principal component analysis (PCA) of the 307 desirable (yellow) and undesirable (blue) NIH probes. 76.6% of the variance was explained with three principal components. B. PCA of the NIH chemical probes (yellow) and 2320 Microsource spectrum compounds (blue). 81.9% of variance was explained by 3 principal components.
The desirable/ and undesirable scores were compared to several other rules and tools used to predict drug-likeness. None of the molecular probes characterized had more than 2 violations of the Rule of Five12, and we found no statistical difference between the desirable and undesirable molecules13. BadApple revealed that the desirable compounds are less likely to be promiscuous, predicting 12% of desirable and 33% of undesirable probes fall into this category (Fisher's exact test, p=0.04). In total, 34 of 322 probes were flagged by the PAINS filters, representing 6.7% of desirable and 25% of undesirable compounds (Fisher's exact test, p>0.0001) (Fig. 3). There was no significant difference in the QED. The mean ligand efficiency of both desirable and undesirable probes was in the “ideal” range defined as greater than 0.3 kcal per mole heavy atom30 (desirable = 0.308 vs. undesirable =0.327, p= 0.000003). All the various scores were used to create a heatmap for ease of comparison (Fig. 4).
Figure 3. Comparison of the expert's evaluation of the NIH chemical probes with PAINS and BadApple Filters.
Desirable NIH chemical probes are less likely to be filtered by PAINS or BadApple as promiscuous than those scored as undesirable. (Fisher's exact test, p>0.0001 for PAINS and p=0.04 for BadApple).
Figure 4. Comparison of desirability scores with Bayesian learning predicted scores for each test set, QED, BadApple, and ligand efficiency metrics.

The colors on the heat map correspond to the value of the indicated metric for each probe, listed vertically. The scale was normalized internally with green corresponding to the optimal condition within each metric.
Bayesian Model 1
Using 57 molecules from the original dataset from 2009. The model had a Receiver Operator Characteristic (ROC) value for leave one out (optimistic) of 0.654, 5 fold cross validation ROC = 0.613 (Table S3) and leave out 50% × 100 ROC of 0.63 (Table 3). The top twenty substructure descriptors consistent with compounds classed as desirable probes contain well known drug like substructures like thiazole, pyridine, and morpholine fragments43, 44 to name just a few representative examples (Fig. S1). The top 20 features in undesirable probes include seleno-organic, compounds prone to facile oxidation such as phenol rich aromatic rings, aniline-rich functionality, and thio-ethers45, 46 (Fig. S2).
Table 3.
Leave out 50% × 100 cross validation analysis of Bayesian models of an expert's evaluation of the NIH chemical probes.
| Dataset size | External ROC Score | Internal ROC Score | Concordance | Specificity | Sensitivity |
|---|---|---|---|---|---|
| N57 | 0.63 ± 0.08 | 0.65 ± 0.14 | 54.30 ± 10.03 | 62.04 ± 29.46 | 51.40 ± 21.98 |
| N170 | 0.59 ± 0.05 | 0.69 ± 0.07 | 53.54 ± 7.62 | 59.30 ± 19.65 | 51.31 ± 17.48 |
| N191 | 0.60 ± 0.05 | 0.69 ± 0.07 | 55.06 ± 8.00 | 57.83 ± 17.81 | 54.10 ± 16.98 |
| N307 | 0.69 ± 0.04 | 0.75 ±0.05 | 64.57 ± 8.06 | 61.89 ± 14.83 | 65.25 ± 13.02 |
ROC = receiver operator characteristic.
Bayesian Model 2
Using 170 molecules the model had an ROC value for leave one out of 0.710, 5 fold cross validation ROC = 0.654 (Table S4) and leave out 50% × 100 ROC of 0.59 (Table 1). In this model, the top twenty substructure descriptors consistent with compounds classed as desirable probes contain drug-like heterocycles, biomimetic amino acid analogs, and charged functionality likely to help with compound solubility at physiologically relevant pH levels (Fig. S3), the top 20 features in undesirable probes include highly conjugated systems with potential Michael acceptors, relatively unstable hydrozones, and easily oxidized heterocycles like the furan derivatives (Fig. S4).
Bayesian Model 3
Using 191 molecules the model had an ROC value for leave one out of 0.707, 5 fold cross validation ROC = 0.671 (Table S5) and leave out 50% × 100 ROC of 0.60. The top twenty substructure descriptors consistent with compounds classed as desirable probes contain drug-like features such as Nitrogen-rich heterocycles, biomimetic amino acid analogs, thioamides, and charged functionality (Fig. S5). The top 20 features in undesirable probes include relatively unstable hydrazones, seleno compounds, heterocycles known to oxidize and a variety of potential electrophiles (Fig. S6).
Bayesian Model 4
Using 307 molecules the model had an ROC value for leave one out of 0.796, 5 fold cross validation ROC = 0.735 (Table S6) and leave out 50% × 100 ROC of 0.69. The top twenty substructure descriptors consistent with compounds classed as desirables probes contain drug-like heterocycles, aromatic fragments with electron withdrawing functionality, and charged functionality (Fig. S7), and the top 20 features in undesirable probes include electron rich ring systems prone to facile oxidation, compounds with adjacent functionality likely to increase functionality (such as two adjacent carbonyl groups or a variety of Michael acceptors), and potentially unstable ring systems (Fig. S8).
Differences are observed between 5 fold cross validation (Tables S1–4) and leave out 50% × 100 cross validation (Table 3). The N307 model has statistics that are slightly higher than the 191 molecule model.
Comparison of machine learning methods
For the n307 dataset a Forest model, a single tree model and an SVM model were all created to compare with the Bayesian model. The 5 fold cross validation ROC values were highest for the Bayesian model followed by the SVM (Table 4).
Table 4.
5 fold cross validation results for n307 Bayesian model of an expert's evaluation of the NIH chemical probes.
| Algorithm | Receiver Operator Characteristic |
|---|---|
| Bayesian | 0.735 |
| Support Vector Machine | 0.638 |
| Recursive partitioning – Single tree | 0.630 |
| Recursive partitioning – Forest | 0.607 |
Rebuilding the Bayesian model in secure CDD Vault with Models
Rebuilding the n307 Bayesian model in CDD Models software with just FCFP_6 fingerprints and using 3 fold cross validation ROC (0.69) suggested that it was comparable to the model developed previously which included additional molecular descriptors (Table 3, 4 Table S4).
External test set predictions
Table 5 summarizes model external testing as additional data were discovered. All four models were compared using the prediction of the final test set of 15 molecules. The largest model with 307 molecules has the best ROC AUC of 0.78 while the model built with 191 molecules comes a close second with an ROC AUC of 0.75. Comparison of the various models and their prediction of test set compounds was enabled via a heatmap (Table 3).
Table 5.
Test set assessment of Bayesian models of an expert's evaluation of the NIH chemical probes, a value of 1 = ideal. Each statistic is colored by the value, where blue is desirable and red is undesirable.
| Model | Test molecules | AUC | Sensitivity | Specificity | Precision | Accuracy |
|---|---|---|---|---|---|---|
| N57 | 114 | 0.58 | 0.7 | 0.42 | 0.75 | 0.62 |
| N57 | 120 | 0.55 | 0.69 | 0.42 | 0.91 | 0.66 |
| N57 | 15 | 0.72 | 0.67 | 0.33 | 0.8 | 0.6 |
| N170 | 21 | 0.55 | 0.38 | 0.6 | 0.75 | 0.43 |
| N170 | 120 | 0.78 | 0.62 | 0.67 | 0.94 | 0.63 |
| N170 | 15 | 0.58 | 0.67 | 0.33 | 0.8 | 0.6 |
| N191 | 120 | 0.8 | 0.67 | 0.75 | 0.96 | 0.68 |
| N191 | 15 | 0.75 | 0.83 | 0.33 | 0.83 | 0.73 |
| N307 | 15 | 0.78 | 0.75 | 0.33 | 0.82 | 0.67 |
We also compared the ability of other tools for predicting the medicinal chemist's desirability scores for the same set of 15 compounds. We found neither the QED, BadApple, or ligand efficiency metrics to be as predictive with ROC AUC of 0.58, 0.36, and 0.29 respectively. Therefore these drug likeness methods do not agree with the medicinal chemist's desirability scores.
DISCUSSION
This work was directly influenced by the earlier study scoring 64 NIH chemical probes by 11 experts1. We hypothesized that we could learn from one of these medicinal chemistry experts (C.A.L.) and use the resultant computational models to predict newer chemical probe scores. While our aim is not to replace the medicinal chemist, we have shown in this study that a machine learning algorithm has the potential to learn from them, and could be used to filter compounds for assessment either alone or alongside other approaches such as rules or filters recently described24, 25,27, 30. Interestingly, ligand efficiency was higher in compounds scored as undesirable, this would echo recent suggestions that ligand efficiency is not a replacement for considering in vitro and in vivo properties of molecules47. Therefore using methods that can learn from the medicinal chemist expert's decisions based on this data may be advantageous.
For accessing and evaluating the public chemical probe data, we followed a standardized process for obtaining prior references. This process had severe limitations caused by a mismatch of privately (commercial) and publicly accessible data, and navigating between these two spheres was problematic which we will discuss elsewhere in due course. In addition, lack of information due to publication bias is another hurdle in medicinal chemistry due diligence. Much of the process of due diligence relies on “soft” skills – such as appropriately combining the literature and making subjective determinations. These aspects of decision making likely will never go away but arguably it would be an advantage if the “soft” aspects of a medicinal chemist's choices could be captured computationally. Much of chemical biology focuses on discovery of tools and probes and sometimes in an environment with considerably more biology than chemistry expertise. We posit that the medicinal chemistry aspect in tool and probe discovery is important and that learning from the thorough documentation of a set of probes that have gone through this process could encode these medicinal chemistry “insights” in an algorithm.
What may have been an early probe learning process could have colored a 2009 published probe assessment1 in the sense that the earlier assessment included compounds with more dubious credentials that would have not been chosen in more recent probe reports (in 2009, for 64 compounds 75% were acceptable, for 307 compounds in this study 78% were acceptable). Surprising to us was the probe use of trifluoroacetate (TFA) salts given the known deleterious effects of TFA on long term cell culture48, the frequent contamination of TFA salts with excess TFA49 and the known biological activity of TFA per se50.
A cheminformatics analysis of the data collected showed that high confidence probe compounds have statistically higher numbers of rotatable bonds, more heavy atoms and a higher molecular weight (Table 1). These and other readily interpretable calculated properties were used as descriptors alongside FCFP_6 fingerprints to develop machine learning models. These models were both internally and externally validated and went through a prospective iterative learning process as we uncovered data on more probes (Table 3–5) which were scored by the medicinal chemist. The results of this study indicate that Naïve Bayesian models represent a potential way to surmount the issues with bridging the public and commercial data sources required to perform the chemical probe due diligence process. They also perform similarly to recently developed heuristics and may be complimentary. Such models may serve as selection criteria for compounds for screening in order to identify future probes that would be acceptable to medicinal chemists.
In a decade and after over half a billion dollars of investment, over 300 chemical probes have come out of NIH funded screening efforts to date. A thorough due diligence process by an experienced medicinal chemist with over 40 years of experience, suggests that out of a total of 322 probes 79% are considered to have desirable qualities. The outputs from this time- consuming process are a moving target but can be effectively used to build algorithms that learn from the data. A comparison versus other molecule quality metrics or filters such as QED, PAINS, BadApple and ligand efficiency indicates that a Bayesian model based on a single medicinal chemist's decisions for a small set of probes not surprisingly can make decisions that are preferable in classifying desirable compounds (based on the expert's a priori definition of desirability). The use of such machine learning methods may be an approach to increase the probability that a chemical probe would be considered useful by a medicinal chemist, and avoid much wasted time and money invested in poor compounds. Our integration of a Bayesian model based on open source FCFP_6 descriptors alone and the 307 NIH chemical probes in the CDD database (Fig. S9) suggests that it could be used readily to score vendor libraries before screening as well as evaluate future chemical probes prior to the extensive due diligence process being performed with commercial tools. This set of NIH chemical probes could also be scored by other in-house medicinal chemistry experts to come up with a customized score that in turn could be used to tailor the algorithm to their own preferences. For example this could be tailored towards CNS or anticancer compounds.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge colleagues at CDD for the development of the CDD Vault, Dr. Alex Clark for assistance and Dr. Jeremy Yang (University of New Mexico) for providing the link to BADAPPLE. We acknowledge that the Bayesian model software within CDD was developed with support from Award Number 9R44TR000942-02 “Biocomputation across distributed private datasets to enhance drug discovery” from the NCATS. SE gratefully acknowledges Biovia (formerly Accelrys) for providing Discovery Studio.
Abbreviations Used
- (AUC)
Area under the curve
- (BadApple)
Bioactivity Data Associative Promiscuity pattern Learning Engine
- (CDD)
Collaborative Drug Discovery
- (CID)
Compound Identifier
- (FCFP)
molecular function class fingerprints
- (MLSCN)
Molecular Libraries Screening Center Network
- (MLPCN)
Molecular Library Probe Production Center Network
- (PAINS)
Pan Assay INterference compoundS
- (PCA)
Principal component analysis
- (SVM)
Support vector machine
- (TFA)
Trifluoroacetate
- (QED)
Quantitative estimate of drug-likeness
- (XV)
cross validated
Footnotes
Supporting Information Available: Additional supplemental data is available in the online version of the paper. This material is available free of charge via the Internet at http://pubs.acs.org.
Data and materials availability: All computational models are available from the authors upon request. All molecules are available in CDD Public (https://app.collaborativedrug.com/register) and at http://molsync.com/demo/probes.php.
Author Contributions The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of Interest NL is an employee and SE is a consultant of CDD Inc. CL is on the scientific advisory board of CDD Inc. BAB is the CEO of CDD Inc.
REFERENCES
- 1.Oprea TI, Bologa CG, Boyer S, Curpan RF, Glen RC, Hopkins AL, Lipinski CA, Marshall GR, Martin YC, Ostopovici-Halip L, Rishton G, Ursu O, Vaz RJ, Waller C, Waldmann H, Sklar LA. A crowdsourcing evaluation of the NIH chemical probes. Nat Chem Biol. 2009;5:441–7. doi: 10.1038/nchembio0709-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy A, McDonald PR, Sittampalam S, Chaguturu R. Open Access High Throughput Drug Discovery in the Public Domain: A Mount Everest in the Making. Curr Pharm Biotechnol. 2010;11:764–778. doi: 10.2174/138920110792927757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser J. National Institutes of Health. Drug-screening program looking for a home. Science. 2011;334:299. doi: 10.1126/science.334.6054.299. [DOI] [PubMed] [Google Scholar]
- 4.Jarvis L. Anatomy of an academic drug discovery program. Chemistry & Engineering News. 2014 Jan 20;:28–29. [Google Scholar]
- 5.Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Lavan P, Weber E, Doak AK, Cote S, Shoichet BK, Urban L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–7. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KL, Edwards DD, Shoichet BK, Roth BL. Predicting new molecular targets for known drugs. Nature. 2009;462:175–81. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 8.Lajiness MS, Maggiora GM, Shanmugasundaram V. Assessment of the consistency of medicinal chemists in reviewing sets of compounds. J Med Chem. 2004;47:4891–6. doi: 10.1021/jm049740z. [DOI] [PubMed] [Google Scholar]
- 9.Cheshire DR. How well do medicinal chemists learn from experience? Drug Discov Today. 2011;16:817–21. doi: 10.1016/j.drudis.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Hack MD, Rassokhin DN, Buyck C, Seierstad M, Skalkin A, ten Holte P, Jones TK, Mirzadegan T, Agrafiotis DK. Library enhancement through the wisdom of crowds. J Chem Inf Model. 2012;51:3275–86. doi: 10.1021/ci200446y. [DOI] [PubMed] [Google Scholar]
- 11.Kutchukian PS, Vasilyeva NY, Xu J, Lindvall MK, Dillon MP, Glick M, Coley JD, Brooijmans N. Inside the mind of a medicinal chemist: the role of human bias in compound prioritization during drug discovery. PLoS One. 2012;7:e48476. doi: 10.1371/journal.pone.0048476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cumming JG, Davis AM, Muresan S, Haeberlein M, Chen H. Chemical predictive modelling to improve compound quality. Nat Rev Drug Discov. 2013;12:948–62. doi: 10.1038/nrd4128. [DOI] [PubMed] [Google Scholar]
- 13.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Del Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 14.Axerio-Cilies P, Castaneda IP, Mirza A, Reynisson J. Investigation of the incidence of “undesirable” molecular moieties for high-throughput screening compound libraries in marketed drug compounds. Eur J Med Chem. 2009;44:1128–34. doi: 10.1016/j.ejmech.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Williams AJ, Tkachenko V, Lipinski C, Tropsha A, Ekins S. Free Online Resources Enabling Crowdsourced Drug Discovery. Drug Discovery World. 2009;10:33–38. Winter. [Google Scholar]
- 16.Walters WP, Murcko MA. Prediction of `drug-likeness'. Adv Drug Del Rev. 2002;54:255–271. doi: 10.1016/s0169-409x(02)00003-0. [DOI] [PubMed] [Google Scholar]
- 17.Hann M, Hudson B, Lewell X, Lifely R, Miller L, Ramsden N. Strategic pooling of compounds for high-throughput screening. J Chem Inf Comput Sci. 1999;39:897–902. doi: 10.1021/ci990423o. [DOI] [PubMed] [Google Scholar]
- 18.Pearce BC, Sofia MJ, Good AC, Drexler DM, Stock DA. An empirical process for the design of high-throughput screening deck filters. J Chem Inf Model. 2006;46:1060–8. doi: 10.1021/ci050504m. [DOI] [PubMed] [Google Scholar]
- 19.Huth JR, Mendoza R, Olejniczak ET, Johnson RW, Cothron DA, Liu Y, Lerner CG, Chen J, Hajduk PJ. ALARM NMR: a rapid and robust experimental method to detect reactive false positives in biochemical screens. J Am Chem Soc. 2005;127:217–24. doi: 10.1021/ja0455547. [DOI] [PubMed] [Google Scholar]
- 20.Huth JR, Song D, Mendoza RR, Black-Schaefer CL, Mack JC, Dorwin SA, Ladror US, Severin JM, Walter KA, Bartley DM, Hajduk PJ. Toxicological evaluation of thiol-reactive compounds identified using a la assay to detect reactive molecules by nuclear magnetic resonance. Chem Res Toxicol. 2007;20:1752–9. doi: 10.1021/tx700319t. [DOI] [PubMed] [Google Scholar]
- 21.Metz JT, Huth JR, Hajduk PJ. Enhancement of chemical rules for predicting compound reactivity towards protein thiol groups. J Comput Aided Mol Des. 2007;21:139–44. doi: 10.1007/s10822-007-9109-z. [DOI] [PubMed] [Google Scholar]
- 22.Baell JB, Holloway GA. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 23.Bruns RF, Watson IA. Rules for identifying potentially reactive or promiscuous compounds. J Med Chem. 2012;55:9763–72. doi: 10.1021/jm301008n. [DOI] [PubMed] [Google Scholar]
- 24.Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. Quantifying the chemical beauty of drugs. Nat Chem. 2012;4:90–8. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anon Probe Reports from the NIH Molecular Libraries Program. http://www.ncbi.nlm.nih.gov/books/NBK47352/ [PubMed]
- 26.Yang JJ, Urso O, Bologna CG, Waller A, Sklar LA. Abstracts of papers of the American Chemical Society. ACS; Indianapolis: 2013. [Google Scholar]
- 27.Hopkins AL, Keseru GM, Leeson PD, Rees DC, Reynolds CH. The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov. 2014;13:105–21. doi: 10.1038/nrd4163. [DOI] [PubMed] [Google Scholar]
- 28.Ekins S, Pottorf R, Reynolds RC, Williams AJ, Clark AM, Freundlich JS. Looking Back To The Future: Predicting In vivo Efficacy of Small Molecules Versus Mycobacterium tuberculosis. J Chem Inf Model. 2014;54:1070–82. doi: 10.1021/ci500077v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bender A, Scheiber J, Glick M, Davies JW, Azzaoui K, Hamon J, Urban L, Whitebread S, Jenkins JL. Analysis of Pharmacology Data and the Prediction of Adverse Drug Reactions and Off-Target Effects from Chemical Structure. ChemMedChem. 2007;2:861–873. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- 30.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–1. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 31.Goldfarb DS. US 20090163545 A1. 2009. Method using lifespan-altering compounds for altering the lifespan of eukaryotic organisms, and screening for such compounds. [Google Scholar]
- 32.Segall M, Champness E, Leeding C, Lilien R, Mettu R, Stevens B. Applying medicinal chemistry transformations and multiparameter optimization to guide the search for high-quality leads and candidates. J Chem Inf Model. 2011;51:2967–76. doi: 10.1021/ci2003208. [DOI] [PubMed] [Google Scholar]
- 33.Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin B. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- 34.Lagorce D, Sperandio O, Galons H, Miteva MA, Villoutreix BO. FAF-Drugs2: free ADME/tox filtering tool to assist drug discovery and chemical biology projects. BMC Bioinformatics. 2008;9:396. doi: 10.1186/1471-2105-9-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anon FAFDrugs2. http://fafdrugs2.mti.univ-paris-diderot.fr/index.html.
- 36.Reynolds CH, Tounge BA, Bembenek SD. Ligand binding efficiency: trends, physical basis, and implications. J Med Chem. 2008;51:2432–8. doi: 10.1021/jm701255b. [DOI] [PubMed] [Google Scholar]
- 37.Bemis GW, Murcko MA. The properties of known drugs 1. molcular frameworks. J Med Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 38.Jones DR, Ekins S, Li L, Hall SD. Computational approaches that predict metabolic intermediate complex formation with CYP3A4 (+b5) Drug Metab Dispos. 2007;35:1466–75. doi: 10.1124/dmd.106.014613. [DOI] [PubMed] [Google Scholar]
- 39.Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models. PLOSONE. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekins S, Reynolds R, Kim H, Koo M-S, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian Models Leveraging Bioactivity and Cytotoxicity Information for Drug Discovery. Chem Biol. 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anon R. http://www.r-project.org/
- 42.Anon MLP probes. http://mli.nih.gov/mli/mlp-probes-2/?dl_id=1352.
- 43.Baumann M, Baxendale IR. An overview of the synthetic routes to the best selling drugs containing 6-membered heterocycles. Beilstein J Org Chem. 2013;9:2265–319. doi: 10.3762/bjoc.9.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goetz AE, Garg NK. Regioselective reactions of 3,4-pyridynes enabled by the aryne distortion model. Nat Chem. 2013;5:54–60. doi: 10.1038/nchem.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sies H. Oxidative stress: oxidants and antioxidants. Exp Physiol. 1997;82:291–5. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 46.Tapiero H, Townsend DM, Tew KD. The antioxidant role of selenium and seleno-compounds. Biomed Pharmacother. 2003;57:134–44. doi: 10.1016/s0753-3322(03)00035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murray CW, Erlanson DA, Hopkins AL, Keseru GM, Leeson PD, Rees DC, Reynolds CH, Richmond NJ. Validity of ligand efficiency metrics. ACS Med Chem Lett. 2014;5:616–8. doi: 10.1021/ml500146d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornish J, Callon KE, Lin CQ, Xiao CL, Mulvey TB, Cooper GJ, Reid IR. Trifluoroacetate, a contaminant in purified proteins, inhibits proliferation of osteoblasts and chondrocytes. Am J Physiol. 1999;277:E779–83. doi: 10.1152/ajpendo.1999.277.5.E779. [DOI] [PubMed] [Google Scholar]
- 49.Hochlowski J, Cheng X, Sauer D, Djuric S. Studies of the relative stability of TFA adducts vs non-TFA analogues for combinatorial chemistry library members in DMSO in a repository compound collection. J Comb Chem. 2003;5:345–9. doi: 10.1021/cc0300107. [DOI] [PubMed] [Google Scholar]
- 50.Tipps ME, Iyer SV, John Mihic S. Trifluoroacetate is an allosteric modulator with selective actions at the glycine receptor. Neuropharmacology. 2012;63:368–73. doi: 10.1016/j.neuropharm.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.