Abstract
Purpose of review
Precision cancer medicine, the use of genomic profiling of patient tumors at the point-of-care to inform treatment decisions, is rapidly changing treatment strategies across cancer types. Precision medicine for advanced prostate cancer may identify new treatment strategies and change clinical practice. In this review, we discuss the potential and challenges of precision medicine in advanced prostate cancer.
Recent findings
Although primary prostate cancers do not harbor highly recurrent targetable genomic alterations, recent reports on the genomics of metastatic castration-resistant prostate cancer has shown multiple targetable alterations in castration-resistant prostate cancer metastatic biopsies. Therapeutic implications include targeting prevalent DNA repair pathway alterations with PARP-1 inhibition in genomically defined subsets of patients, among other genomically stratified targets. In addition, multiple recent efforts have demonstrated the promise of liquid tumor profiling (e.g., profiling circulating tumor cells or cell-free tumor DNA) and highlighted the necessary steps to scale these approaches in prostate cancer.
Summary
Although still in the initial phase of precision medicine for prostate cancer, there is extraordinary potential for clinical impact. Efforts to overcome current scientific and clinical barriers will enable widespread use of precision medicine approaches for advanced prostate cancer patients.
Keywords: BRCA2, castration-resistant prostate cancer, DNA repair, genomics, precision medicine, prostate cancers
INTRODUCTION
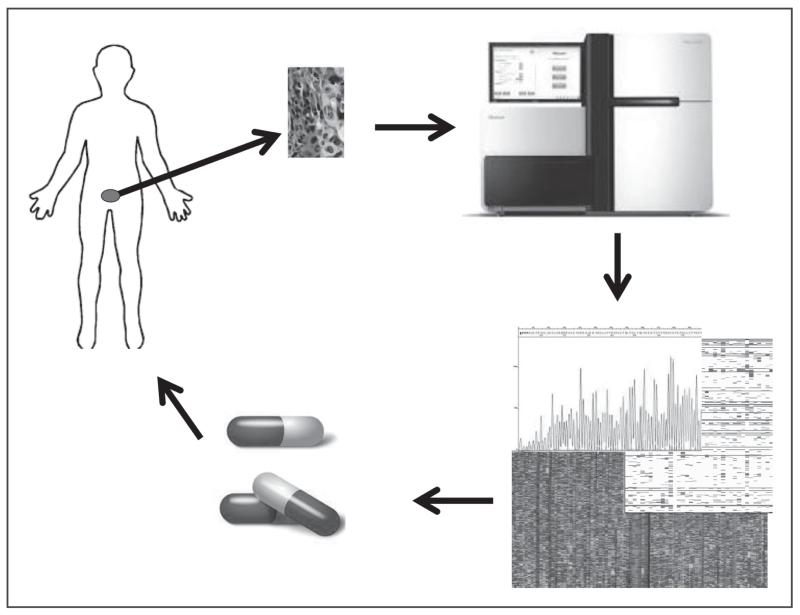
Precision cancer medicine, the use of genomic profiling at the point-of-care to inform treatment decisions (Fig. 1), is changing cancer care by enabling more accurate and efficient prediction of therapies for individual cancer patients. This revolution is the result of numerous studies identifying key cancer drivers, their alterations, and therapies to specifically target these alterations [1]. More recently, multiple cancer landscape studies have provided further insight into the alterations within and between tumor types [2]. That success, along with the increased affordability and reliability of sequencing, and development of computational tools for clinical genomic analysis, has led to the integration of genome science directly into clinical practice. Tight networks and collaboration between clinicians, genome scientists, and pharmaceuticals companies will continue to advance precision medicine [3–5].
FIGURE 1.
Precision medicine in advanced prostate cancer revolves around the ability to take a tumor sample, ideally metastatic tumor, sequence the tumor, and assign therapy to the patient. This involves biopsy of tumor tissue, DNA/RNA extraction, WES/WGS/Transcriptome sequencing, bioinformatics interpretation of results, and assignment of treatment based on therapeutic profile. This process may be repeated as needed when patients progress. WES, whole-exome sequencing; WGS, whole genome sequencing.
Prostate cancer is the most common solid tumor in men in the USA [6]. Prostate cancer is a hormone-dependent tumor, demonstrated by recurrent alterations in the androgen receptor and its pathway [7]. Castration-resistant prostate cancer (CRPC) is a lethal clinical state in which the tumor has developed resistance to androgen deprivation therapy. This occurs in the majority of advanced or metastatic prostate cancer patients. The genomic landscape of localized prostate cancer has been well defined [8–11,12■■,13–15]. Multiple studies have highlighted the lack of highly recurrent clinically actionable alterations, as well as the high level of tumor intraheterogeneity even in the primary setting [15,16]. In contrast with the primary prostate cancer genome, the extensive mutational landscape of metastatic CRPC lesions has exposed the possibility of targeted therapies and precision medicine in CRPC [17■■]. In this review, we discuss the potential of genomics to impact the clinical management of CRPC, and consider the challenges that must be overcome to enable wide implementation in the clinic.
CURRENT THERAPEUTIC APPROACHES FOR CASTRATION-RESISTANT PROSTATE CANCER
In the CRPC setting, the primary successful therapeutic target remains the androgen receptor. The discovery of androgen receptor, along with the persistent unearthing of androgen receptor resistance mechanisms, has enabled additional effective treatments in CRPC patients [7,18–21]. Approved therapies in this space include new androgen synthesis pathway agents, such as abiraterone, and direct inhibitors of androgen receptor, such as enzalutamide [22–28]. However, the majority of CRPC patients ultimately develop resistance to androgen receptor-focused therapies, despite multiple new agents reaching the clinic. Furthermore, there are many patients who never respond to these therapies and manifest intrinsic resistance to this therapeutic approach. Generally, almost all patients with CRPC ultimately succumb to the disease.
Genomics and lethal prostate cancer
Studies examining the genomic alterations involved in lethal prostate cancer from autopsy cohorts revealed the underlying biology behind the disease and the evolutionary processes driving advanced disease [29,30■■,31,32■■]. Despite these crucial insights, these studies did not have clinical cohorts to explore the landscape of potential clinically actionable targets. Recently, a study examined 150 metastatic site biopsies from living patients with CRPC through integrative whole-exome sequencing (WES) and whole transcriptome sequencing [17■■]. A high number of targetable mutations, defined as predicting response or resistance to a therapy, or having a diagnostic or prognostic utility, were found in these patients, in contrast to localized prostate cancer. Not including androgen receptor, 65% of patients had a targetable mutation, many of which have been linked to ongoing clinical trials (Table 1). The details of these findings are described below.
Table 1.
Ongoing clinical trials in advanced CRPC with potential precision medicine applications
| Gene | Potential therapeutic | Current clinical trials/therapies* | Phase | |
|---|---|---|---|---|
| Androgen receptor | Androgen receptor | Mifepristone (RU-486) | NCT00140478 | Phase 2 |
| Mifepristone/enzalutamide | NCT02012296 | Phases 1, 2 | ||
| Galeterone | NCT02438007 | Phase 3 | ||
| GTx-758 | NCT01615120 | Phase 2 | ||
| VT-464 | NCT02445976 | Phase 2 | ||
| Trilostane | NCT00181597 | Phase 2 | ||
| AZD3514 | NCT01162395 | Phase 1 | ||
| Orteronel (TAK-700) |
NCT00569153, NCT01809691, NCT01809691 |
Phases 1, 2, 3 | ||
| Triamcinalone | NCT00186108 | Phase 1 | ||
|
| ||||
| Immunotherapy | Ipilimimab + ADT | NCT01377389 | Phase 2 | |
| AR DNA Vaccine | NCT02411786 | Phase 1 | ||
| Ipilimimab | NCT00170157 | Phase 2 | ||
| BNIT-PR-001 | Phase 1 | |||
|
| ||||
| PIK3CA | PIK3CA | Buparlisib (BKM120) | NCT02487823 | Phase 1 |
| PIK3CB | AZD8186 | NCT01884285 | Phase 1 | |
| PIK3CB | GSK2636771/enzalutamide | NCT02215096 | Phase 1 | |
| PIK3CB | GDC-0068/abiraterone | NCT01485861 | Phase 2 | |
|
| ||||
| Cell Cycle | BCL-2 | Navitoclax/abiraterone | NCT01828476 | Phase 2 |
| CDK4/6 | Ribociclib | NCT02555189 | Phases 1, 2 | |
| CDK4/6 | PD 0332991 | NCT02059213 | Phase 2 | |
|
| ||||
| DNA damage | PARP | Niraparib/enzalutamide | NCT02500901, NCT00749502 | Phase 1 |
| Olaparib/enzalutamide | NCT01972217 | Phase 2 | ||
| BMN 673 | NCT01286987 | Phase 1 | ||
| Veliparib | NCT00892736 | Phase 1 | ||
|
| ||||
| WNT | WNT | Foxy-5 | NCT02020291 | Phase 1 |
| OMP-54F28 | NCT01608867 | Phase 1 | ||
List collected October 2015.
Androgen receptor
Numerous studies have addressed the androgen receptor and its pathway alterations in metastatic CRPC [7,29,30■■,31,32■■]. Despite reduced androgen circulation in CRPC, androgen receptor is still activated through various mechanisms, including androgen receptor amplification or overexpression, activating androgen receptor mutations, alterative androgen production, androgen receptor coactivator overexpression, and indirect androgen receptor activation [33]. Robinson et al. presented that 34% of CRPC patients still have only androgen receptor as a clinically relevant mutation, indicating they may be differentially sensitive to existing and novel androgen receptor-directed therapies. However, the clinical significance of these androgen receptor mutations for predicting response or resistance to these agents remains to be determined. Although several drugs have shown promise targeting androgen receptor in this space, including abiraterone and enzalutamide, most patients eventually develop resistance to these agents [23,25,26]. Recently, it was reported that patients with androgen receptor-V7 splice variant, in the transcriptomic data, may be resistant to enzalutamide but respond to galeterone, a novel androgen receptor therapy currently in phase III trials [34■,35], potentially demonstrating the first genomically driven therapy in CRPC. Furthermore, there are numerous experimental agents that target androgen receptor or its pathway in novel ways (Table 1) that may augment the ability to effectively inhibit this dominant pathway in patients with tumors that are still wholly dependent on androgen receptor signaling.
DNA repair pathway
Beyond the androgen receptor pathway, the most striking result from clinical genomic profiling of CRPC patients was that 19% of patients have a DNA repair pathway alterations, including 12.7% of patients with a putative pathogenic BRCA2 germ line mutation [36,37]. Additional somatic and germ line DNA repair alterations were found in ATM, BRCA1, CDK12, FANCA, RAD51B, and RAD51C. Many of these alterations are associated with platinum response in other cancer types [38–40]. In addition, PARP inhibition demonstrated great efficacy in patients with BRCA2 mutations and other DNA repair alterations in CRPC and other tumor types [41■■]. Mateo et al. conducted a phase II trial of olaparib plus genomic correlates in 50 CRPC patients. A total of 16 of the 50 patients harbored DNA repair gene inactivation alterations, and 14 out of those 16 patients responded to olaparib [41■■], highlighting another potential genomically driven therapy in CRPC. Based on these findings there are currently multiple clinical trials testing the effects of PARP inhibitors with or without androgen receptor-targeted therapies in CRPC patients, demonstrating the rapid impact of this DNA repair genomic discovery on realizing precision medicine for prostate cancer.
Phosphoinositide 3-kinase pathway
The phosphoinositide 3-kinase (PI3K) pathway is recurrently mutated in CRPC, commonly through loss of PTEN, amplification of PIK3CA/B, and activating mutation of PIK3CA/B and AKT1 [42]. In Robinson et al., PI3K pathway was altered in 49% of patients, making it the second most frequently altered pathway after androgen receptor. In the past, many PI3K monotherapies have had a lack of efficacy, thought to be because of lack of specificity, coexisiting alterations, and signaling feedback [43■]. Recently, multiple inhibitors of specific PI3K isoforms have begun testing in clinical trials, potentially increasing the specificity of these agents. In CRPC, there are recurrent mutations in PIK3CB and frequent loss of PTEN, which may activate PIK3CB over PIK3CA [44], emphasizing the need for these specific PI3K isoforms inhibitors [45,46] to effectively clinically target this pathway. There has also been evidence that there is cross-pathway interaction between PI3K and homologous recombination pathway, indicating that patients with PI3K pathway alterations may respond to PARP inhibitors as well [47–49].
WNT pathway
In Robinson et al., 18% of metastatic CRPC patient are presented with mutation in WNT pathway, including activating CTNNB1, APC, RNF43, RSPO2, and ZNRF3 mutations. Furthermore, a recent study of CRPC patients’ circulating tumor cells demonstrated an upregulation of WNT signaling in this clinical setting [50]. Historically, the WNT pathway has been extremely difficult to target because of the multitude of receptors, ligands, and downstream pathways [51]. The WNT pathway has many imperative biologic functions from embryonic development to tissue homeostasis and is activated by proteins secreted by tumor cells as part of an autocrine loop, or they may be produced by surrounding stromal cells, increasing the difficulty of targeting this pathway. The WNT pathway is also thought to be activated in cancer stem cells, which are thought to drive resistance to many therapies [52■]. Although it is known that the WNT signaling pathway is altered in CRPC, it is unknown whether an antagonist or agonist would work better to inhibit growth. There is contradicting evidence in multiple tumor types whether activation or repression of the pathway increases survival. Evidence also supports β-catenin signaling in dictating tissue-specific predisposition to APC-driven tumorigenesis [53], helping to indicate whether a repressor or activator would work best. This is again demonstrated by the contradicting therapeutics currently in clinical trials (Table 1): Foxy-5 activates the WNT pathway, whereas OMP-54F28 inhibits the pathway. Efficacy data will inform their utility in this clinical setting. Additional preclinical and clinical studies are needed to determine how to best target this pathway in CRPC. It is also known that all of the WNT pathway therapeutics have had significant toxicities associated because of the wide range of functions of the WNT pathway [54].
Cell cycle pathway
Loss of RB1 was seen in 21% of metastatic biopsies versus 1% in localized prostate cancer [15,17■■]. Less common cell cycle pathway alterations including mutations in CDKN2A/B, CDKN1B, and amplifications of CCND1, were seen. The major role of the cell cycle pathway is to stop mitosis to allow for DNA repair via the inhibition of cyclin-dependent kinases (CDKs) and RB1 phosphorylation [55]. CDK4/6 inhibition can induce cell cycle arrest and cancer cell senescence. Currently, there is one CDK4/6 inhibitor approved in breast cancer, and multiple other CDK4/6 and pan-CDK inhibitors in clinical trials [56]. In CRPC, there are multiple trials for CDK4/6 inhibitors (Table 1). There are known resistance to these drugs, including Rb-negativity and a lack of codeletion of CDKN2A/CDKN2B in glioblastoma, indicating any trial with these compounds must be genomically driven and may be relevant in CRPC [57].
Immunotherapy
The most mature effort for CRPC immunotherapy is sipuleucel-T, a cell-based immunotherapy, which is Federal Drug Administration approved [22]. It is created using mature, autologous antigen-presenting cells obtained from patients. However, this approach has thus far not resulted in clinical benefit in stratified patient subsets. In order to expand immunotherapy approaches in the setting of success in other tumor types, multiple checkpoint inhibitors have been testing in CRPC, although thus far, these therapies have had limited clinical results [58]. Notably, multiple studies have recently demonstrated a correlation between response to immune checkpoint inhibitors and mutational burden [59■■–61■■]. A subset of CRPC patients has a high mutation load because of alterations in mismatch repair genes, MLH1 and MSH2. Thus, CRPC patients with a high mutational burden may benefit greatly from immunotherapy, such as a checkpoint inhibitor, and future efforts geared toward determining whether this relationship holds across CRPC patients may inform the clinical utility of checkpoint inhibitors in the CRPC setting.
TECHNOLOGIES FOR ENHANCING PRECISION MEDICINE IN CASTRATION-RESISTANT PROSTATE CANCER
Despite continuous advances in high-throughput genomic sequencing technologies and their utilization in cancer, there are several challenges in successful implementation of precision medicine in CRPC (Table 2). For example, access to tumor tissue for profiling is especially complicated because of the need to obtain metastatic biopsies, including bone metastasis [62] and the low percentage of cancer cells in many of these samples. Although it has been demonstrated that sequencing tumor from bone biopsies is feasible, these approaches are difficult and require much expertise [63■]. There is also the reality that not all patients can get a biopsy or that the capacity of interventional radiology facilities will permit biopsy sampling in all patients. Moreover, a single biopsy may not capture the extent of disease. As demonstrated in localized prostate cancer and by studying multiple metastatic sites from individual patients, there is significant intra-tumor heterogeneity in prostate cancer [30■■]. If a patient has multiple metastatic sites, only sampling one, may not demonstrate the biology of the whole tumor. In addition, one must also consider the possibility of not finding any targetable genetic alteration in a patient’s tumor. With additional genomics research and novel therapeutics, this possibility will decrease.
Table 2.
Logistical and scientific challenges in CRPC precision medicine initiatives
| Challenge | Description | Potential solutions | |
|---|---|---|---|
| Logistical challenges | Genomic testing infrastructure |
Setting up genomic testing in a hospital requires infrastructure with pathology, clinician, bioinformatics, information systems, and many more |
Have key academic centers for testing and distribute results to community centers |
| Cost of genomic testing | In nonacademic centers, genomic testing is run by private companies |
Allow testing to be covered by insurance or make low cost testing available |
|
| Metastatic biopsies | Biopsy is not feasible in all patients | Collection and sequencing of CTCs | |
| Testing target therapeutics in CRPC |
Although we know there are therapeutics that work with many of these targets in other diseases, these therapeutics must be testing in CRPC |
Designing basket and bucket trials to test multiple targets or therapeutics at the same time |
|
| Scientific challenges | Tumor intraheterogeneity | CRPC is known to have a lot of heterogeneity in the primary specimen, by biopsying one metastatic lesion, it is unknown if we see the entire genomic picture. |
Future studies should focus on heterogeneity in the metastatic setting |
| Lack of targetable mutations |
Not all patients will have an oncogenic driver because of a low number of genes screened |
Perform WES, RNA or protein based assays on these patients |
|
| Secondary resistance | Although patients may respond to a targeted therapy, most will develop resistance. Posttreatment biopsy tumor samples may have a different genomic profile than pretreatment |
Track patient progress using ctDNA or CTCs. Test and provide combination therapy for patients |
|
| Identification of driver mutation |
Some patients may present with multiple targetable mutations |
Use heuristic and predictive modeling to determine which mutation is best to initially target. |
CRPC, castration-resistant prostate cancer; CTCs, circulating tumor cells; ctDNA, Circulating tumor DNA; WES, whole-exome sequencing.
A potential technology that may address difficulty in obtaining biopsies, and tumor heterogeneity in CRPC is the use of liquid biopsy techniques. One such approach involves the use of circulating tumor cells (CTCs) to identify genomic alterations of CRPC. CTCs have been demonstrated to have a reasonable readout of the tumor genomic landscape in patients with CRPC [64]. Circulating tumor DNA (ctDNA) is another novel way to identify genomic alterations and track patient’s genomic landscape over time [65■]. In other tumor types, WES has been performed from ctDNA [66■■], indicating metastatic biopsies may not be needed in the future. These technologies may also help us identify patients who are developing resistance earlier than a radiological scan [64,67,68]. As seen in multiple other cancer types, the genomic profile of tumors after treatment is often very different [69■,70■]. From a clinical standpoint, early detection of resistance is crucial to optimizing therapy, and the use of ultradeep sequencing in multiple regions of a biopsy, together with monitoring of tumor evolution using ctDNA could provide this information [71■].
CLINICAL STRATEGIES TO IMPLEMENT PRECISION MEDICINE
A major logistical challenge toward implementing precision medicine in CRPC and across cancer types is building an infrastructure for genomic sequencing in cancer, including collecting tissue, genomic alterations testing, genomic analysis, and reporting results back to patients [72■■,73■■,74–77]. Another limitation to genomic sequencing is cost. Although the cost of genomic sequencing is continuously decreasing, it is rarely covered by insurance [78], causing limited access to many patients [79]. In addition, once genomic alterations are identified, it is often difficult for patients to get access to affordable medication [80]. Finally, owing to the small percentage of patients with particular genomic alterations, new clinical trial schemas have been developed to support ongoing precision medicine therapeutics.
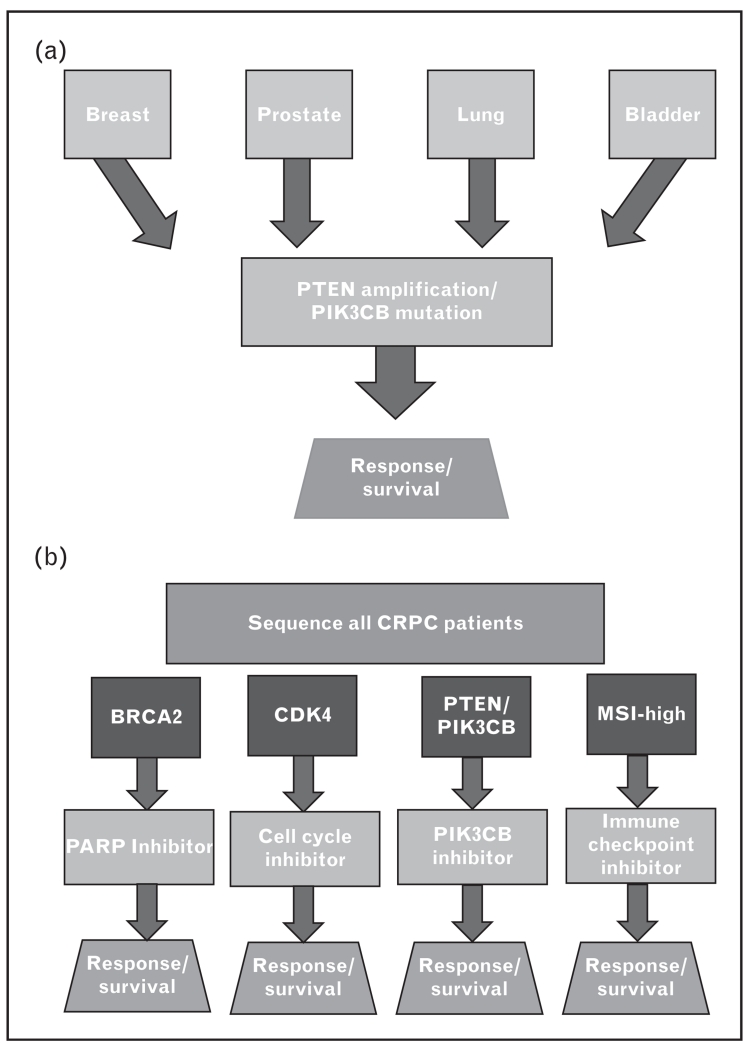
Two efficient ways to test the effects of multiple drugs is through a basket or umbrella trial (Fig. 2a and b). These trial schemas have been well demonstrated in lung cancer [81] and are currently a declared initiative of the National Institutes of Health (NIH). The National Cancer Institute program, a large basket trial initiative, will include 3000 patients with different types of cancer to find early signals of a response to targeted therapies. Basket trials test the effect of a single drug targeting a specific molecular alteration in a variety of cancers. This design not only allows for a faster identification of candidate patient, but also to assess the potential value of this targeted therapy across different tumor types. An umbrella trial assesses the effect of different drugs in different molecular alterations either in one or several tumors. This would involve sequencing the tumors of all men with CRPC, and placed in the appropriate slot based on the genomic profile. One additional challenge to umbrella trials is identifying the correct driver mutation. Ongoing efforts to identify and rank known driver mutations [72■■] should help place patients in the appropriate slot. These trial schemas will also help drug development for less common alterations. Implementing either of these trial schemas for CRPC will allow expedited identification of targeted therapies that work well in this setting.
FIGURE 2.
Basket trials (a) and umbrella trials (b) are two approaches to precision medicine with novel therapeutics in advanced prostate cancer. (a) Basket trials take patients with multiple different tumor types with the same genetic alteration to test a single therapeutic. This allows for fast identification of potential patients and tests the drug/alteration across different diseases quickly. (b) Umbrella trials test the effect of different drugs on different genetic alterations within the same disease type. This allows for increase accrual of all advanced prostate patients and tests multiple drug/genomic alteration combinations at the same time.
CONCLUSION
The concept of precision medicine driven by genomics for CRPC is appealing; however, it is in its infancy. We must continue to obtain additional genomic information and correlate this with therapeutic response. Developing catalogues of CRPC cancer-related genes, together assessments of pathway activation could enable a better identification of additional driver in the future. The characterization of the genomic landscape of tumors and of the activated protein network will guide combination therapies to optimize therapeutic effects. Finally, logistics and operational challenges need to be addressed.
KEY POINTS.
Precision medicine shows great promise in advanced prostate cancer, but it is still in the initial stages.
Advancement in prostate cancer precision medicine is dependent on continuous research in prostate cancer genomics and correlation with response to current and novel therapies.
Expansion of liquid biopsy techniques, such as circulating tumor cells or cell-free DNA, may be especially impactful in precision medicine for CRPC.
Acknowledgements
None.
Financial support and sponsorship
This study was supported by the Prostate Cancer Foundation Young Investigator Award (E.M.V.) and NIH K08CA188615 (E.M.V.).
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Yuan Y, Van Allen EM, Omberg L, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol. 2014;32:644–652. doi: 10.1038/nbt.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumor types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciardiello F, Arnold D, Casali PG, et al. Delivering precision medicine in oncology today and in future-the promise and challenges of personalised cancer medicine: a position paper by the European Society for Medical Oncology (ESMO) Ann Oncol. 2014;25:1673–1678. doi: 10.1093/annonc/mdu217. [DOI] [PubMed] [Google Scholar]
- 4.Schwaederle M, Zhao M, Lee JJ, et al. Impact of precision medicine in diverse cancers: a meta-analysis of phase II clinical trials. J Clin Oncol. 2015;33:3817–3825. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 7.Taplin ME, Bubley GJ, Shuster TD, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 8.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pflueger D, Terry S, Sboner A, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XS, Shankar S, Dhanasekaran SM, et al. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 2011;1:35–43. doi: 10.1158/2159-8274.CD-10-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlins SA, Rubin MA, Chinnaiyan AM. Integrative biology of prostate cancer progression. Annu Rev Pathol. 2006;1:243–271. doi: 10.1146/annurev.pathol.1.110304.100047. [DOI] [PubMed] [Google Scholar]
- 12■■.Cooper CS, Eeles R, Wedge DC, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. The article demonstrates the existence of branching evolution within a single prostate tumor as well as abnormal mutational processes in surrounding normal prostate tissue.
- 13.Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. The article profiles multiple localized prostate tumors, demonstrating the highly heterogeneous. They also show evidence of divergent tumor evolution in multifocal cancer and, in some cases, tumors of independent clonal origin.
- 15.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. Electronic address: schultz@c-bio.mskcc.org; Cancer Genome Atlas Research Network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 17■■.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. The article describes the mutational landscape from 150 metastatic prostate cancer patient biopsies. Many more details on the mutation and genetic alterations discussed in our study are presented in this article.
- 18.Crook JM, O’Callaghan CJ, Duncan G, et al. Intermittent androgen suppression for rising PSA level after radiotherapy. N Engl J Med. 2012;367:895–903. doi: 10.1056/NEJMoa1201546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain M, Tangen CM, Berry DL, et al. Intermittent versus continuous androgen deprivation in prostate cancer. N Engl J Med. 2013;368:1314–1325. doi: 10.1056/NEJMoa1212299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambeaud JJ. Intermittent complete androgen blockade in metastatic prostate cancer. Eur Urol. 1999;35(Suppl 1):32–36. [PubMed] [Google Scholar]
- 21.Akakura K, Bruchovsky N, Goldenberg SL, et al. Effects of intermittent androgen suppression on androgen-dependent tumors. Apoptosis and serum prostate-specific antigen. Cancer. 1993;71:2782–2790. doi: 10.1002/1097-0142(19930501)71:9<2782::aid-cncr2820710916>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 22.Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 23.Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 24.Beer TM, Tombal B. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:1755–1756. doi: 10.1056/NEJMc1410239. [DOI] [PubMed] [Google Scholar]
- 25.de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan CJ, Smith MR, de Bono JS, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrylak DP, Tangen CM, Hussain MH, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 28.Tannock IF, de Wit R, Berry WR, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 29.Beltran H, Yelensky R, Frampton GM, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30■■.Hong MK, Macintyre G, Wedge DC, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun. 2015;6:6605. doi: 10.1038/ncomms7605. The article demonstrates the heterogenetity of spread of metastatic prostate cancer. Multiple tumors from one patient were biopsied and WES was performed. Blood was also sequenced, identifying both metastatic and primary tumor clones.
- 31.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32■■.Gundem G, Van Loo P, Kremeyer B, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. The article presents evidence for polyclonal seeding in androgen-deprived metastatic prostate cancer. Whole genome sequencing from multiple metastatic sites from 10 patients identified evolution from primary tumors and multiple metastatic sites. Metastasis-to-metastasis spread was found to be common as well as de novo monoclonal seeding in metastatic sites.
- 33.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34■.Kwegyir-Afful AK, Ramalingam S, Purushottamachar P, et al. Galeterone and VNPT55 induce proteasomal degradation of AR/AR-V7, induce significant apoptosis via cytochrome c release and suppress growth of castration resistant prostate cancer xenografts in vivo. Oncotarget. 2015;6:27440–27460. doi: 10.18632/oncotarget.4578. The article discusses the ability for galeterone, a novel androgen receptor inhibitor to deplete full length androgen receptor and androgen receptor splice variants, unlike other androgen receptor inhibitors currently approved.
- 35.Antonarakis ES, Lu C, Luber B, et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015;1:582–591. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundararajan S, Ahmed A, Goodman OB. The relevance of BRCA genetics to prostate cancer pathogenesis and treatment. Clin Adv Hematol Oncol. 2011;9:748–755. [PubMed] [Google Scholar]
- 37.Agalliu I, Kwon EM, Zadory D, et al. Germline mutations in the BRCA2 gene and susceptibility to hereditary prostate cancer. Clin Cancer Res. 2007;13:839–843. doi: 10.1158/1078-0432.CCR-06-2164. [DOI] [PubMed] [Google Scholar]
- 38.Van Allen EM, Mouw KW, Kim P, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carey LA. Targeted chemotherapy? Platinum in BRCA1-dysfunctional breast cancer. J Clin Oncol. 2010;28:361–363. doi: 10.1200/JCO.2009.24.0838. [DOI] [PubMed] [Google Scholar]
- 40.Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41■■.Mateo J, Carreira S, Sandhu S, et al. DNA-Repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. The article presents the findings of a phase II clinical trial of olaparib, a PARP-1 inhibitor, in metastatic prostate cancer. A total of 14/16 patients with DNA repair alterations achieved a clinical response to olaparib.
- 42.Sarker D, Reid AH, Yap TA, de Bono JS. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin Cancer Res. 2009;15:4799–4805. doi: 10.1158/1078-0432.CCR-08-0125. [DOI] [PubMed] [Google Scholar]
- 43■.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. The article reviews the multiple PI3K isoforms, therapeutic targets for the isoforms, and their impact in the clinic.
- 44.Wee S, Wiederschain D, Maira SM, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni J, Liu Q, Xie S, et al. Functional characterization of an isoform-selective inhibitor of PI3K-p110β as a potential anticancer agent. Cancer Discov. 2012;2:425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta S, Ramjaun AR, Haiko P, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 47.Juvekar A, Burga LN, Hu H, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2:1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ibrahim YH, García-García C, Serra V, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.González-Billalabeitia E, Seitzer N, Song SJ, et al. Vulnerabilities of PTEN-TP53-deficient prostate cancers to compound PARP-PI3K inhibition. Cancer Discov. 2014;4:896–904. doi: 10.1158/2159-8290.CD-13-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical WNT signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–532. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52■.Yun EJ, Zhou J, Lin CJ, et al. Targeting cancer stem cell in castration resistant prostate cancer. Clin Cancer Res. 2015;22:670–679. doi: 10.1158/1078-0432.CCR-15-0190. The article presents evidence of cancer stem cells in prostate cancer. It also shows evidence of potential increased therapeutic activity of chemotherapy if cancer stem cells are targeted as well.
- 53.Bakker ER, Hoekstra E, Franken PF, et al. β-Catenin signaling dosage dictates tissue-specific tumor predisposition in APC-driven cancer. Oncogene. 2013;32:4579–4585. doi: 10.1038/onc.2012.449. [DOI] [PubMed] [Google Scholar]
- 54.Sakata T, Chen JK. Chemical ‘Jekyll and Hyde’s: small-molecule inhibitors of developmental signaling pathways. Chem Soc Rev. 2011;40:4318–4331. doi: 10.1039/c1cs15019g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manning AL, Dyson NJ. RB: mitotic implications of a tumour suppressor. Nat Rev Cancer. 2012;12:220–226. doi: 10.1038/nrc3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner NC, Huang Bartlett C, Cristofanilli M. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373:1672–1673. doi: 10.1056/NEJMc1510345. [DOI] [PubMed] [Google Scholar]
- 57.Wiedemeyer WR, Dunn IF, Quayle SN, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 2010;107:11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slovin SF, Higano CS, Hamid O, et al. Ipilimumab alone or in combination with radiotherapy in metastatic castration-resistant prostate cancer: results from an open-label, multicenter phase I/II study. Ann Oncol. 2013;24:1813–1821. doi: 10.1093/annonc/mdt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59■■.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. The article presents findings on correlates with response to anti-CTLA4 from 110 metastatic melanoma patients. Overall mutational load, neoantigen load, and expression of cytolytic markers in the immune microenvironment were significantly associated with clinical benefit.
- 60■■.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in nonsmall cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. The article presents findings from two independent cohorts on the correlation between nonsynonymous mutational burden, neoantigen burdern, and DNA repair pathways with mutations with response to anti-PD1 therapy in patients with metastatic nonsmall cell lung cancer.
- 61■■.Snyder A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade. N Engl J Med. 2015;372:783. doi: 10.1056/NEJMc1415938. The article presents findings on correlates with response to anti-CTLA4 a cohort of metastatic melanoma patients.
- 62.Bubendorf L, Schöpfer A, Wagner U, et al. Metastatic patterns of prostate cancer: an autopsy study of 1589 patients. Hum Pathol. 2000;31:578–583. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 63■.Van Allen EM, Foye A, Wagle N, et al. Successful whole-exome sequencing from a prostate cancer bone metastasis biopsy. Prostate Cancer Prostatic Dis. 2014;17:23–27. doi: 10.1038/pcan.2013.37. The article presents evidence that prostate bone biopsies can be used for successful WES and potential actionable genomic alterations can be identified.
- 64.Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65■.Romanel A, Tandefelt DG, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re10. doi: 10.1126/scitranslmed.aac9511. The article analyzes circulating tumor DNA samples from 97 CRPC patients and identified emerging mutations during therapy with abiraterone.
- 66■■.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. The article presents a novel sequencing method for ctDNA. ctDNA was found to be highly correlated with tumor volume and clinical imaging.
- 67.Konieczkowski DJ, Garraway LA. Resistance to EGFR blockade in colorectal cancer: liquid biopsies and latent subclones. Cell Res. 2013;23:13–14. doi: 10.1038/cr.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murtaza M, Dawson SJ, Tsui DW, et al. Noninvasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 69■.Wagle N, Grabiner BC, Van Allen EM, et al. Response and acquired resistance to everolimus in anaplastic thyroid cancer. N Engl J Med. 2014;371:1426–1433. doi: 10.1056/NEJMoa1403352. The article presents evidence of mutations in one patient which lead to an exceptional response to everolimus as well as potential mutations that confer resistance to everolimus.
- 70■.Ahronian LG, Sennott EM, Van Allen EM, et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 2015;5:358–367. doi: 10.1158/2159-8290.CD-14-1518. The article presents the initial characterization of clinical acquired resistance mechanisms to RAF inhibitors by several MAPK pathway alterations, reactivating MAPK signaling.
- 71■.De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. It is a proof of principle study that demonstrates high depth sequencing of plasma-derived ctDNA for de novo mutation identification and monitoring of other somatic genetic alterations over treatment of therapy.
- 72■■.Van Allen EM, Wagle N, Stojanov P, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. doi: 10.1038/nm.3559. The article describes a WES platform for archival formalin-fixed, paraffin-embedded tumor samples, including clinical analysis and interpretation of WES data from both a retrospective and prospective cohorts.
- 73■■.Sboner A, Elemento O. A primer on precision medicine informatics. Brief Bioinform. 2015;17:145–153. doi: 10.1093/bib/bbv032. The article reviews the key components of a computation infrastructure for precision medicine program including reporting, integration into the electronic Health records, and regulatory aspects.
- 74.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;371:1170. doi: 10.1056/NEJMc1408914. [DOI] [PubMed] [Google Scholar]
- 75.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov. 2013;12:358–369. doi: 10.1038/nrd3979. [DOI] [PubMed] [Google Scholar]
- 76.Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB. Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol. 2013;31:1849–1857. doi: 10.1200/JCO.2012.45.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chantrill LA, Nagrial AM, Watson C, et al. Precision medicine for advanced pancreas cancer: the individualized molecular pancreatic cancer therapy (IMPaCT) trial. Clin Cancer Res. 2015;21:2029–2037. doi: 10.1158/1078-0432.CCR-15-0426. [DOI] [PubMed] [Google Scholar]
- 78.Chakradhar S. Tumor sequencing takes off, but insurance reimbursement lags. Nat Med. 2014;20:1220–1221. doi: 10.1038/nm1114-1220. [DOI] [PubMed] [Google Scholar]
- 79.Tran B, Dancey JE, Kamel-Reid S, et al. Cancer genomics: technology, discovery, and translation. J Clin Oncol. 2012;30:647–660. doi: 10.1200/JCO.2011.39.2316. [DOI] [PubMed] [Google Scholar]
- 80.Lewin J, Siu LL. Cancer genomics: the challenge of drug accessibility. Curr Opin Oncol. 2015;27:250–257. doi: 10.1097/CCO.0000000000000185. [DOI] [PubMed] [Google Scholar]
- 81.Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]