Abstract
The kelch-like ECH-associated protein 1 (Keap1)-nuclear factor erythroid 2-related factor 2 (Nrf2) signaling axis serves as a “master regulator” in response to oxidative/electrophilic stresses and chemical insults through the coordinated induction of a wide array of cytoprotective genes. Therefore, activation of Nrf2 is considered to be an important approach for preventing chronic diseases triggered by stresses and toxins, including cancer. Despite extensive studies suggested that the Keap1-Nrf2 signaling pathway is subject to multiple layers of regulation at the transcriptional, translational, and post-translational levels, the potential epigenetic regulation of Nrf2 and Keap1 has begun to be recognized only in recent years. Epigenetic modifications, heritable alterations in gene expression that occur without changes in the primary DNA sequence, have been reported to be profoundly involved in oxidative stress responses. In this review, we discuss the latest findings regarding the epigenetic regulation of Keap1-Nrf2 signaling by DNA methylation, histone modification, and microRNAs. The crosstalk among these epigenetic modifications in the regulation of Keap1-Nrf2 signaling pathways is also discussed. Studies of the epigenetic modification of Nrf2 and Keap1 have not only enhanced our understanding of this complex cellular defense system but have also provided potential new therapeutic targets for the prevention of certain diseases.
Keywords: Nrf2, Keap1, Epigenetic, DNA methylation, histone modification, microRNAs
1. Introduction
Mammalian cells are constantly exposed to oxidative stresses that are regarded as some of the most important and ubiquitous causes of neoplastic, metabolic, cardiovascular, neurodegenerative, and many other chronic diseases 【Molecular Basis of the Keap1-Nrf2 system function by Masayuki Yamamoto in this issue】. To deal with the deleterious effects of oxidative stresses, cells have evolved an elaborate and powerful cellular defense machinery against reactive oxygen species (ROS). Central to this cellular defensive machinery is the transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) and its negative regulator kelch-like ECH-associated protein 1 (Keap1). Under basal conditions, Keap1 acts as an adaptor between Nrf2 and the ubiquitination ligase Cullin-3 (Cul3) and promotes the proteasomal degradation of Nrf2. Upon modification of specific thiols, Keap1 allows Nrf2 to translocate into nucleus and activate the expression of a wide array of antioxidative metabolizing/detoxifying and many other genes by binding to the antioxidant response element (ARE) in their regulatory regions 【Structural Basis of KEAP1 Interactions with Nrf2 by Alex Bullock in this issue】[1]. In addition to the Keap1-Nrf2 interaction, the transcriptional activity of Nrf2 is regulated by a complex signaling network, which is discussed in detail elsewhere in the present issue 【Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues and energy status, and its contribution to the regulation of antioxidant status, detoxification and metabolism by J.D. Hayes】[2].
The Keap1-Nrf2 signaling axis is the pivotal coordinator of cytoprotective responses towards oxidative/electrophilic stimuli and protects cells against chemical insults. Therefore, activation of Keap1-Nrf2 signaling has been widely accepted as an important strategy to prevent oxidative damage-related chronic diseases, including cancer [3]. However, upregulated Nrf2 activity in cancerous cells leads to resistance to radio- and chemotherapies [4]. Furthermore, activation of Nrf2 confers neoplastic cells with growth and survival advantages during their transformation and progression [5]. Indeed, although Nrf2-knockout mice were more susceptible to chemical-induced carcinogenesis than control mice, high expression of Nrf2 in tumors predicts a poor prognosis, and inhibition of Nrf2 sensitizes cancer cells to chemotherapeutic drugs [6]. Such apparently paradoxical roles of Nrf2 in different stages of cancer initiation and progression are essentially arose from the “double-edged sword” nature of ROS in cancer, and have been extensively investigated and reviewed [5]. However, the roles of Nrf2 in other ROS-related diseases such as neurodegenerative diseases, diabetes, and cardiovascular disease are simply protective. Thus, the mechanisms regulating Keap1-Nrf2 signaling are expected to produce different even opposite outcomes, and the preventive or therapeutic applications of Keap1-Nrf2 signaling modulators have to be carefully evaluated according to the context.
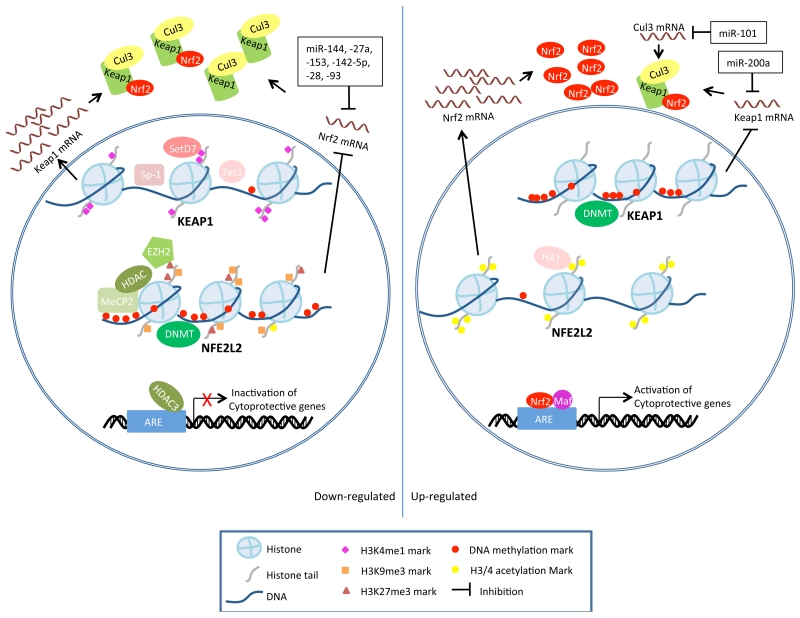
Importantly, very different expression levels and activities of Keap1 and Nrf2 have been observed at different stages of different pathological processes. Functional somatic mutations or single nucleotide polymorphisms (SNPs) of Keap1 or Nrf2 occur in many types of cancer and have been utilized to explain the variations in expression and activity of Keap1 and Nrf2 【Functional Polymorphisms in NRF2: Implications for Human, by Steven Kleeberger in this issue】 [4]. However, although the expression of Keap1 and Nrf2 exhibits significant inter-individual variations in these cancers, somatic mutations exist in only a small portion of cancer tissues [7, 8]. For example, Solis et al. detected nuclear Nrf2 expression in 26% and low or absent Keap1 expression in 56% of non-small cell lung cancers (NSCLCs), and found that this expression correlated with clinicopathologic characteristics; nevertheless, mutations in the NFE2L2 and KEAP1 genes were very uncommon in the examined samples [8]. Therefore, alternative mechanisms must exist to regulate Keap1 and Nrf2 expression. Recently, a body of evidence is emerging that shows that the Keap1-Nrf2 signaling can be regulated by epigenetic mechanisms in cancers as well as in other diseases, and the present review will focus on the epigenetic regulation of Keap1-Nrf2 signaling as schematically depicted in Figure 1.
Figure 1.
Schematic model depicting epigenetic modifications of Nrf2 and Keap1. Left and right panel shows epigenetic modifications that lead to down- and up-regulation of Keap1-Nrf2 signaling, respectively.
2. Epigenetic modifications
The term epigenetics refers to the study of heritable alterations in gene expression that are not due to changes in the primary DNA sequence. DNA-based mechanisms (DNA methylation and histone modification) and RNA-based mechanisms (non-coding RNA) are known to mediate these heritable gene expression alterations [9]. The addition of a methyl group to cytosine bases and covalent modifications of histones at a given promoter can modulate DNA accessibility and chromatin structure, ultimately regulating gene transcription [9]. By targeting mRNA degradation, translation inhibition, and chromatin architecture, non-coding RNAs interfere with various levels of gene expression [10]. Furthermore, interactions between DNA methylation, histone modification, and non-coding RNAs in controlling the epigenome landscape have been recognized; however, the regulatory network remains elusive.
2.1. Epigenetic modifications and human diseases
Epigenetic mechanisms, together with genetic factors, are fundamental for maintaining cellular differentiation and mammalian development [11]. However, the disruption of either epigenetic modifications or genetic functions is associated with abnormalities in various signaling pathways and can lead to the pathogenesis of many human disorders. Unlike genetic changes, aberrant epigenetic marks tend to be acquired in a gradual process [12]. Long-lasting effects of environmental factors and the aging process introduce alterations in the landscape of the epigenome [13, 14]. Thus, studies of epigenetic disruptions are primarily focused on chronic diseases, especially cancer. Studies of epigenetic abnormalities associated with carcinogenesis suggest that epigenetic alterations may interact with genetic dysregulation at all stages to initiate and promote cancer [15, 16]. Global DNA hypomethylation, regional hypermethylation at specific promoters, global reduction of monoacetylated H4K16, and overall microRNA (miRNA) down-regulation are characteristics of cancer cells [17, 18]. In addition, the important role of epigenetic modifications in diabetes [19], autoimmune diseases [20], cardiovascular diseases [21], and neurodegenerative diseases [22] has been recognized.
2.2. Epigenetic therapy
Unlike genetic mutations, epigenetic disruptions in diseases are potentially reversible. For example, genes that have been transcriptionally silenced by epigenetic modifications can be reactivated through epigenetic mechanisms because these genes remain intact, whereas genetic mutations are permanent. Given that epigenetic abnormalities play an important role in human diseases, including cancer, increasing efforts have been focused on the development of agents that target epigenetic mechanisms. Successful examples of the use of epigenetic therapies in the treatment of cancer include hypomethylating drugs and histone deacetylase inhibitors that have been approved by the US Food and Drug Administration (FDA) [23]. In addition, the second generation of novel potential epigenetic therapies, such as histone methyltrasferase inhibitors and epigenetic reader protein inhibitors, have been discovered and are currently under investigation [24]. Furthermore, numerous studies have suggested that the consumption of dietary phytochemicals may alter epigenetic modifications and reverse abnormal gene transcription, thereby preventing certain diseases, including cancer [25].
2.3. Epigenetic modifications and oxidative stress
Oxidative stresses are involved in almost all chronic diseases including ageing. Interestingly, epigenetic mechanisms have been reported to be profoundly involved in oxidative stress responses. ROS, such as hydroxyl radicals, can cause serious DNA lesions and lead to mutagenesis; such lesions can also result in global DNA hypomethylation [26, 27]. For instance, the DNA oxidization product 8-OHdG strongly inhibits the methylation of adjacent cytosines and impairs the binding of methyl-CpG-binding domain proteins (MBDs) [27]. Moreover, demethylation of methyl-CpG by Ten-eleven translocation (TET) enzymes, a family of Fe(II)/α-ketoglutarate-dependent dioxygenases, is a highly oxidative stress labile process. It involves serial oxidation of 5-methylcytosine to 5-hydroxymethylcytosine, which inhibits DNA methyltransferase 1 (DNMT1) recognition, then to 5-formylcytosine and 5-carboxylcytosine, and finally be excised from DNA by glycosylases [26]. Furthermore, histone modifications can also be modulated by oxidative stresses [28]. In addition, many Nrf2-activating chemopreventive compounds have been identified as epigenetic modulators, and the expression of several Nrf2-target genes has been found to be regulated epigenetically [29-32]. Therefore, it is expectable that complex interactions exist between Keap1-Nrf2 signaling and epigenetic modifications.
3. Regulation of the Keap1-Nrf2 signaling pathway by DNA methylation
DNA methylation is widely observed in organisms ranging from prokaryotic bacteria to vertebrates; however, in vertebrates, heritable methylation only occurs at the 5 position of the cytosine pyrimidine ring in CpG dinucleotides. CpG methylation serves as an epigenetic mechanism to memorize the transcriptional state [33]. In mammalian cells, DNA methylation patterns are established during embryogenesis and development or under certain physiological and pathological conditions by de novo DNMT3a and DNMT3b, then maintained by DNMT1 during DNA replication. On the other hand, DNA demethylation can occur through active demethylation by TET enzymes or through passive demethylation caused by the absence of DNMT1 activity during DNA replication [34]. Approximately 60% of human genes contain clusters of CpG sites called CpG islands in their GC-rich promoter regions, and their expression can be epigenetically regulated by DNA methylation [35]. The methyl moiety lies in the major groove of the DNA helix and can potentially interact with many DNA-binding proteins. CpG methylation in the binding sequences can inhibit the binding of transcription factors and the initiation of transcription, or it can attract MBDs, such as MBD1, MBD2, and MeCP2, which can recruit co-repressor complexes to silence gene transcription [35]. In addition, DNA methylation also collaborates with histone modifications to regulate chromatin accessibility and gene transcription. MBDs often associate with histone deacetylases (HDACs) and histone lysine methyltransferases (HKMTs) to regulate histone modifications [33].
The hypermethylation of several genes regulated by Keap1-Nrf2 signaling has been investigated for decades. For example, the hypermethylation of CpG islands in the promoter region and expression silencing of pi class glutathione S-transferase (GSTPi) have been observed in prostate cancers [31]. NAD(P)H:quinone oxidoreductase 1 (NQO1), UDP-glucuronosyltransferases 1A1 (UGT1A1), glutathione peroxidase (GPX), and manganese superoxide dismutase (MnSOD) have also been reported to be regulated by promoter methylation [30, 36-38]. Regulation of Keap1 and Nrf2 expression by DNA methylation has been investigated in recent years and is discussed below and summarized in Table 1.
Table 1.
DNA methylation regulates Keap1-Nrf2 signaling pathway.
| Target | Diseases | Methylation | Experimental Model | Impact on Keap1-Nrf2 | Outcomes | Ref |
|---|---|---|---|---|---|---|
| Nrf2 | Prostate cancer | ↑ | TRAMP mice and TRAMP-C1 cells | ↓ Nrf2, NQO1, GST mu | Prostate carcinogenesis | [40-42] |
| ↑ | Human prostate cancer tissues | ↓ Nrf2 | Advanced stages | [43] | ||
| ↓ | TRAMP-C1 or LNCap cells treated by 5-Aza/TSA |
↑ Nrf2, NQO1, HO-1 ↓ DNMTs, HDACs |
Not Applicable | [42, 43] | ||
| ↓ | TRAMP-C1 cells or TRAMP mice treated by γ-TmT, curcumin, sulforaphane, 3,3′- diindolylmethane, or Z-Ligustilide |
↑ Nrf2, NQO1, HO-1 ↓ DNMT, HDAC |
Inhibition of carcinogenesis |
[44, 49- 52] |
||
| Skin cancer | ↓ | Mouse skin epidermal JB6 P+ cells treated by sulforaphane, apigenin, or Tanshinone IIA |
↑ Nrf2, NQO1, HO-1 ↓ DNMT, HDAC |
Inhibition of TPA-induced transformation |
[53-55] | |
|
| ||||||
| Keap1 | Lung cancer | ↑ | Human lung cancer tissues and cell lines |
↓ Keap1 | Not Applicable | [56, 57] |
| Human non-small cell lung cancer | Not Applicable | Worse prognosis | [58] | |||
| Gliomas | ↑ | Human malignant gliomas | ↓ Keap1 | Better prognosis | [59] | |
| Breast cancer | ↑ | Primary breast cancers and pre- invasive lesions |
↓ Keap1 | Higher mortality in triple- negative; reduced relapse |
[60] | |
| Colorectal cancer |
↑ | Colorectal cancer cell lines and surgical specimens |
↓ Keap1 ↑ Nrf2, NQO1, AKR1C1 |
Not Applicable | [61] | |
| Prostate cancer | ↑ | Prostate cancer cell lines | ↓ Keap1; ↑ Nrf2, HO-1, NQO1, Gclc |
Increased tumor growth, enhanced chemo- and radio-resistance |
[63] | |
| Thyroid cancers | ↑ | Papillary thyroid carcinoma | ↓ Keap1 ↑ Nrf2-regulated genes |
Not Applicable | [64] | |
| Age-related cataracts |
↓ | Cataractous lenses with increasing age or in diabetes patients; human lens epithelial cells treated by homocysteine, valproic acid, methylglyoxal or selenite |
↑ Keap1, TET1 ↓ Nrf2, CAT, GST, DNMT |
Elevated ROS, age-related cataracts |
[65, 67- 72] |
|
| Diabetic cardiomyopathy |
↓ | Myocardial biopsies of non- diabetic and type-2 diabetic cardiomyopathy patients |
↓ Keap1 ↑ Nrf2-regulated genes |
Failure of Nrf2 mediated antioxidant system |
[73] | |
3.1. Regulation of Nrf2 expression by DNA methylation
The protein expression of Nrf2 and the Nrf2-targeted gene heme oxygenase 1 (HO-1) was abolished in skin tumors in a skin cancer mouse model [39]. Similar results were obtained in a transgenic adenocarcinoma of mouse prostate (TRAMP) model, in which the expression of Nrf2 and its downstream target genes, such as UGT1A1, glutathione S-transferase Mu 1 (GSTM1), and NQO1, were gradually down-regulated in prostate tumors during tumorigenesis [40]. Frolich et al. also reported that the expression of Nrf2 and GST mu family genes was significantly decreased in TRAMP prostate tumors [41]. More importantly, the expression of Nrf2 and several downstream genes such as GST and NQO1 has been found to be decreased in human prostate cancers compared with normal epithelia or localized adenoma [41, 42]. Yu et al. identified CpG islands in the promoter regions of human, rat, and mouse NFE2L2 genes and demonstrated that the suppression of Nrf2 expression in TRAMP prostate tumors and TRAMP C1 cells was mediated by the hypermethylation of specific CpG sites in the Nrf2 promoter [41, 42]. Further study by Khor et al. using human prostate cancer samples identified three specific CpG sites in the Nrf2 promoter that were hypermethylated during prostate cancer progression [43]. Moreover, treatment of TRAMP cells with the DNMT inhibitor 5-aza-2′-deoxycytidine (5-aza) and the HDAC inhibitor trichostatin A (TSA) could restore Nrf2 expression, which was accompanied by the dissociation of MBD2, MeCP2, and methylated histones [42].
Interestingly, inhibition of methylation or demethylation of the Nrf2 promoter has been found to be involved in the action of many chemopreventive chemicals. Dietary feeding of a γ-tocopherol–rich mixture of tocopherols (γ-TmT) dose-dependently suppressed prostate tumorigenesis and hypermethylation of the Nrf2 promoter in TRAMP mice and was associated with higher Nrf2 and NQO1 protein levels. γ-TmT treatment inhibited the protein expression of DNMT1, DNMT3a, and DNMT3b in the prostate of TRAMP mice, suggesting that γ-TmT inhibited both de novo and sustained methylation [44]. It has been well documented that many dietary cancer chemopreventive compounds, including curcumin [45], isothiocyanates [46], tea polyphenols [47], and genistein [48], are epigenetic modifiers [29]. Some of these compounds, such as curcumin, sulforaphane, 3,3′-diindolylmethane, and Z-Ligustilide (from the traditional Chinese medicine Radix Angelicae Sinensis), were also found to demethylate the Nrf2 promoter and re-activate Nrf2 signaling in the prostate of TRAMP mice or TRAMP C1 cells, possibly through the inhibition of DNMT and HDAC expression [49-52]. The CpG sites in the promoter region of Nrf2 are heavily methylated in mouse skin epidermal JB6 P+ cells and could be demethylated by sulforaphane, apigenin, or Tanshinone IIA. Such demethylation was associated with suppression of TPA-induced transformation, reactivation of Nrf2 signaling, and expression of Nrf2 target genes, along with the inhibition of protein expression of DNMTs and HDACs [53-55]. These findings suggest that Nrf2 expression during carcinogenesis can be epigenetically regulated through DNA methylation at specific CpG sites in its promoter and that such mechanisms could be targeted for cancer prevention. However, given the paradoxical roles of Nrf2 in the process of carcinogenesis, the exact impact of Nrf2 modulators on cancer would be context-sensitive.
3.2. Regulation of Keap1 expression by DNA methylation
Loss of Keap1 function has been observed in many cancer tissues and is regarded as the main cause of Nrf2 over-activation. In addition to somatic mutations, the epigenetic regulation of Keap1 has been investigated in human tissues and cells of different diseases. Wang et al. first showed that Keap1 is highly expressed in BEAS-2B human normal bronchial epithelial cells but is down-regulated in a series of lung cancer cell lines and human lung cancer tissues. This down-regulation was accompanied by the hypermethylation of CpG sites in the Keap1 promoter region and could be restored by 5-aza treatment [56]. Further studies by the same group suggested that hypermethylation of the Keap1 promoter abrogated the binding of stimulating protein-1 (SP-1), and 5-aza treatment restored SP-1 binding to the Keap1 promoter [57]. In another study, using 47 pairs of NSCLC tissues and normal specimens, promoter methylation was detected in 47% of NSCLCs but in none of the normal tissues, whereas somatic mutations were detected in 15% of NSCLCs; patients harboring both alterations had the worst prognosis [58].
Similar results have been obtained in other cancers, including malignant gliomas and breast, colorectal, prostate, thyroid, and head and neck cancer cells. Frequent promoter hypermethylation and correlated down-regulation of Keap1 expression were observed in malignant gliomas and contributed to resistance to therapies and disease progression [59]. Aberrant Keap1 promoter methylation was detected in more than half of primary breast cancers and pre-invasive lesions but not in normal breast tissues, whereas no Keap1 mutations were detected in examined breast cancer cases. Methylation was more frequent in ER-positive, HER2-negative than in triple-negative breast cancers, and Keap1 promoter hypermethylation predicted higher mortality risk in triple-negative patients [60]. Keap1 promoter methylation was also observed in 53% of colorectal cancer tissues, in 25% of adjacent normal mucosa, and in 8 out of 10 colorectal cancer cell lines analyzed [61, 62]. Loss of Keap1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. In addition to point mutations of Keap1 in various prostate cancer cell lines, down-regulation of Keap1 expression by promoter hypermethylation was identified in DU-145 prostate cancer cells [63]. Hypermethylation is the major inactivating mechanism of Keap1 in thyroid cancers (70.6%) and head and neck cancers (29.3%) and is associated with a worse prognosis [64]. Here again we saw the uncertainty of the outcomes produced by epigenetic regulation of Keap1-Nrf2 signaling in cancers: while silencing of Nrf2 by DNA methylation is implicated in carcinogenesis, activation of Nrf2 signaling by hypermethylation of Keap1 promoter is also associated with tumor progression and resistance to therapies.
On the other hand, Keap1 promoter hypermethylation in oxidative stress-related diseases other than cancers plays mainly protective roles. Increased oxidative stress during chronic aging is a major pathological factor of age-related cataracts (ARCs), especially in diabetes patients; therefore, impaired Keap1-Nrf2 signaling is proposed to be involved in the pathogenesis of ARCs [65]. Although no SNPs in Nrf2 or Keap1 were found to be associated with Alzheimer’s disease or age-related cataracts [66], demethylation of the Keap1 promoter accompanied by increased Keap1 and decreased Nrf2 expression was identified in cataractous lenses with increasing age or in diabetes patients, which may lead to failure of the cytoprotective system and increased oxidative stress [67, 68]. Exposure to homocysteine resulted in endoplasmic reticulum (ER) stress and the suppression of Keap1-Nrf2 signaling by ER-associated degradation (ERAD) and demethylation of the Keap1 promoter, elevated ROS generation and lens oxidation [65]. Treatment of human lens epithelial cells (HLEC) with acetyllcarnitine prevented the effects of homocysteine and significantly increased the levels of Nrf2 and downstream antioxidant genes [69]. Sodium selenite has been employed to induce cataracts in animal models and can suppress Keap1-Nrf2 signaling in HLECs by ERAD and Keap1 promoter demethylation, possibly by both reducing DNMT1/3a protein levels and inducing Tet1 expression [70]. Methylglyoxal and valproic acid promote lenticular protein oxidation and cataract formation by almost the same mechanisms as selenite [71, 72]. In addition, demethylation of the Keap1 promoter and increased Keap1 expression have been observed in diabetic cardiomyopathy, thus suppressing Nrf2 activity and disturbing the redox balance [73].
According to the observations described above, it would be of therapeutic interest to determine whether demethylation of the Keap1 promoter in neoplastic tissues could suppress tumor progression and resistance to therapies or whether the activation of Nrf2 signaling in lens epithelial cells could prevent cataract formation or the onset of other oxidative stress-initiated diseases.
4. Histone modifications and the Keap1-Nrf2 signaling pathway
Eukaryotic DNA is wrapped by octomers of four core histone proteins into repeating nucleosomes, which are further folded into chromatin fibers [74]. This highly organized and dynamic protein-DNA complex has two structurally and functionally distinguishable configurations, namely, heterochromatin and euchromatin. Heterochromatin represents a highly condensed structure with repressed gene transcription as a result of low accessibility of transcription factors and RNA polymerase II to their recognition sequences, whereas euchromatin is loosely packed and more easily transcribed [75]. It is suggested that posttranslational modifications of specific residues on the N-terminal tails of histones play a pivotal role in the modulation of the chromatin structure; ultimately, they regulate the transcriptional activity of a wide variety of genes. Here, we review and discuss the mutual effects of histone modifications and the Keap1-Nrf2 signaling pathway. Histone modifications shown to regulate the Keap1-Nrf2 signaling are summarized in Table 2.
Table 2.
Histone modifications regulate Keap1-Nrf2 signaling pathway.
| Target | Diseases | Modifications | Enzymes | Experimental Model | Impact on Keap1-Nrf2 and outcomes | Ref |
|---|---|---|---|---|---|---|
| Nrf2 | Neuroinflammation and neurodegenerative diseases |
Deacetylation of histones H3 and H4 |
HDACs | Astrocyte-rich cultures exposed to conditioned medium |
High level of HDAC activity leads to: ↓Nrf2 and γGCL-M; ↓Nrf2-mediated antioxidant defense |
[78] |
| Chronic obstructive pulmonary disease |
Histone acetylation |
HDAC2 | Human airway epithelial BEAS2B cells, monocyte- derived macrophages from COPD patients |
Treatment of HDAC inhibitor leads to: ↓Nrf2 stability; ↓Nrf2-regulated HO-1 expression; ↑sensitivity to oxidative stress |
[80] | |
| Human non-small cell lung cancer |
H3K27me3 | EZH2 | A549 cells, human non-small cell lung cancer patients, nu/nu mice |
Low expression of EZH2 leads to: ↑Nrf2, NQO1, and HO-1 |
[93] | |
|
| ||||||
| Keap1 | Cerebral ischemic injury | Histone acetylation |
HDACs | Permanent middle cerebral artery occlusion model in mice, cortical neuronal cells, and RAW 264.7 cells |
Treatment of HDAC inhibitor leads to: ↓Keap1; ↑Nrf2 nuclear translocation; ↑Nrf2-ARE binding; ↑HO-1, NQO1, and GCLC; ↑neuronal cell viability; and ↓cerebral ischemic injury |
[79] |
| Diebetic retinopathy | H3K4me1 | SetD7 | Bovine retinal endothelial cells, retina from rats and human donors |
Hyperglycemia leads to: ↑SetD7; ↑binding of Sp1 at Keap1; ↑Keap1; and ↓Nrf2, Gclc, and HO-1. |
[96] | |
|
| ||||||
| HO-1 E1 enhancer |
Pathological processes of inflammation and cancer |
Histone hypoacetylation |
HDAC3 | HepG2, HEK293, and L929 cells |
↓ ARE-dependent gene expression | [77] |
|
| ||||||
| Sod2 | Diebetic retinopathy | H3K4 methylation |
LSD1 | Bovine retinal endothelial cells, retina from rats and human donors |
Hyperglycemia leads to: ↑binding of LSD1 and Sp1 at Sod2; ↓Sod2 expression. |
[97] |
|
| ||||||
| Gclc-ARE4 | Diebetic retinopathy | H3K4 methylation |
LSD1, KDM5A |
Bovine retinal endothelial cells, retina from rats and human donors |
Hyperglycemia leads to: ↓binding of Nrf2 at Gclc-ARE4 and Gclc transcripts |
[98] |
4.1. Histone acetylation and the Keap1-Nrf2 signaling pathway
There is compelling evidence that the acetylation of histones neutralizes the positive charge, destabilizes the nucleosome structure, and promotes the accessibility of transcriptional factors to a genetic locus, thereby activating gene transcription, whereas histone deacetylation leads to gene silencing [76]. Histone acetyltransferases (HATs) and HDACs, which add and remove the acetyl groups, respectively, constitute a group of enzymes that dynamically regulate histone acetylation/deacetylation and gene transcriptional activity. Liu et al. reported that class 1 HDACs (1, 2, and 3) inhibit ARE-dependent gene expression. Furthermore, this study demonstrated that the Nf-kB subunit p65 suppresses the Nrf2-ARE pathway via selective deprivation of CREB-binding protein (CBP, a member of HAT) from Nrf2 and promotion of the recruitment of HDAC3 to ARE. Specifically, p65 enhances the interaction of HDAC3 with MafK (a known dimerization partner with Nrf2), facilitates the recruitment of endogenous HDAC3 to the ARE element, helps to maintain the histone hypoacetylation state in the local chromosome and hence represses ARE-dependent gene expression [77]. This mechanism provides direct evidence regarding the involvement of HDAC3 in the negative regulation of the Nrf2 pathway by NF-κB in response to inflammatory-related stimuli. Similarly, the impact of HDACs on the inhibition of Nrf2-mediated antioxidant defense in neuroinflammation has been investigated. Exposure to conditioned medium from lipopolysaccharide (LPS)-treated microglia (MCM10) induced HDAC activity in astrocyte-rich cultures, which correlated with decreased acetylation in histones (H3 and H4) and reduced expression of Nrf2 and its target gene γ-glutamyl cysteine ligase modulatory subunit (γGCL-M). Notably, treatment with HDAC inhibitors, such as valproic acid and TSA, markedly elevated acetylation in H3 and H4, restored the Nrf2-mediated antioxidant responses, and thus resulted in an increased resistance to oxidative stress (H2O2) in astrocyte-rich cultures exposed to MCM10 [78]. In addition to protecting against neuroinflammation, the HDAC inhibitor also exhibited a promising effect in the protection of neuronal cell viability from oxygen-glucose deprivation and the attenuation of cerebral ischemic injury in the ischemic stroke mouse model via Nrf2 activation. Experimental evidence clearly showed that HDAC inhibitors activated the Nrf2 signaling pathway and up-regulated the Nrf2 downstream targets HO-1, NQO1, and glutamate-cysteine ligase catalytic subunit (GCLC) by suppressing Keap1 and promoting dissociation of Keap1 from Nrf2, Nrf2 nuclear translocation, and Nrf2-ARE binding. Importantly, the protective effect of HDAC inhibitors in cerebral ischemia was abolished in Nrf2-deficient mice [79]. Therefore, activation of Nrf2 through HDAC inhibition may provide a promising therapeutic strategy for preventing neural damage in ischemic stroke.
However, inhibition of HDAC does not always lead to Nrf2 activation. Mercado et al. reported that Nrf2 activity is impaired as a result of decreased Nrf2 stability in the presence of TSA (an HDAC inhibitor) in BEAS2B (human airway epithelial) cells or in HDAC2-knockdown cells. TSA treatment also significantly ameliorated the elevation of HO-1 expression in mice exposed to cigarette smoke. In addition, a significant correlation between the expression of HDAC2 and Nrf2 was found in monocyte-derived macrophages obtained from chronic obstructive pulmonary disease (COPD) patients [80]. Thus, a vicious circle in the pathogenesis of COPD is proposed: reduced HDAC2 activity observed in COPD as a result of oxidative stress could suppress Nrf2 stability and activity, thereby increasing oxidative stress due to limited antioxidant responses, which then further impairs HDAC2 activity [80]. In fact, several studies have suggested that Nrf2 plays an important role in lung inflammation by modulating HDAC activity. For example, Nrf2-deficient mice were found to have diminished HDAC2 levels in the lungs and increased susceptibility to chronic cigarette smoke- and LPS-induced lung inflammation, which were not reversed by steroid therapy [81]. HDAC6, a critical regulator of autophagy-mediated airway inflammatory responses, was elevated in the lungs of Nrf2-deficient mice in response to cigarette smoke exposure [82]. Taken together, these findings show that the activity of the Keap1-Nrf2 signaling pathway is epigenetically regulated by HDACs; conversely, Nrf2-mediated oxidative stress responses may have an epigenetic impact on other signaling pathways through the modulation of HDAC activity. The involvement of the Nrf2-HDAC axis in the pathogenesis of human disorders, especially in inflammatory diseases, requires further investigation.
The detailed mechanisms underlying the regulation of the Keap1-Nrf2 pathway by HDAC/HAT are not yet fully understood. It appears that HDAC and HAT and their inhibitors not only regulate Nrf2 activity and ARE-dependent gene expression via the adjustment of histone acetylation in the promoter regions [77, 78] but also selectively modulate the acetylation of Nrf2 independently of histones [83, 84]. Lysine residues within the Nrf2 Neh1 DNA-binding domain can be acetylated directly by HAT (p300/CBP) in response to sodium arsenite-induced stress, and this acetylation is followed by the elevated expression of ARE-dependent genes [84]. hMOF, the HAT required for histone H4K16 acetylation, can acetylate Nrf2 at Lys588. In human NSCLC tissues, hMOF-mediated acetylation of Nrf2 increased its nuclear retention and the transcription of its downstream genes, subsequently modulating tumor growth and drug resistance [85]. In addition, the acetylation of Lys588 and Lys 591 in the Neh3 domain by a selective inhibitor of sirtuin 1 (SIRT1, a class III HDAC) favors the nuclear localization of Nrf2, resulting in enhanced binding of Nrf2 to ARE and thereby increasing Nrf2-mediated gene expression. By contrast, a SIRT1 activator induces deacetylation, suppressing Nrf2 signaling accordingly [83].
The regulation of phase II detoxification enzymes by histone acetylation/deacetylation has also been investigated. It was previously reported that the acetylation of histones H3 and H4 on the chromatin of the promoter regions of the glutathione S-transferase placental form (GSTP) and GSTP enhancer 1 (GPE1) occurred in the H4IIE hepatoma cell line, where GSTP expression is activated, but not in the normal liver [86]. Monocytic leukemia zinc-finger protein (MOZ), a member of HAT, stimulates GSTP promoter activity in the presence of Nrf2. Although the precise mechanism by which histone acetylation regulates the gene transcription of GSTP remains unclear, the elevation of both MOZ and Nrf2 levels may be required [87]. UGT1A is another example of phase II enzymes regulated by histone acetylation. Gender-specific repression of UGT1A is controlled via chromatin remodeling through the recruitment of estrogen receptor alpha (ERα), HDAC1, and HDAC 2 to the xenobiotic response element (XRE) sites [88].
4.2. Histone methylation and the Keap1-Nrf2 signaling pathway
Histone methylation is also a critical player in the regulation of chromatin compaction and gene expression. The methylation of histones occurs on all basic residues, including arginines, lysines, and histidines. Different lysine sites can be mono (me1), di (me2) or tri (me3) methylated [89]. Depending on which residue is methylated and the degree of methylation, histone methylation can lead to either gene activation or suppression. For example, enhancer of zeste homolog 2 (EZH2) specifically catalyzes the trimethylation of histone H3 lysine 27 (H3K27me3) and leads to transcription repression [90], whereas histone-lysine N-methyltransferase (SetD7) monomethylates histone H3 lysine 4 (H3K4me1) and favors the binding of the transcription factor [91]. Abnormal expression of histone methyltransferases (HMT) and histone demethyltransferases (HDMs) may write an aberrant epigenetic mark on the histone tail, influencing gene expression and resulting in disease. The proteasome inhibitor MG132 increases the degradation of EZH2 through a compensatory Nrf1- and Nrf2-dependent increase in the proteasome subunit level [92]. Li and coworkers showed that decreased EZH2 expression significantly correlated with the elevated expression of Nrf2, NQO1, and HO1 in lung cancer tissues and cell lines, which was mainly attributed to a decrease in H3K27me3 in the Nrf2 promoter but not the NQO1 or HO1 promoter. Interestingly, the inhibitory effect of EZH2 on lung cancer growth in vitro and in vivo was abolished by Nrf2 deficiency [93]. Collectively, these data suggest that EZH2 suppresses lung cancer growth by inhibiting Nrf2 expression via H3K27 trimethylation in the promoter region. However, the up-regulated EZH2 level in a number of cancers, including prostate cancer, breast cancer, lymphomas, gastric cancer, hepatocellular carcinoma, and bladder cancer (see review [90]), suggests that EZH2 can be tumorigenic. The correlation of overexpression of EZH2 and the Keap1-Nrf2 signaling pathway in cancer tissues needs to be investigated. Furthermore, given the promising effect of EZH2 inhibitors in the treatment of cancer [94, 95], it will be interesting to explore the effect of EZH2 inhibitors on the Nrf2 pathway in cancer cells.
The modification of the Keap1-Nrf2 signaling pathway by methylation of histone 3 lysine 4 in diabetic retinopathy has been identified [96-98]. The diabetic environment induces oxidant production in the retina and its capillary cells and decreases the antioxidant response [99]. Mitochondrial superoxide dismutase (Sod2), an Nrf2 downstream target, becomes subnormal in diabetes due to lysine-specific demethylase-1 (LSD1)-mediated reduction of H3K4me1 and - me2 levels at the retinal Sod2 promoter. Sod2 inhibition may result in increased mitochondrial superoxide and the development of diabetic retinopathy [97]. In addition to Sod2, suppression of GCLC, an enzyme that is important for the biosynthesis of GSH, has been implicated in the progression of diabetic retinopathy. Specifically, reduced H3K4me1 and H3K4me3 and increased H3K4me2 at Gclc-ARE4 in the retina in diabetes results in impaired binding of Nrf2 at Gclc-ARE4. One possible reason for such an increase in H3K4me2 could be that activated JARID family protein (KDM5A), an H3K4me3 demethylase, demethylates H3K4me3, resulting in elevated H3K4me2 [98]. As an upper regulator, the association of Keap1 and Nrf2 could be regulated through epigenetic mechanisms, further impairing the antioxidant responses in diabetes. Indeed, hyperglycemia increases the binding of Sp1 at the Keap1 promoter through enrichment of H3K4me1 due to the activation of SetD7. In line with this finding, SetD7 knockdown leads to a lower H3K4me1 level at the Keap1 promoter and reduced Sp1 binding, accompanied by the restoration of Nrf2 in high-glucose conditions. Following Nrf2 restoration, the binding of Nrf2 at the Gclc promoter and the expression of GCLC and HO-1 were enhanced, which is beneficial to rebalance the oxidative stress in a diabetic environment [96]. In addition, as epigenetic modifications can persist in a system, the above-mentioned alterations of the histone methylation level may continue to suppress the expression of antioxidant genes, resulting in high oxidative stress, even after the termination of the hyperglycemic challenge, known as metabolic memory phenomenon. The reestablishment of normal glucose conditions failed to reverse the abnormal methylation marks of H3K4 at the Sod2, Gclc-ARE4, and Keap1 promoters; therefore, the activity of Nrf2 and its downstream genes continues to be compromised [96-98]. According to these studies, the knockdown of the key enzymes that catalyze histone methylation appears to be effective in restoring the antioxidant defense system; thus, specific inhibitors of these enzymes may potentially protect diabetic retinopathy by regulating the Keap1-Nrf2 signaling pathway.
4.3. Histone readers and the Keap1-Nrf2 signaling pathway
Histone readers are the proteins that recognize the histone modifications that are deposited or removed by histone writers or erasers [100]. These readers play an essential role in the translation of a “histone code” for gene transcription. The bromodomain and extraterminal (BET) proteins are perhaps the most thoroughly characterized acetyl-lysine readers [101]. After binding to acetylated lysine residues, BET proteins may interact with transcription factors and chromatin remodeling complexes, recruiting them to gene promoters and thus activating or inactivating gene transcription [102]. It was recently found that BET proteins are involved in the regulation of antioxidant gene expression [103, 104]. BET proteins act as negative regulators of Nrf2 signaling; inhibition of BET proteins by genetic knockdown or the specific inhibitor JQ1 activates Nrf2-dependent transcription, increases the expression of the antioxidant genes HO-1, NQO1, and GCLC, and further ameliorates the ROS production induced by H2O2. BET proteins may interact directly with Nrf2 and are constitutively present at Nrf2-binding sites on the promoters of HO-1 and NQO1 [103]. In addition, Hussong et al. showed that BRD4, a member of the BET protein family, is a key mediator of Keap1 transcription under stress. However, under normal conditions, BRD4 appears to modulate anti-oxidative responses by directly targeting Sp-1 binding sites in the inducible heme oxygenase 1 (HMOX1) promoter [104]. However, the roles of histone readers in the interpretation of the histone code, chromatin remodeling, and recruitment of the repressive complex or co-activators to the Nrf2-regulated gene promoter remain unclear and need to be investigated in depth in the future.
5. Interaction of miRNAs and the Keap1-Nrf2 signaling pathway
miRNAs are endogenous short non-coding RNAs that usually contain 20-22 nucleotides. By complementary pairing with mRNA sequences, miRNAs inhibit the translation of mRNAs in ribosomes and/or facilitate the degradation of mRNA molecules. Thus, miRNAs represent another category of epigenetic mechanism, which regulates gene expression at the post-transcriptional level, mostly in a “fine-tuning” manner. During recent decades, increasing efforts have been made to profile miRNA expression patterns and characterize miRNA functions to identify novel diagnostic markers and therapeutic targets. Among these studies, a number of miRNAs have been reported to affect the Keap1-Nrf2 signaling pathway at several nodes (summarized in Table 3).
Table 3.
miRNAs regulate Keap1-Nrf2 signaling pathway.
| Target | miRNA | Experimental model | Impact on Keap1-Nrf2 | Outcomes | Ref |
|---|---|---|---|---|---|
| Nrf2 | miR-144 | K562 cell line, primary erythroid progenitor cells |
↓Nrf2 levels, ↓glutathione regeneration ↓antioxidant capacity |
Associated with anemia severity in sickle cell disease |
[105] |
| miR-27a,-142-5p,- 144, -153 |
SH-SY5Y cells | ↓Gclc, Gsr levels | Not applicable | [106] | |
| miR-28 | MCF-7 cell line | ↓Nrf2 mRNA | Increased anchorage-independent growth |
[109] | |
| miR-93 | E2-induced breast carcinognesis in ACI rats |
↓Nrf2 and Nrf2 regulated genes |
Decreased apoptosis, increased DNA damage |
[110] | |
|
| |||||
| Keap1 | miR-200a | MDA-MB-231 cell line |
Keap1 mRNA degradation, ↑Nrf2 nuclear accumulation, ↑NQO1 |
Inhibits anchorage-independent growth |
[111] |
|
| |||||
| Cul3 | miR-101 | hypoxic condition | ↑Nrf2 nuclear accumulation, ↑HO-1 |
Improves neovascularization and blood flow in ischemia |
[112] |
|
| |||||
| Bach1 | let-7 | Huh7 cell line | ↑HO-1 | Increased resistance against oxidant injury against tBuOOH |
[115] |
| miR-155 | primary HUVECs | ↑HO-1 | Cytoprotective during inflammation | [116] | |
5.1. miRNAs regulate Nrf2 activity by directly targeting the mRNA of Nrf2
As miRNAs function as post-transcriptional repressors of gene expression, miRNAs that directly target Nrf2 usually negatively regulate the Keap1-Nrf2 pathway. Notably, inefficient activation of Nrf2 results in alteration of Nrf2-dependent redox homeostasis, thus potentially triggering disease outcomes. Erythrocytes from patients with homozygous sickle cell disease (HbSS) have a reduced tolerance for oxidative stress. In a subset of HbSS patients with more severe anemia, higher erythrocytic miR-144 expression has been observed [105]. In the same study, Sangokoya et al. found that the 3′ UTR of Nrf2 is directly targeted by miR-144 in K562 cells and primary erythroid progenitor cells. Therefore, increased miR-144 may contribute to the attenuated Nrf2 levels in HbSS erythrocytes, which could account for the decrease in glutathione regeneration and impaired oxidative stress tolerance. By employing bioinformatic analysis of the human Nrf2 3′ UTR sequences for miRNA binding sites, Narasimhan et al. reported an in-silico prediction of 4 different miRNAs targeting human Nrf2, including hsa-miR27a, hsa-miR153, hsa-miR142-5p, as well as the already reported hsa-miR144 [106]. The direct interaction between the four identified miRNAs and Nrf2 was further validated using luciferase constructs carrying either the 3′ UTR of human Nrf2 or mutated miRNA binding sites within the Nrf2 3′ UTR. Moreover, ectopic expression of the corresponding miRNA mimics affected cellular Nrf2 mRNA levels as well as the nucleo-cytoplasmic concentration of the Nrf2 protein in a Keap1-independent manner, which consequently lessens GCLC and glutathione reductase (GSR) expression.
In the context of cancer, a variety of miRNAs have been implicated in cell differentiation, cell proliferation/apoptosis, and tumor suppression [107, 108]. Specifically in the Keap1-Nrf2 pathway, miR-28 expression has been reported to be reversibly correlated with Nrf2 mRNA levels in human mammary epithelial cells and the breast cancer MCF-7 cell line [109]. Yang et al. also demonstrated that miR-28 regulates the Nrf2 pathway by targeting the 3′ UTR region, thereby facilitating the degradation of Nrf2 mRNA. In addition, Nrf2 shRNA or ectopic expression of miR-28 inhibited the anchorage-independent cell growth of MCF-7 cells, suggesting that miR-28 might influence breast cancer motility and growth by regulating the Nrf2 pathway. Similarly, in 17β-estradiol (E2)-induced rat breast carcinogenesis, an E2-mediated increase in miR-93 levels was associated with decreased expression of Nrf2 [110]. Furthermore, in human breast cell lines, miR-93 has been shown to have oncogenic potential, including the ability to increase colony formation, mammosphere formation, cell migration, and DNA damage and to decrease apoptosis.
5.2. miRNAs regulate Nrf2 activity by interacting with cellular Nrf2 regulators
On the other hand, those miRNAs that interact with cellular Nrf2 regulators are expected to influence Nrf2/ARE signaling as well. Keap1 has been most extensively studied as a cellular suppressor of Nrf2. In human breast cancer MDA-MB-231 cells, Eades et al. reported that miR-200a could interact with the Keap1 3′UTR, facilitating its mRNA degradation [111]. Therefore, the decreased miR-200a levels in breast cancer may provide a novel mechanistic explanation for the deregulation of the Nrf2 pathway. By inducing the re-expression of miR-200a, the reduction in Keap1 levels subsequently enhances Nrf2 nuclear accumulation and NQO1 gene transcription. Furthermore, this study demonstrates that Nrf2 activation consequently inhibits the anchorage-independent growth of breast cancer cells in vitro and carcinogen-induced mammary hyperplasia in vivo. Cul3 is an important component of the Keap-1 protein complex that promotes Keap1-dependent Nrf2 ubiquitination and proteasomal degradation [112]. Kim et al. demonstrated that Cul3 is a target of miR-101. Under hypoxic conditions, miR-101 is up-regulated in a HIF-1a-dependent manner, thereby stabilizing Nrf2 protein and inducing HO-1. Local overexpression of miR-101 improves neovascularization and blood flow in a mouse model of hindlimb ischemia. A mechanistic study indicated that a positive feedback loop between the Nrf2/HO-1 and VEGF/eNOS axes is implicated in miR-101-mediated post-ischemic vascular remodeling and angiogenesis. Bach1 is a MAF-related transcription factor that plays critical role in specific of HO-1 gene regulation. In cells naïve to oxidative stress, Bach1 conceals the ARE sequences; thus, it antagonizes Nrf2 binding and represses HO-1 gene transcriptional activation [113]. In an earlier report, MacLeod et al. demonstrated the specificity attributes to that only HO-1 contains the necessary multiple cis-elements required for efficient Bach1 binding among human ARE-driven gene battery [114]. Hou et al. reported that let-7 miRNAs (let-7b, 7c) enhanced HO-1 gene transcription by down-regulating Bach1 protein levels [115]. Ectopic expression of the let-7 miRNA in Huh-7 cells resulted in increased resistance against oxidant injury induced by tert-butyl-hydroperoxide (tBuOOH), whereas the enhanced anti-oxidative capacity was counteracted by Bach1 over-expression. It is worth mentioning that the pro-inflammatory miR-155 also targets Bach1 degradation and induces subsequent elevation of HO-1 expression in endothelial cells [116]. It has been proposed that the cytoprotective response to inflammation results from the miR-155-mediated regulation of HO-1 rather than direct induction via the NF-κB pathway. This study provides a novel mechanistic insight by introducing miRNA in the cross-talk between the inflammatory and oxidative stress pathways. However, in lipopolysaccharide-stimulated murine RAW264.7 macrophages, either sulforaphane or allyl-isothiocyanate treatment leads to decrease in miR-155 levels, accompanied by HO-1 induction [117]. Given that each miRNA could have multiple target genes while being controlled by a variety of upstream signals, the precise mechanisms by which miR-155 affects the Nrf2-mediated cellular protective system remain to be fully elucidated.
5.3. Transcriptional regulation of miRNAs by Nrf2
The biogenesis of miRNAs is similar to that of other RNA molecules and starts with the transcription of the genes that encode immature primary miRNA (pri-miRNA) by RNA polymerase II. It is possible that the transcription of pri-miRNAs could be regulated by transcription factors (TFs) such as Nrf2. Indeed, recent studies provide evidence that a number of miRNAs can be regulated by Nrf2, reviewed by [118]. In addition, a systematic analysis of the interactors and regulators of Nrf2 conducted by Papp et al. predicted 85 miRNA-Nrf2 mRNA interactions [119]. Interestingly, 35 TFs regulated by Nrf2 could increase the levels of 63 out of the 85 miRNAs mentioned above. This model indicates that miRNAs are involved in the fine-tuning feedback loops in Nrf2 signaling.
6. Cross-talk of epigenetic mechanisms in the modification of the Keap1-Nrf2 signaling pathway
Circumstantial evidence suggests that different epigenetic layers may be engaged in complex crosstalk to establish and maintain different chromatin states. Thus, the above-mentioned epigenetic modifications may not function alone, but they may be linked to each other and work in combination to regulate gene transcription [120]. Studies in our laboratory suggested that hypermethylated CpG islands in TRAMP C1 cells were associated with MBD2 and histone modifications, indicating interplay between DNA methylation and histone modification in the regulation of Nrf2 transcription activity. Chromatin immunoprecipitation assays showed that MBD2 and tri-methylated histone 3-lys9 (H3K9me3) are enriched in methylated CpGs in the Nrf2 promoter, whereas acetylated histone 3 (H3Ac) is associated with unmethylated CpGs [42]. Additionally, recent research has reported that nutritional phytochemicals, including sulforaphane, apigenin, 3,3′-diindolylmethane, and tanshinone IIA, epigenetically re-activate the expression of Nrf2 through the inhibition of both DNMT and HDAC [50, 51, 53-55]. Although these studies suggest an intimate communication and confounding actions between DNA methylation and histone acetylation/methylation in the silencing/activation of the Nrf2 gene, the fundamental question regarding which epigenetic event initiates and steers the crosstalk and Nrf2 silencing remains to be answered. Two different sequential models have been proposed to describe the interplay of DNA methylation and histone modification in gene silencing (reviewed in [121]). In one scenario, partial DNA methylation trigged by environmental and intrinsic signals attracts the binding of MePC2 and HDAC to the CpG sites, leads to deacetylated histone and inactive chromatin configuration, and further recruits DNMT1 to amplify the silencing signals. In another scenario, imbalanced HAT and HDAC activities induce chromatin hypoacetylation, a chromatin state that will be recognized by de novo DNMTs and result in a local hypermethylation state. Future studies are necessary to understand the mechanisms underlying the combination of the epigenetic events in the regulation of Keap1-Nrf2 signaling pathways.
These epigenetic modifications not only work in combination to interact with Keap1-Nrf2 signaling pathways, but are also known to cross-regulate each other in a manner that diversifies their functions and ultimately influences cellular activity. For example, the miR-200 family was previously shown to be aberrantly silenced by epigenetic mechanisms in breast cancer [122], and impaired miR-200a activity led to the overexpression of SIRT1 (class III HDAC) [123]. It is noteworthy that the epigenetic silencing of miR-200a by histone acetylation might contribute to the overexpression of Keap1 and loss of the Nrf2-dependent antioxidant pathway in breast cancer cells. Furthermore, Eades and coworkers found that treatment with the HDAC inhibitor suberoylanilide hydroxamic acid re-expressed miR-200a, which corresponded to decreased Keap1 expression, increased Nrf2 translocation, and elevated Nrf2-dependent NQO1 expression [111]. More recently, another study reported similar results: the HDAC inhibitor MS-275 efficiently reduces the deacetylation in the miR-200a promoter region and reactivates miR-200a. As a consequence, mature miR-200a destabilizes Keap1 mRNA, leads to enhanced translocation and binding of Nrf2 to the polyamine-responsive element of the spermidine/spermine N1-acetyltransferase (SSAT) promoter, and ultimately results in reduced polyamine synthesis and growth inhibition [124]. These studies suggested that cross-regulation of epigenetic modifications modulates Keap1-Nrf2 signaling. Therefore, more in-depth understanding will be necessary to exploit this complex regulatory network to combat dysregulation of the Keap1-Nrf2 pathway in chronic diseases.
7. Conclusions and perspectives
Keap1-Nrf2 signaling plays important roles in a variety of physiological, pathological, pharmacological, and toxicological processes and is subjected to multiple layers of regulation at transcriptional, translational, and post-translational levels. In recent years, the epigenetic regulation of Keap1 and Nrf2 expression in various oxidative stress-related diseases has begun to be unveiled. As depicted schematically in Figure 1, Keap1 and Nrf2 expression could be regulated by methylation/demethylation of CpGs in the promoter regions, acetylation/deacetylation and methylation/demethylation of histones, or targeting of mRNAs by miRNAs. In addition, Nrf2 has also been implicated in the transcriptional regulation of certain non-coding RNAs. However, although oxidative stresses are profoundly engaged in epigenetic modifications and chromatin organization, it is not clear whether Keap1-Nrf2 signaling directly or indirectly regulates epigenetic processes other than miRNA transcription.
To date, most epigenetic regulation of Keap1-Nrf2 signaling has been identified in the context of cancer. Because oxidative stresses and Keap1-Nrf2 signaling are widely involved in almost all major chronic diseases, the participation of epigenetic mechanisms in the regulation of Keap1-Nrf2 signaling in these diseases will be interesting to elucidate. Indeed, investigations have revealed the important roles of the demethylation of the Keap1 promoter in ARCs and cardiomyopathy and the reactivation of Nrf2 by HDAC inhibitors in neuroinflammation and cerebral ischemic injury, and further investigations in other oxidative stress-related diseases are guaranteed. Moreover, given the complexity of the crosstalk between genetic/epigenetic modifications and the Keap1-Nrf2 signaling networks, the exact mechanisms of epigenetic regulation of Keap1-Nrf2 signaling and their physiological significance remain open for further investigation.
Acknowledgments
This work was supported in part by institutional funds and by the National Natural Science Foundation of China (Grant No.81272468 and 81472657), R01-CA118947 and R01-CA152826 from the National Cancer Institute (NCI), R01AT007065 from the National Center for Curr Pharmacol Rep Complementary and Alternative Medicines (NCCAM), and the Office of Dietary Supplements (ODS). We thank all the members of the laboratory for the discussion and preparation of this review.
References
- [1].Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. When NRF2 talks, who’s listening? Antioxidants & redox signaling. 2010;13:1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jeong WS, Jun M, Kong AN. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxidants & redox signaling. 2006;8:99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- [4].Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends in biochemical sciences. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- [5].Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nature reviews. Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS medicine. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, Minna JD, Stewart DJ, Wistuba Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- [10].Mattick JS, Makunin IV. Non-coding RNA. Human molecular genetics. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- [11].Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature reviews. Genetics. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- [12].Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- [13].Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Human molecular genetics. 2013;22:R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nature reviews. Genetics. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- [15].Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Current opinion in genetics & development. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- [16].You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature reviews. Genetics. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- [18].Portela A, Esteller M. Epigenetic modifications and human disease. Nature biotechnology. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- [19].Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martino D, Kesper DA, Amarasekera M, Harb H, Renz H, Prescott S. Epigenetics in immune development and in allergic and autoimmune diseases. Journal of reproductive immunology. 2014;104-105:43–48. doi: 10.1016/j.jri.2014.05.003. [DOI] [PubMed] [Google Scholar]
- [21].Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marques SC, Oliveira CR, Pereira CM, Outeiro TF. Epigenetics in neurodegeneration: a new layer of complexity. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35:348–355. doi: 10.1016/j.pnpbp.2010.08.008. [DOI] [PubMed] [Google Scholar]
- [23].Gronbaek K, Hother C, Jones PA. Epigenetic changes in cancer. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2007;115:1039–1059. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- [24].Dhanak D, Jackson P. Development and classes of epigenetic drugs for cancer. Biochemical and biophysical research communications. 2014;455:58–69. doi: 10.1016/j.bbrc.2014.07.006. [DOI] [PubMed] [Google Scholar]
- [25].Guo Y, Su Z-Y, Kong A-NT. Current Perspectives on Epigenetic Modifications by Dietary Chemopreventive and Herbal Phytochemicals. Current Pharmacology Reports. 2015:1–13. doi: 10.1007/s40495-015-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annual review of biochemistry. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer letters. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- [28].Niu Y, DesMarais TL, Tong Z, Yao Y, Costa M. Oxidative stress alters global histone modification and DNA methylation. Free radical biology & medicine. 2015;82:22–28. doi: 10.1016/j.freeradbiomed.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Su ZY, Shu L, Khor TO, Lee JH, Fuentes F, Kong AN. A perspective on dietary phytochemicals and cancer chemoprevention: oxidative stress, nrf2, and epigenomics. Topics in current chemistry. 2013;329:133–162. doi: 10.1007/128_2012_340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tada M, Yokosuka O, Fukai K, Chiba T, Imazeki F, Tokuhisa T, Saisho H. Hypermethylation of NAD(P)H: quinone oxidoreductase 1 (NQO1) gene in human hepatocellular carcinoma. Journal of hepatology. 2005;42:511–519. doi: 10.1016/j.jhep.2004.11.024. [DOI] [PubMed] [Google Scholar]
- [31].Meiers I, Shanks JH, Bostwick DG. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review 2007. Pathology. 2007;39:299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- [32].Belanger AS, Tojcic J, Harvey M, Guillemette C. Regulation of UGT1A1 and HNF1 transcription factor gene expression by DNA methylation in colon cancer cells. BMC molecular biology. 2010;11:9. doi: 10.1186/1471-2199-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- [34].Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- [35].Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harbor perspectives in biology. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gagnon JF, Bernard O, Villeneuve L, Tetu B, Guillemette C. Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:1850–1858. doi: 10.1158/1078-0432.CCR-05-2130. [DOI] [PubMed] [Google Scholar]
- [37].Hurt EM, Thomas SB, Peng B, Farrar WL. Molecular consequences of SOD2 expression in epigenetically silenced pancreatic carcinoma cell lines. British journal of cancer. 2007;97:1116–1123. doi: 10.1038/sj.bjc.6604000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mohamed MM, Sabet S, Peng DF, Nouh MA, El-Shinawi M, El-Rifai W. Promoter hypermethylation and suppression of glutathione peroxidase 3 are associated with inflammatory breast carcinogenesis. Oxidative medicine and cellular longevity. 2014;2014:787195. doi: 10.1155/2014/787195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer research. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- [40].Barve A, Khor TO, Nair S, Reuhl K, Suh N, Reddy B, Newmark H, Kong AN. Gamma-tocopherol-enriched mixed tocopherol diet inhibits prostate carcinogenesis in TRAMP mice. International journal of cancer. Journal international du cancer. 2009;124:1693–1699. doi: 10.1002/ijc.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Frohlich DA, McCabe MT, Arnold RS, Day ML. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27:4353–4362. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- [42].Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, Foster BA, Kan YW, Kong AN. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS one. 2010;5:e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Khor TO, Fuentes F, Shu L, Paredes-Gonzalez X, Yang AY, Liu Y, Smiraglia DJ, Yegnasubramanian S, Nelson WG, Kong AN. Epigenetic DNA Methylation of Antioxidative Stress Regulator NRF2 in Human Prostate Cancer. Cancer prevention research. 2014;7:1186–1197. doi: 10.1158/1940-6207.CAPR-14-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Huang Y, Khor TO, Shu L, Saw CL, Wu TY, Suh N, Yang CS, Kong AN. A gamma-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. The Journal of nutrition. 2012;142:818–823. doi: 10.3945/jn.111.153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guo Y, Shu L, Zhang C, Su ZY, Kong AN. Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochemical pharmacology. 2015;94:69–78. doi: 10.1016/j.bcp.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gerhauser C. Epigenetic impact of dietary isothiocyanates in cancer chemoprevention. Current opinion in clinical nutrition and metabolic care. 2013;16:405–410. doi: 10.1097/MCO.0b013e328362014e. [DOI] [PubMed] [Google Scholar]
- [47].Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer research. 2003;63:7563–7570. [PubMed] [Google Scholar]
- [48].Xie Q, Bai Q, Zou LY, Zhang QY, Zhou Y, Chang H, Yi L, Zhu JD, Mi MT. Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes, chromosomes & cancer. 2014;53:422–431. doi: 10.1002/gcc.22154. [DOI] [PubMed] [Google Scholar]
- [49].Khor TO, Huang Y, Wu TY, Shu L, Lee J, Kong AN. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochemical pharmacology. 2011;82:1073–1078. doi: 10.1016/j.bcp.2011.07.065. [DOI] [PubMed] [Google Scholar]
- [50].Wu TY, Khor TO, Su ZY, Saw CL, Shu L, Cheung KL, Huang Y, Yu S, Kong AN. Epigenetic modifications of Nrf2 by 3,3′-diindolylmethane in vitro in TRAMP C1 cell line and in vivo TRAMP prostate tumors. The AAPS journal. 2013;15:864–874. doi: 10.1208/s12248-013-9493-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang C, Su ZY, Khor TO, Shu L, Kong AN. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85:1398–1404. doi: 10.1016/j.bcp.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Su ZY, Khor TO, Shu L, Lee JH, Saw CL, Wu TY, Huang Y, Suh N, Yang CS, Conney AH, Wu Q, Kong AN. Epigenetic reactivation of Nrf2 in murine prostate cancer TRAMP C1 cells by natural phytochemicals Z-ligustilide and Radix angelica sinensis via promoter CpG demethylation. Chemical research in toxicology. 2013;26:477–485. doi: 10.1021/tx300524p. [DOI] [PubMed] [Google Scholar]
- [53].Paredes-Gonzalez X, Fuentes F, Su ZY, Kong AN. Apigenin reactivates Nrf2 anti-oxidative stress signaling in mouse skin epidermal JB6 P + cells through epigenetics modifications. The AAPS journal. 2014;16:727–735. doi: 10.1208/s12248-014-9613-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Su ZY, Zhang C, Lee JH, Shu L, Wu TY, Khor TO, Conney AH, Lu YP, Kong AN. Requirement and epigenetics reprogramming of Nrf2 in suppression of tumor promoter TPA-induced mouse skin cell transformation by sulforaphane. Cancer Prev Res (Phila) 2014;7:319–329. doi: 10.1158/1940-6207.CAPR-13-0313-T. [DOI] [PubMed] [Google Scholar]
- [55].Wang L, Zhang C, Guo Y, Su ZY, Yang Y, Shu L, Kong AN. Blocking of JB6 cell transformation by tanshinone IIA: epigenetic reactivation of Nrf2 antioxidative stress pathway. The AAPS journal. 2014;16:1214–1225. doi: 10.1208/s12248-014-9666-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochemical and biophysical research communications. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- [57].Guo D, Wu B, Yan J, Li X, Sun H, Zhou D. A possible gene silencing mechanism: hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochemical and biophysical research communications. 2012;428:80–85. doi: 10.1016/j.bbrc.2012.10.010. [DOI] [PubMed] [Google Scholar]
- [58].Muscarella LA, Parrella P, D’Alessandro V, la Torre A, Barbano R, Fontana A, Tancredi A, Guarnieri V, Balsamo T, Coco M, Copetti M, Pellegrini F, De Bonis P, Bisceglia M, Scaramuzzi G, Maiello E, Valori VM, Merla G, Vendemiale G, Fazio VM. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics : official journal of the DNA Methylation Society. 2011;6:710–719. doi: 10.4161/epi.6.6.15773. [DOI] [PubMed] [Google Scholar]
- [59].Muscarella LA, Barbano R, D’Angelo V, Copetti M, Coco M, Balsamo T, la Torre A, Notarangelo A, Troiano M, Parisi S, Icolaro N, Catapano D, Valori VM, Pellegrini F, Merla G, Carella M, Fazio VM, Parrella P. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics : official journal of the DNA Methylation Society. 2011;6:317–325. doi: 10.4161/epi.6.3.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Barbano R, Muscarella LA, Pasculli B, Valori VM, Fontana A, Coco M, la Torre A, Balsamo T, Poeta ML, Marangi GF, Maiello E, Castelvetere M, Pellegrini F, Murgo R, Fazio VM, Parrella P. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics : official journal of the DNA Methylation Society. 2013;8:105–112. doi: 10.4161/epi.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, Ishiguro A, Kijima H, Mimura J, Itoh K, Fukuda S, Saijo Y. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC cancer. 2012;12:66. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jazirehi AR, Wenn PB, Arle D. Is there a decrease in Keap1 RNA expression in colorectal cancer cells, and is this decrease in expression due to hypermethylation? Epigenomics. 2012;4:253–254. [PubMed] [Google Scholar]
- [63].Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Molecular cancer therapeutics. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Martinez VD, Vucic EA, Pikor LA, Thu KL, Hubaux R, Lam WL. Frequent concerted genetic mechanisms disrupt multiple components of the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in thyroid cancer. Mol Cancer. 2013;12:124. doi: 10.1186/1476-4598-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Elanchezhian R, Palsamy P, Madson CJ, Lynch DW, Shinohara T. Age-related cataracts: homocysteine coupled endoplasmic reticulum stress and suppression of Nrf2-dependent antioxidant protection. Chemico-biological interactions. 2012;200:1–10. doi: 10.1016/j.cbi.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].von Otter M, Landgren S, Nilsson S, Zetterberg M, Celojevic D, Bergstrom P, Minthon L, Bogdanovic N, Andreasen N, Gustafson DR, Skoog I, Wallin A, Tasa G, Blennow K, Nilsson M, Hammarsten O, Zetterberg H. Nrf2-encoding NFE2L2 haplotypes influence disease progression but not risk in Alzheimer’s disease and age-related cataract. Mechanisms of ageing and development. 2010;131:105–110. doi: 10.1016/j.mad.2009.12.007. [DOI] [PubMed] [Google Scholar]
- [67].Palsamy P, Ayaki M, Elanchezhian R, Shinohara T. Promoter demethylation of Keap1 gene in human diabetic cataractous lenses. Biochemical and biophysical research communications. 2012;423:542–548. doi: 10.1016/j.bbrc.2012.05.164. [DOI] [PubMed] [Google Scholar]
- [68].Gao Y, Yan Y, Huang T. Human agerelated cataracts: Epigenetic suppression of the nuclear factor erythroid 2related factor 2mediated antioxidant system. Molecular medicine reports. 2015;11:1442–1447. doi: 10.3892/mmr.2014.2849. [DOI] [PubMed] [Google Scholar]
- [69].Yang SP, Yang XZ, Cao GP. Acetyllcarnitine prevents homocysteineinduced suppression of Nrf2/Keap1 mediated antioxidation in human lens epithelial cells. Molecular medicine reports. 2015;12:1145–1150. doi: 10.3892/mmr.2015.3490. [DOI] [PubMed] [Google Scholar]
- [70].Palsamy P, Bidasee KR, Shinohara T. Selenite cataracts: activation of endoplasmic reticulum stress and loss of Nrf2/Keap1-dependent stress protection. Biochimica et biophysica acta. 2014;1842:1794–1805. doi: 10.1016/j.bbadis.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Palsamy P, Bidasee KR, Ayaki M, Augusteyn RC, Chan JY, Shinohara T. Methylglyoxal induces endoplasmic reticulum stress and DNA demethylation in the Keap1 promoter of human lens epithelial cells and age-related cataracts. Free radical biology & medicine. 2014;72:134–148. doi: 10.1016/j.freeradbiomed.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Palsamy P, Bidasee KR, Shinohara T. Valproic acid suppresses Nrf2/Keap1 dependent antioxidant protection through induction of endoplasmic reticulum stress and Keap1 promoter DNA demethylation in human lens epithelial cells. Experimental eye research. 2014;121:26–34. doi: 10.1016/j.exer.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Liu ZZ, Zhao XZ, Zhang XS, Zhang M. Promoter DNA demethylation of Keap1 gene in diabetic cardiomyopathy. International journal of clinical and experimental pathology. 2014;7:8756–8762. [PMC free article] [PubMed] [Google Scholar]
- [74].Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- [75].Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115:110–122. doi: 10.1007/s00412-006-0052-x. [DOI] [PubMed] [Google Scholar]
- [76].Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe AP. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. The EMBO journal. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liu GH, Qu J, Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochimica et biophysica acta. 2008;1783:713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- [78].Correa F, Mallard C, Nilsson M, Sandberg M. Activated microglia decrease histone acetylation and Nrf2-inducible anti-oxidant defence in astrocytes: restoring effects of inhibitors of HDACs, p38 MAPK and GSK3beta. Neurobiology of disease. 2011;44:142–151. doi: 10.1016/j.nbd.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang B, Zhu X, Kim Y, Li J, Huang S, Saleem S, Li RC, Xu Y, Dore S, Cao W. Histone deacetylase inhibition activates transcription factor Nrf2 and protects against cerebral ischemic damage. Free radical biology & medicine. 2012;52:928–936. doi: 10.1016/j.freeradbiomed.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mercado N, Thimmulappa R, Thomas CM, Fenwick PS, Chana KK, Donnelly LE, Biswal S, Ito K, Barnes PJ. Decreased histone deacetylase 2 impairs Nrf2 activation by oxidative stress. Biochemical and biophysical research communications. 2011;406:292–298. doi: 10.1016/j.bbrc.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Adenuga D, Caito S, Yao H, Sundar IK, Hwang JW, Chung S, Rahman I. Nrf2 deficiency influences susceptibility to steroid resistance via HDAC2 reduction. Biochemical and biophysical research communications. 2010;403:452–456. doi: 10.1016/j.bbrc.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, An CH, Shan B, Franks JM, Haley KJ, Owen CA, Tesfaigzi Y, Washko GR, Quackenbush J, Silverman EK, Rahman I, Kim HP, Mahmood A, Biswal SS, Ryter SW, Choi AM. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. The Journal of clinical investigation. 2013;123:5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kawai Y, Garduno L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. The Journal of biological chemistry. 2011;286:7629–7640. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Molecular and cellular biology. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen Z, Ye X, Tang N, Shen S, Li Z, Niu X, Lu S, Xu L. The histone acetylranseferase hMOF acetylates Nrf2 and regulates anti-drug responses in human non-small cell lung cancer. British journal of pharmacology. 2014;171:3196–3211. doi: 10.1111/bph.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ikeda H, Nishi S, Sakai M. Transcription factor Nrf2/MafK regulates rat placental glutathione S-transferase gene during hepatocarcinogenesis. The Biochemical journal. 2004;380:515–521. doi: 10.1042/BJ20031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ohta K, Ohigashi M, Naganawa A, Ikeda H, Sakai M, Nishikawa J, Imagawa M, Osada S, Nishihara T. Histone acetyltransferase MOZ acts as a co-activator of Nrf2-MafK and induces tumour marker gene expression during hepatocarcinogenesis. The Biochemical journal. 2007;402:559–566. doi: 10.1042/BJ20061194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kalthoff S, Winkler A, Freiberg N, Manns MP, Strassburg CP. Gender matters: estrogen receptor alpha (ERalpha) and histone deacetylase (HDAC) 1 and 2 control the gender-specific transcriptional regulation of human uridine diphosphate glucuronosyltransferases genes (UGT1A) Journal of hepatology. 2013;59:797–804. doi: 10.1016/j.jhep.2013.05.028. [DOI] [PubMed] [Google Scholar]
- [89].Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature reviews. Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chase A, Cross NC. Aberrations of EZH2 in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- [91].Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Molecular cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- [92].Balasubramanian S, Kanade S, Han B, Eckert RL. A proteasome inhibitor-stimulated Nrf1 protein-dependent compensatory increase in proteasome subunit gene expression reduces polycomb group protein level. The Journal of biological chemistry. 2012;287:36179–36189. doi: 10.1074/jbc.M112.359281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Li Z, Xu L, Tang N, Xu Y, Ye X, Shen S, Niu X, Lu S, Chen Z. The polycomb group protein EZH2 inhibits lung cancer cell growth by repressing the transcription factor Nrf2. FEBS letters. 2014;588:3000–3007. doi: 10.1016/j.febslet.2014.05.057. [DOI] [PubMed] [Google Scholar]
- [94].Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Waters NJ, Smith JJ, Porter-Scott M, Chesworth R, Moyer MP, Copeland RA, Richon VM, Uenaka T, Pollock RM, Kuntz KW, Yokoi A, Keilhack H. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Molecular cancer therapeutics. 2014;13:842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- [95].Crea F, Fornaro L, Bocci G, Sun L, Farrar WL, Falcone A, Danesi R. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer metastasis reviews. 2012;31:753–761. doi: 10.1007/s10555-012-9387-3. [DOI] [PubMed] [Google Scholar]
- [96].Mishra M, Zhong Q, Kowluru RA. Epigenetic modifications of Keap1 regulate its interaction with the protective factor Nrf2 in the development of diabetic retinopathy. Investigative ophthalmology & visual science. 2014;55:7256–7265. doi: 10.1167/iovs.14-15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhong Q, Kowluru RA. Epigenetic modification of Sod2 in the development of diabetic retinopathy and in the metabolic memory: role of histone methylation. Investigative ophthalmology & visual science. 2013;54:244–250. doi: 10.1167/iovs.12-10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mishra M, Zhong Q, Kowluru RA. Epigenetic modifications of Nrf2-mediated glutamate-cysteine ligase: implications for the development of diabetic retinopathy and the metabolic memory phenomenon associated with its continued progression. Free radical biology & medicine. 2014;75:129–139. doi: 10.1016/j.freeradbiomed.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Miranda M, Muriach M, Johnsen S, Bosch-Morell F, Araiz J, Roma J, Romero FJ. Oxidative stress in a model for experimental diabetic retinopathy: treatment with antioxidants. Archivos de la Sociedad Espanola de Oftalmologia. 2004;79:289–294. doi: 10.4321/s0365-66912004000600007. [DOI] [PubMed] [Google Scholar]
- [100].Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- [101].Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Current opinion in drug discovery & development. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- [102].Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. The Journal of biological chemistry. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- [103].Michaeloudes C, Mercado N, Clarke C, Bhavsar PK, Adcock IM, Barnes PJ, Chung KF. Bromodomain and extraterminal proteins suppress NF-E2-related factor 2-mediated antioxidant gene expression. Journal of immunology. 2014;192:4913–4920. doi: 10.4049/jimmunol.1301984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Hussong M, Borno ST, Kerick M, Wunderlich A, Franz A, Sultmann H, Timmermann B, Lehrach H, Hirsch-Kauffmann M, Schweiger MR. The bromodomain protein BRD4 regulates the KEAP1/NRF2-dependent oxidative stress response. Cell death & disease. 2014;5:e1195. doi: 10.1038/cddis.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Narasimhan M, Patel D, Vedpathak D, Rathinam M, Henderson G, Mahimainathan L. Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PloS one. 2012;7:e51111. doi: 10.1371/journal.pone.0051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer research. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- [108].Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- [109].Yang M, Yao Y, Eades G, Zhang Y, Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast cancer research and treatment. 2011;129:983–991. doi: 10.1007/s10549-011-1604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Singh B, Ronghe AM, Chatterjee A, Bhat NK, Bhat HK. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis. 2013;34:1165–1172. doi: 10.1093/carcin/bgt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. The Journal of biological chemistry. 2011;286:40725–40733. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kim JH, Lee KS, Lee DK, Kim J, Kwak SN, Ha KS, Choe J, Won MH, Cho BR, Jeoung D, Lee H, Kwon YG, Kim YM. Hypoxia-responsive microRNA-101 promotes angiogenesis via heme oxygenase-1/vascular endothelial growth factor axis by targeting cullin 3. Antioxidants & redox signaling. 2014;21:2469–2482. doi: 10.1089/ars.2014.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Reichard JF, Motz GT, Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic acids research. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].MacLeod AK, McMahon M, Plummer SM, Higgins LG, Penning TM, Igarashi K, Hayes JD. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30:1571–1580. doi: 10.1093/carcin/bgp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hou W, Tian Q, Steuerwald NM, Schrum LW, Bonkovsky HL. The let-7 microRNA enhances heme oxygenase-1 by suppressing Bach1 and attenuates oxidant injury in human hepatocytes. Biochimica et biophysica acta. 2012;1819:1113–1122. doi: 10.1016/j.bbagrm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Pulkkinen KH, Yla-Herttuala S, Levonen AL. Heme oxygenase 1 is induced by miR-155 via reduced BACH1 translation in endothelial cells. Free radical biology & medicine. 2011;51:2124–2131. doi: 10.1016/j.freeradbiomed.2011.09.014. [DOI] [PubMed] [Google Scholar]
- [117].Wagner AE, Boesch-Saadatmandi C, Dose J, Schultheiss G, Rimbach G. Anti-inflammatory potential of allyl-isothiocyanate--role of Nrf2, NF-(kappa) B and microRNA-155. Journal of cellular and molecular medicine. 2012;16:836–843. doi: 10.1111/j.1582-4934.2011.01367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shah NM, Rushworth SA, Murray MY, Bowles KM, MacEwan DJ. Understanding the role of NRF2-regulated miRNAs in human malignancies. Oncotarget. 2013;4:1130–1142. doi: 10.18632/oncotarget.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Papp D, Lenti K, Modos D, Fazekas D, Dul Z, Turei D, Foldvari-Nagy L, Nussinov R, Csermely P, Korcsmaros T. The NRF2-related interactome and regulome contain multifunctional proteins and fine-tuned autoregulatory loops. FEBS letters. 2012;586:1795–1802. doi: 10.1016/j.febslet.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kondo Y. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei medical journal. 2009;50:455–463. doi: 10.3349/ymj.2009.50.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutation research. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- [122].Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PloS one. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. The Journal of biological chemistry. 2011;286:25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Murray-Stewart T, Hanigan CL, Woster PM, Marton LJ, Casero RA., Jr. Histone deacetylase inhibition overcomes drug resistance through a miRNA-dependent mechanism. Mol Cancer Ther. 2013;12:2088–2099. doi: 10.1158/1535-7163.MCT-13-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]