Abstract
Focal malformations of cortical development (FMCD) account for the majority of drug-resistant pediatric epilepsy. Postzygotic somatic mutations activating the PI3K-AKT-mTOR pathway are found in a wide range of brain diseases, including FMCD. It remains unclear how a mutation in a small fraction of cells can disrupt the architecture of the entire hemisphere. We show that, within human FMCD brain, cells showing activation of this pathway were enriched for the mutation. Introducing the FMCD mutation into mouse brain resulted in electrographic seizures and impaired hemispheric architecture. Mutation-expressing neural progenitors showed reelin misexpression, which led to a non-cell autonomous migration defect in neighboring cells, due at least in part to FOXG1-mediated de-repression of reelin transcription. Treatments aimed at blocking downstream AKT signaling or inactivating reelin restored migration. These findings suggest a central AKT-FOXG1-Reelin signaling pathway in FMCD, and support pathway inhibitors as potential treatments or therapies for some forms of focal epilepsy.
Keywords: Postzygotic, somatic mosaic, neuropsychiatric, seizures, epilepsy, AKT, PI3K, MTOR, malformations of cortical development
Malformations of cortical development (MCD) are increasingly recognized as an important cause of autism, neurodevelopmental delay and epilepsy, especially medically-intractable ‘catastrophic’ epilepsy. Disruption at various critical stages of cortical development – neuronal proliferation, migration, and organization – leads to characteristic MCD. Focal MCDs (FMCD) are characterized by localized cortical and subcortical lesions 2. At the most severe end of the spectrum is hemimegalencephaly (HME), a clinically devastating pediatric FMCD characterized by enlargement of most or all of one entire cerebral hemisphere. Individuals with HME typically present with epilepsy, psychomotor disability and contralateral hemiparesis 3. Most affected individuals display progressive and catastrophic epilepsy, which invariably requires radical hemispherectomy (i.e. removal of the affected cerebral hemisphere) as a surgical treatment to relieve epilepsy and to prevent further psychomotor disability.
FMCDs are characterized by cortical dyslamination and ectopic neurons in the subcortical white matter, and in the most severe form of disease, are associated with the presence of dysmorphic, cytomegalic neurons and ‘balloon cells’ 4,5. The lesions are consistent with recent models of brain mosaicism 6. Because some FMCD lesions show pathological features similar to tuberous sclerosis complex (TSC)-related cortical tubers (i.e. presence of phosphorylated ribosomal protein S6 (pS6), a downstream component of mTOR activation) 3, mutations in the mTOR pathway were anticipated.
Recently, postzygotic brain-specific somatic mosaic mutations in PIK3CA, AKT3, and MTOR were identified in FMCDs 1,7–10. Mutations were found in 8–30% of cells and were some of the same gain-of-function activating mutations seen in solid malignancies including glioblastoma multiforme (GBM). Sequencing at the single-cell level identified a mutation burden both in neural and non-neural cells, indicating that mutations occur in progenitors 11. However, the mechanisms of disease and potential for treatment remain poorly defined. Moreover, how a relatively small percent of cells with mutation can disrupt the organization of the entire hemisphere is largely unknown 12.
RESULTS
Mosaic mutation is restricted to cells with mTOR activation
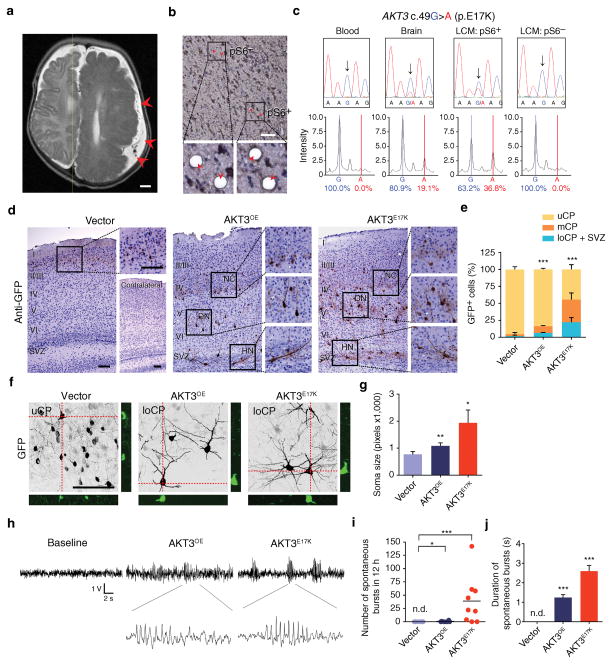
We focused on AKT3, a commonly mutated gene in FMCD 1,7,8,13. To test whether pS6+ cells were enriched for the mosaic mutation, we studied a human brain tissue with a somatic gain-of-function mutation (c.49G>A; p.E17K) which was present in 19.1% of the measured alleles based upon mass spectrometric analysis (Fig. 1a–c) 1. Laser-captured microdissection of pS6+ vs. pS6− neural cells was followed by PCR, then capillary sequencing and single base-extension mass spectrometry to test for the mutant allele. Analysis of captured cells showed that the mutant allele was undetectable in pools of pS6− cells whereas pS6+ cells of the same brain section were enriched for the mutation, detected in 36.8% of measured alleles. We conclude that pS6 is a sensitive but not specific marker for cells carrying the mutation, suggesting that activation of the pathway is relevant to the pathogenesis of disease.
Figure 1. Modeling FMCD mutation in developing brain.
(a) Representative presurgical axial T2 MRI of subject with hemimegalencephaly (previously reported in ref 1) with FMCD due to somatic mosaic AKT3 p.E17K mutation with dysorganization across the entire hemisphere (arrowheads), compared with healthy contralateral hemisphere. (b) Representative images of brain section with somatic mosaic AKT3 c.49G>A (p.E17K) mutation showing laser-captured microdissection (LCM) of pS6+ or pS6− cells. Postcapture of indicated cells (arrowheads) below. (c) Total DNA from blood or brain tissue and microdissected nuclei analyzed by capillary (Sanger) sequencing (top) and mass spectrometry (bottom). Arrows AKT3 c.49G>A. (d–g) Immunostained (GFP) images of mouse brain sections at postnatal day 20 (d), in utero electroporated at E14.5 with GFP-alone vector (n = 4) or GFP along with AKT3 wildtype (AKT3OE, n = 5) or AKT3 p.E17K (AKT3E17K, n = 5) expression vectors. Insets: neuronal crowding (NC), dysmorphic neurons (DN) and heterotopic neurons (HN). Contralateral side showed no structural defect. Localization of GFP+ cells quantified in upper (uCP, layer I–IV); middle (mCP, layer V), lower cortical plate (loCP, layer VI)-subventricular zone (SVZ) (e). Confocal z-stack images of P20 25 μm sections, showing orthogonal views (f) for quantification of cell size (g). (h–j) Representative surface EEG recordings from 1 month-old mice (h). Number (i) and duration (j) of spontaneous electroencephalographic bursts in 12 h window (n = 4, 6 and 9 for each condition). Values: mean ± s.d. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.d., not detected, G-test of goodness-of-fit (e); Student’s t-test (g, i, j). Scale bars: 1 cm (a); 100 μm (b, d, f).
Mouse model of focal malformations of cortical development
To test whether introduction of the FMCD mutation is sufficient to cause neurodevelopmental and neuropathological features, we electroporated a vector encoding the gain-of-function mutation of AKT3 (AKT3E17K), found in at least two persons with FMCD, into the right hemisphere of embryonic day (E) 14.5 mice. We chose this model because we could limit the expression of AKT3 to the neural progenitors of a single hemisphere, mimicking the somatic postzygotic mosaicism found in the embryonic brain of FMCD cases (Supplementary Fig. 1a). Electroporations were timed to target predominantly neurons destined for upper cortical layer (II/III), the cells proposed most severely affected in FMCD 14,15.
Histological examinations at postnatal day (P) 20 revealed key features observed in FMCD, including neuronal heterotopias, dysmorphic neurons with enlarged soma, and neuronal crowding (Fig. 1d–g) 3. While only 5.6% ± 3.9% of cells electroporated with control vector (i.e. GFP+) failed to reach upper cortical layers (uCP, layers II/III and IV), 16.3% ± 1.6% and 55.5% ± 7.0% of cells electroporated with human wildtype AKT3 (i.e. overexpression; AKT3OE) or AKT3E17K, respectively, were misplaced either in the middle cortical layer (mCP, layer V), the lower cortical layer (loCP, layer VI), or the subventricular zone (SVZ). Misplaced cells in the SVZ expressed neuronal (NeuN) but not glial markers (GFAP). No anatomical changes in the contralateral hemisphere were found.
The most devastating pathologies of FMCD are early onset seizures, which are often medically intractable 3. Mice electroporated with wildtype AKT3OE or AKT3E17K showed higher perinatal lethality (11.9%, vector control, n = 42; 24.3%, AKT3OE, n = 37 and 67.0%, AKT3E17K, n = 103), suggesting seizures might be a cause. Electroencephalogram (EEG) recording showed recurrent and unprovoked excessive activity in AKT3E17K (Fig. 1h–j). Control mice in contrast showed no activity, whereas AKT3OE mice showed only occasional bursts of lesser duration. An inflammatory response in the form of activated microglia was not apparent, suggesting disrupted neuronal circuitry or activity as a cause.
Developmental neuropathology results from AKT3 activation
Although clinical epilepsy may have numerous underlying cellular and molecular mechanisms, the investigation of epileptogenesis in FMCD has largely focused on intracortical and corticothalamic projections 16. Examination of axonal projections of electroporated neurons in mice showed only typical subcortical projections (data not shown). Further, we found no gross changes in overall inhibitory neurons evidenced by number or distribution of GAD1/2+ cells in electroporated regions.
We next examined whether the cell fate was altered by AKT3E17K expression. We utilized layer-specific markers SATB2 and CTIP2 to examine cortical cell positioning in developing brains and confirmed defective migration at E18.5 (Supplementary Fig. 1b, c). We found no changes in radial glia integrity detected by BLBP expression in electroporated regions (data not shown). In control vector-electroporated brains, 96% of GFP+ cells showed SATB2+ uCP identity, and were CTIP2−, a mCP marker. AKT3OE- or AKT3E17K-electroporated neurons were also predominantly SATB2+CTIP2− (93% or 96%, respectively) but were localized in the loCP (Supplementary Fig. 1d, e). These data suggest that despite the migration defect, birthdate-specified identity was not altered.
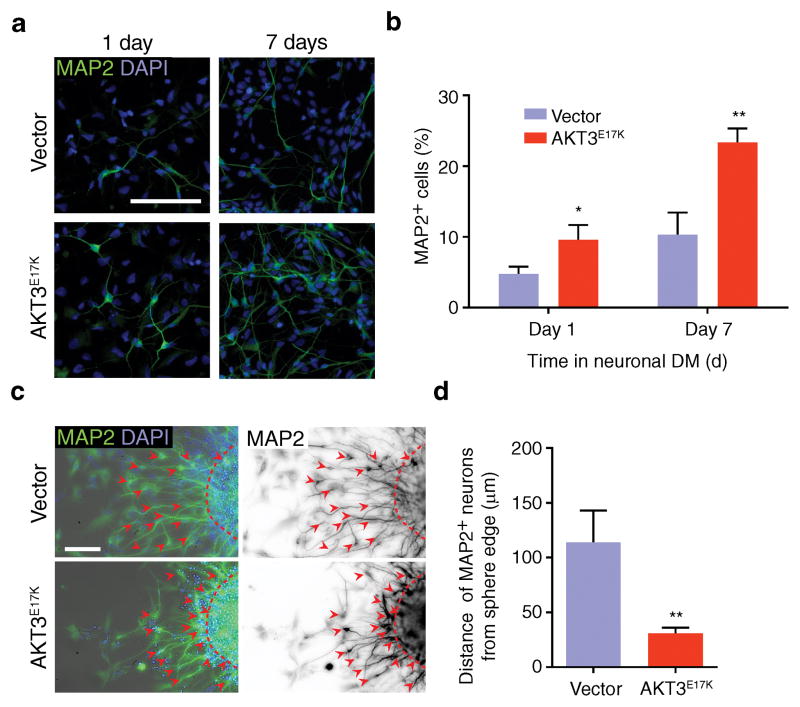
To test neuronal migration defects in human, we utilized human neural progenitor cells (hNPCs) differentiated from healthy donor induced-pluripotent stem cells (iPSCs). We selected these cells because they derive from human, they have been well studied and they express markers of both differentiating and migrating neurons depending upon the type of media used for culture 17,18. Cells were transduced with lentivirus expressing either RFP alone or AKT3E17K with RFP, then fluorescence-assisted cell sorted (FACS) for similar fluorescent intensity. In propagation media, approximately 4.6% ± 1.1% of control hNPCs expressed the neuronal differentiation marker MAP2, while 10.3% ± 3.1% of AKT3E17K-expressing hNPCs were MAP2+. When cultured in differentiation media, again about twice the AKT3E17K hNPCs were MAP2+ compared with controls (Fig. 2a, b), suggesting the mutation leads to premature differentiation. This was confirmed in E18.5 mouse brains, where GFP+MAP2+ neurons were found below the subplate in AKT3E17K-electroporated brains but not in control (Supplementary Fig. 1f).
Figure 2. Cellular pathology from AKT3 activation in human neural progenitor cells.
(a) Neural differentiation assayed by MAP2 staining (green) in human neural progenitor cells (hNPCs) expressing GFP-alone vector or AKT3E17K at 1 and 7 days in differentiation media (DM), quantified (b) (n = 5 cultures for both). (c) Neuronal migration of FAC-sorted lentiviral transduced hNPCs expressing GFP-alone vector or AKT3E17K in neurosphere assays. MAP2 (green) and DAPI (blue) on left used to define the edge of the neurosphere; MAP2 (black) on right used to measure migration distance. Dashed lines: boundary of neurosphere; arrowheads: cell body position. (d) Migration was quantified by measuring the distance of MAP2+ cells from the neurosphere boundary (n = 15 neurospheres from 5 cultures for both). Values: mean ± s.d. *, P < 0.05; **, P < 0.01, Student’s t-test (b, d). Scale bars: 100 μm.
In neurosphere migration assays, control neurons migrated 113.8 μm ± 29.0 μm from the neurosphere edge after two days whereas neurons of AKT3E17K-expressing neurospheres were densely located near the edge, migrating 30.9 μm ± 5.1 μm (Fig. 2c, d). Moreover, MAP2+ neurons in AKT3E17K-expressing neurospheres were often misoriented and failed to display typical migration morphology. Thus the premature differentiation may contribute to the migration defects.
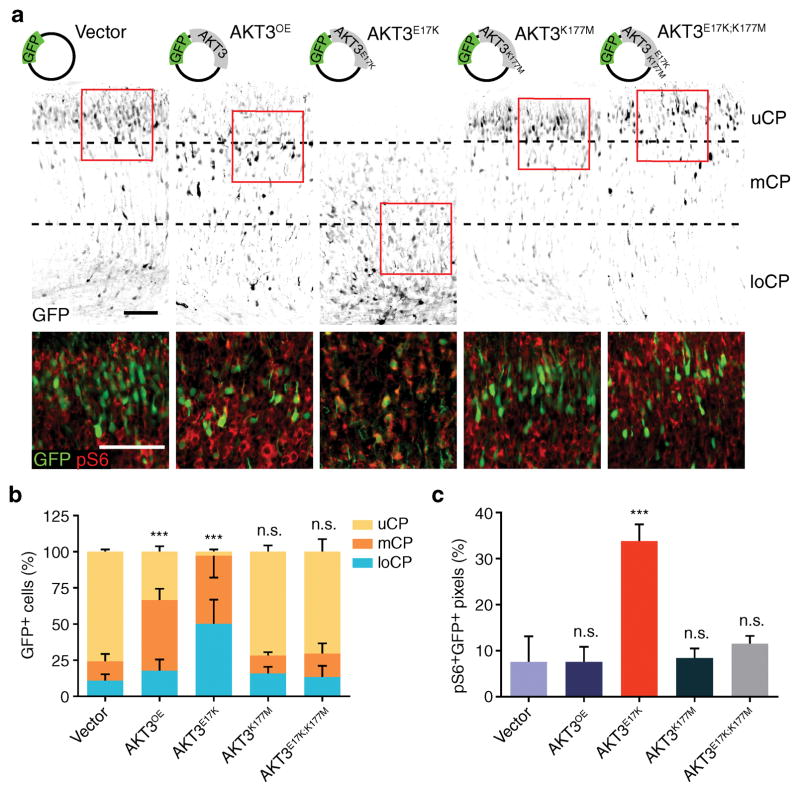
Requirement of AKT3 kinase activity
To investigate the requirement of AKT3 kinase activity in mediating neuronal migration deficits in vivo, we employed in utero electroporation of AKT3E17K constructs with additional p.K177M mutation in the same vector. This mutation renders AKT3 kinase null and thus the membrane localization-inducing p.E17K mutation has no effect on activating kinase activity. The kinase-deficient mutation abolished not only neuronal migration defects but also the hyperphosphorylation of the downstream target pS6, (Fig. 3a–c) suggesting a requirement for kinase activity. Furthermore introducing p.S472A mutation, a phosphorylation site critical for AKT3 activation, into the vector also prevented pathology (Supplementary Fig. 2a, b). Conversely, a pseudo-phosphorylation mutation (p.S472E), which activates downstream signaling, caused migration defects similar to that of AKT3E17K. We additionally tested the effect of overexpression of human wildtype PIK3CA (PIK3CAOE) or PIK3CAE545K mutation, a gene we and others have reported mutated in FMCD and other segmental overgrowth syndromes 1,7, and observed similar neuronal migration defects, and dysmorphic neurons with enlarged soma (Supplementary Fig. 2c–f). Activation of PI3K in developing cortex by inducing the expression of PIK3CAH1047R germline knock-in allele 19 by in utero hemispheric electroporation of Cre plasmid also caused similar neurodevelopmental defects (data not shown). Importantly, the neuronal migration defect in brains electroporated with PIK3CAE545K was almost completely prevented by co-expression of the kinase-null AKT3K177M (Supplementary Fig. 2c–d), consistent with a linear signaling pathway in the etiology of FMCD. Therefore the migration defect is not allele or gene specific but rather due to a general pathway hyperactivation.
Figure 3. AKT3 kinase activity is essential for aberrant migration phenotype.
(a) In utero electroporation at E14.5 of GFP vector encoding AKT3 variants, harvested at E18.5. Images were inverted for visibility, co-stained with phospho-S6 (pS6) of boxed regions below, enhanced contrast for visibility. (b) Localization of GFP+ cells quantified in upper (uCP); middle (mCP), and lower cortical plate (loCP). (c) Quantification of pS6+GFP+ pixels for the images described in (a). Values: mean ± s.d. (n = 3, 3, 6, 5 and 5 for each condition). n.s., not significant; ***, P < 0.001, G-test of goodness-of-fit (b); Student’s t-test (c). Scale bars: 100 μm.
Developmental rescue of neuronal defects
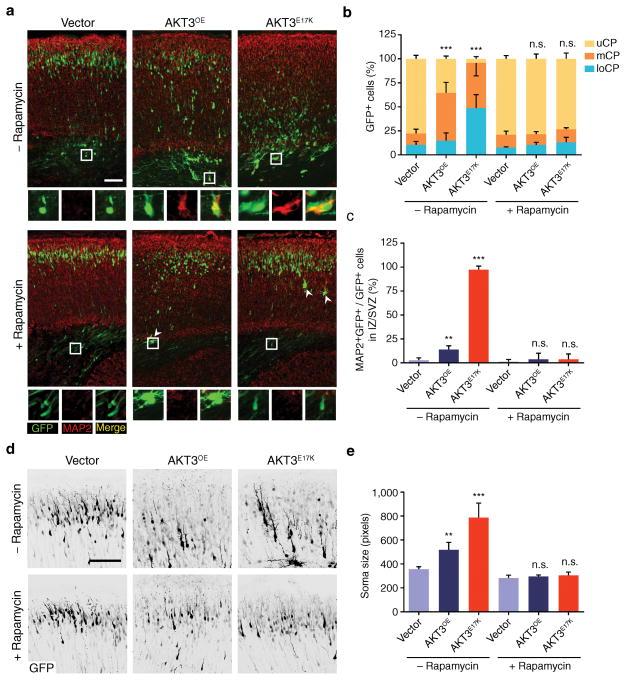
To test whether inhibition of the pathway could prevent the migration defect, we administered rapamycin, a potent inhibitor of mTOR signaling, to pregnant dams for four days following electroporation of AKT3 constructs (Supplementary Fig. 3a). Rapamycin treatment effectively rescued AKT downstream activation in developing cortex as assessed by pS6 immunostaining. It also largely rescued the cortical dyslamination resulting from aberrant neuronal migration, assessed by MAP2 immunostaining, as well as enlarged soma size (Fig. 4a–e, Supplementary Fig. 3b, c). However, postnatal treatment with rapamycin at P1 and P3 did not restore neuronal migration defect caused by AKT3E17K (data not shown), suggesting a specific developmental effectiveness window.
Figure 4. Pharmacological rescue of AKT3E17K-induced phenotypes.
(a) In utero electroporation at E14.5 followed by daily administration of rapamycin (3 μg/gram-body-weight/day), visualized at E18.5. MAP2 positivity of electroporated cells in lower cortex (insets). Arrowheads: rare giant neurons in rapamycin-treated embryos. (b, c) Quantified localization of GFP+ cells in upper (uCP); middle (mCP), and lower cortical plate (loCP), and MAP2+GFP+ cells in intermediate zone (IZ)/subventricular zone (SVZ). (d) Representative images used to calculate soma size of neurons from mice electroporated with GFP-alone, AKTOE, or AKTE17K expression construct, with or without rapamycin treatment. Tissue for analysis was harvested at E18.5. GFP immunofluorescence is shown in black. (e) Quantification of neuronal soma size for the groups described in (d). Values: mean ± s.d. (n = 3, 3, 3, 5, 3, 5). **, P < 0.01; ***, P < 0.001, G-test of goodness-of-fit (b); Student’s t-test (c, e). Scale bars: 100 μm.
Non-cell autonomous effect on migration
In AKTOE- and AKTE17K-electroporated hemispheres, we noticed an ectopic layer of SATB2+ neurons in the intermediate zone (IZ) or SVZ also included cells that were GFP− (Supplementary Fig. 1b, e), suggesting an effect of the mutant cells on neighboring wild-type cells. To explore potential non-cell autonomous migration defects we performed sequential in utero electroporation experiments 20. Serial electroporation of RFP followed by AKT3E17K construct also expressing GFP showed that AKT3E17K-expressing cells (i.e. RFP−GFP+) affected the migration of neighboring cells (i.e. RFP+GFP−) (Fig. 5a, b). BrdU birthdate analysis from E14.5 injections demonstrated dual-labeled ectopic AKT3OE or AKT3E17K cells in mCP or loCP, suggesting that the misplaced cells underwent DNA synthesis concomitant with other cells destined for upper cortical layers (Supplementary Fig. 4a, b), and no significant difference in proliferation or apoptosis rates were detected in migrating neurons (data not shown). Increased GFP− cells that were BrdU+ in loCP in brains expressing either AKT3 or AKT3E17K was also observed (Supplementary Fig. 4c), together suggesting an effect of AKT3E17K-expressing cells on the migration of neighboring cells.
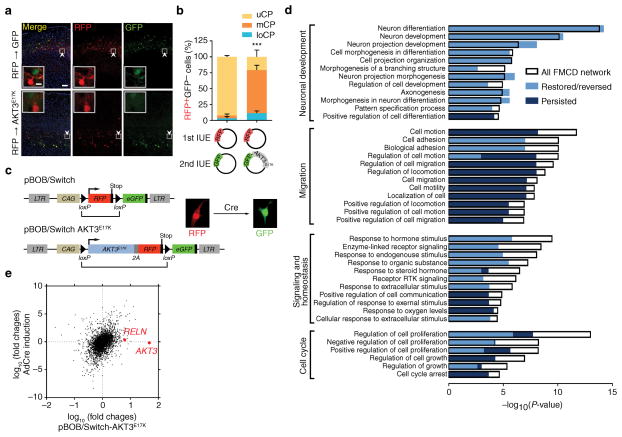
Figure 5. Genetic recombination of AKT3E17K defines reversibility of FMCD networks.
(a) In utero electroporation at E14.5 of RFP vector, followed by second electroporation after 15 min of GFP-alone (n = 5) or GFP along with AKT3E17K (n = 3) harvested at P14. Insets: singularity of plasmid expression. (b) Quantified localization of RFP+GFP− cells. (c) pBOB/Switch vectors, empty (top) or with AKT3E17K linked (bottom). RFP expressed from a bicistronic 2A sequence, allowing stoichiometric RFP and AKT3E17K expression. LTR, long terminal repeat; CAG, chicken actin promoter. RFP-expression cassette with stop codon is removed upon Cre recombination with adenoviral transduction (bracket), providing color switch from RFP to GFP. (d) GO analysis identified four major enriched categories (Neuronal development, Migration, Signaling and homeostasis, and Cell cycle), and several subcategories as indicated. P-values for GO enrichment for each subcategory are shown (x-axis). Colors indicate P-values of the GO enrichment for the portion of the genes within each subcategory that persisted (dark blue), or were restored (from high to background or from low to background, light blue) or reversed (from high to low or from low to high, light blue), overlaid on each subcategory. (e) Comparison of mRNA abundance after AKT3E17K overexpression (x-axis) vs. genetic removal of AKT3E17K by Cre recombination (y-axis). Note that the expression of AKT3 and RELN were initially elevated with AKT3E17K overexpression but were restored upon Cre recombination. Values: mean ± s.d. ***, P < 0.001, G-test of goodness-of-fit (b); Scale bars: 100 μm (a); 10 μm (inset in a).
Functional networks underlying FMCD
The non-cell autonomous migration defect may be due to physical blockade of wildtype cells trying to migrate past mutant cells, or may have more specific signaling mechanisms. The AKT3E17K mutation was introduced to hNPCs via lentiviral transduction using the pBOB/Switch vector, which allows Cre-mediated recombination of the cDNA cassette (Fig. 5c). To obtain homogeneous AKT3E17K-expressing hNPCs, we collected cells expressing RFP at similar intensity by FACS (Supplementary Fig. 5a), and then immediately isolated RNA and performed RNA-seq expression profiling. We identified 835 genes whose expression was significantly altered (false-discovery-rate-corrected P-value < 0.05) in AKT3E17K compared with empty vector. These included several genes previously reported downstream of AKT signaling such as CDKN2A (encoding p16INK4a and p14ARF), CDKN2B (encoding p15INK4b) and CCND1 (encoding cyclin D1), suggesting robust methodology. Network analysis of the 835 genes using gene ontology (GO) enrichment analysis revealed four major categories: 1] Neuronal development, 2] Migration 3] Signaling and homeostasis and 4] Cell cycle (Fig. 5d, Supplementary Fig. 5b).
Genetic rescue identifies reversible FMCD network
To determine reversibility of molecular changes induced by FMCD mutations, Cre recombinase was introduced by adenoviral transduction into hNPCs, to remove the AKT3E17K-viral 2A-RFP cassette. Cre recombination also removed the stop codon located 5′ of the GFP start site, enabling visualization of recombined cells by GFP expression (Supplementary Fig. 5c). Upon Cre expression, AKT3 transcript level was restored to basal level (Fig. 5e) suggesting efficient genetic recombination that correlated with color switching from red to green. Repeating the RNA-seq analysis identified 275 of the 835 genes (i.e. 1/3) persistently misexpresssed, whereas the majority 560 (i.e. 2/3) saw expression changes restored or moved in the opposite direction (i.e. reversed).
Critical role of reelin in developmental pathogenesis
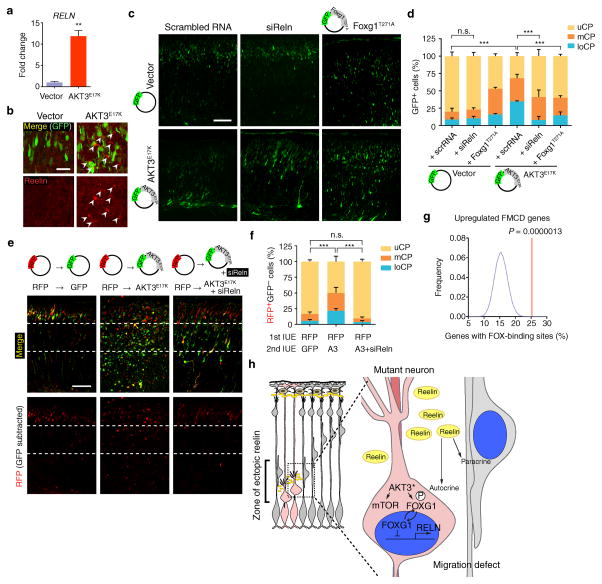
The rescue of the migration defect with rapamycin suggested that the relevant pathway disrupted by AKT3E17K expression was reversible, and thus we focused on the set of genes that restored expression following Cre recombination. Among these, RELN, encoding the reelin secreted glycoprotein 20 essential for neuronal migration and lamination during cortical development 21–24, was the most significantly changed gene (P < 0.00005), and was dramatically upregulated.
Loss of RELN leads to profound defects in neuronal positioning and dendritogenesis in human and mouse 25, whereas ectopic expression in migrating neurons induces neuronal ectopia 26,27. We confirmed increased expression of RELN in AKT3E17K hNPCs, increased about ten-fold (Fig. 6a). In AKT3E17K-expressing brains, reelin was detected in excessive amounts, localized to perisomal regions close to ectopic neurons (Fig. 6b). Reelin misexpression by AKT3E17K was not detected at postnatal day 20, suggesting that the pathogenic mechanisms are limited to a specific developmental window (Supplementary Fig. 6a). We also examined the expression of RELN in pS6+ vs. pS6− cells microdissected from surgically resected FMCD human brain lesions tested for somatic AKT3E17K mutation and confirmed RELN expression in only one of four pool of the pS6+ cells (Supplementary Fig. 6b). We conclude that reelin misexpression resulting from AKT3E17K is mostly limited to developing rather than mature neurons.
Figure 6. Neuronal migration defects rescued by Reln siRNA.
(a) Quantitative real-time PCR was performed to measure RELN expression in hNPCs expressing AKT3E17K and compared to vector-expressing hNPCs. Three independent experiments were quantified in triplicate. (b) Mouse embryos at E14.5 were electroporated with indicated DNA constructs. Brains isolated at E18.5 used for reelin expression. Arrowheads: reelin immunostaining in pericellular area of ectopic neurons (intensity-per-pixel: 622.55 ± 45.80 compared to 4092.77 ± 9.46, marginal zone). (c) E14.5 embryos were co-electroporated with indicated constructs or RNAs. (d) Localization of GFP+ neurons at E18.5 was quantified in each cortical region (n = 3 for each). (e, f) Developing cortices were in utero electroporated (IUE) at E14.5 with RFP vector. After 15 min, embryos were sequentially electroporated with either GFP vector (n = 3) or GFP also expressing AKT3E17K (A3) with or without Reln siRNA (n = 6 and 4, respectively). At E18.5, brains were sectioned for imaging. Localization of RFP+GFP− cells was quantified in each cortical region. Values: mean ± s.d. n.s., not significant; **, P < 0.01; ***, P < 0.001, Student’s t-test (a); G-test of goodness-of-fit (d, f). Scale bars: 25 μm (b); 100 μm (c, e). (g) Enrichment of FOX-binding site. See Online Methods for details. (h) Model of effect of AKT3 mutation on surrounding cells. Mutant cell, pink; surrounding cells, gray. AKT3* activating somatic mutation leads to mTOR activation and phosphorylation and cytoplasmic sequestration of FOXG1, relieving repression of RELN, which is then secreted by mutant cells to act in an autocrine (cell autonomous) and paracrine (non-cell autonomous) fashion.
Reelin is initially expressed throughout in the cortical anlage at the preplate stage (E9.5), but after E11.5 expression is mainly restricted to the Cajal-Retzius cells of the marginal zone. Therefore, we tested the degree to which defective migration in AKT3E17K-expressing cells and neighboring cells was due to the misexpression of reelin in ectopic regions by knockdown of Reln in AKT3E17K-electroporated cells. In utero electroporation of Reln siRNA constructs into cortical progenitors alone produced no defect in migration, supporting its role in specialized Cajal-Retzius cells at this time point. However, the migration defect of AKT3E17K-expressing neurons was partially but significantly restored (P < 0.001) by the co-electroporation of siRNA targeting Reln (Fig. 6c, d) suggesting its overexpression in AKT3E17K cells leads to an autocrine or paracrine effect. To test specifically for a paracrine effect of reelin on neuronal migration, sequential electroporation of RFP followed by a mixture of AKT3E17K and Reln siRNA was electroporated such that RFP-expressing cells should thus no longer be exposed to ectopic reelin (i.e. GFP+). This nearly fully rescued the non-cell autonomous migration defects (Fig. 6e, f) suggests a central role for ectopic reelin expression in the etiology of the migration defect in AKT3E17K brains.
Reelin misregulation by activation of AKT3-FOXG1 pathway
To investigate the mechanism by which AKT3 regulates reelin expression, we looked for commonalities in genes altered by AKT3E17K but restored after Cre recombination. We found that among the genes meeting this criterion, there was over-representation of those with FOX-binding sites within 5kb upstream of the transcription start site (P = 1.3 × 10−6) (Fig. 6g). The forkhead box (FOX) transcription factors mediate downstream signaling from the AKT pathway. Despite the established role of the FOXO class mediating AKT downstream effects 28, expression of constitutively active FOXO1 or FOXO3A in hNPCs, did not repress RELN expression (data not shown).
In mice, Foxg1 controls the morphogenesis of the telencephalon 29, and mutants show increased numbers of reelin-expressing Cajal-Retzius cells 30. Under basal condition, FOX factors act as transcriptional repressors, but after phosphorylation by AKT, there is cytoplasmic translocation, inhibiting transcriptional repressor activity 31. Thus, we searched for conserved FOX-binding sequences and identified four potential sites (R1–R4) upstream of RELN. Chromatin immunoprecipitation demonstrated that FOXG1 was significantly enriched (P < 0.001) in two of the sites that have two FOX-binding sites (Supplementary Fig. 6c). Next, we confirmed cytoplasmic localization of FOXG1 in hNPCs expressing AKT3E17K (Supplementary Fig. 6d, e). In NIH 3T3 cells, Foxg1-GFP fusion protein predominantly localized to the nucleus, however, co-expression with AKT3E17K induced cytoplasmic translocation (Supplementary Fig. 6f, g). Mutation of the putative T271 phosphorylation site to non-phosphorylatable alanine (Foxg1T271A) abolished AKT3E17K-induced cytoplasmic translocation of Foxg1. Conversely, the phosphomimetic glutamic acid (Foxg1T271E) increased cytoplasmic localization. Furthermore, increased RELN transcription in AKT3E17K hNPCs was repressed by the expression of Foxg1T271A (Supplementary Fig. 6h). Similar to Reln siRNA, co-electroporation of Foxg1T271A in developing brain partially rescued migration defects caused by AKT3E17K expression (Fig. 6c, d). These findings suggest that AKT3 mediates FOXG1 phosphorylation-dependent de-repression of RELN transcription.
DISCUSSION
A major focus in neuroscience is improved care for patients with intractable epilepsy. The fact that the vast majority of patients undergoing surgical resection of FMCDs become seizure-free and show cognitive stabilization or improvement 32 indicates that the focus for the neurocognitive defects most frequently resides within the lesion.
Here we demonstrate that postzygotic mutations identified in FMCDs are restricted to cells reporting activation of the pathway, and are sufficient to recapitulate the disrupted neuronal migration, excessive neuronal size, altered differentiation, and epileptiform activity observed in FMCDs. The question became how somatic mutations in just a small percentage of cells could produce such severe and widespread defects in brain development. Profiling mRNAs pointed to reelin ectopic misexpression which was due at least in part to AKT3-mediated phosphorylation-dependent inactivation of Foxg1, leading to cytoplasmic localization and reelin derepression in developing brain (Fig. 6h).
A role for the mTOR pathway in brain development was first proposed from work in tuberous sclerosis complex 33, and later demonstrated through conditional removal of key regulators like PTEN and TSC1/2 34–36. The emerging consensus is that activation of the pathway leads to neuronal hypertrophy, excessive dendritic branching, with a cell-type specific effect on excitability, whereas inhibition shows opposite effects 37–39. Recently focus has been on the mTOR pathway in autism, both because of the striking autistic features displayed by persons with monogenic syndromic mTORopathies, as well as evidence of mutations in the mTOR pathway in individuals with non-syndromic forms of autism 40. This connection is supported by animal models with forebrain-specific deletion of Pten or cerebellar-specific deletion of Tsc1 resulting in marked abnormalities of social behavior 41,42. The data suggest that germline or widespread mutations may result in overt structural brain defects, whereas subtle, cell-type specific or mosaic mutations may result in restricted phenotypes.
De novo heterozygous mutations in FOXG1 present with a congenital variant of Rett syndrome with a generalized cortical malformation 43, suggesting haploinsufficiency as a disease mechanism, whereas complete deletion in mouse results in near absence of the telencephalon 29. Interestingly, whereas humans with homozygous RELN mutations present with a form of lissencephaly 25, RELN genetic variants in the 5′ UTR predispose to autistic disorder 44. A conceptual advance in our data is that it was not so much reelin misexpression but rather ectopic misexpression in cortical zones not normally exposed to reelin that led to the migration defect during developmental stage. Knockdown of Reln rescued migration not just for the neighboring cells but also the cells expressing the AKT3 mutation, suggesting autocrine or paracrine effect of reelin on neuronal migration. Rapamycin rescued the non-cell autonomous migration defect probably because of its effect on cell autonomous migration defects, thus hindering accumulation of reelin in this ectopic location. The ectopic RELN misexpression was seen in developing brain but not in mature brain, suggesting that RELN expression will not be a sensitive marker for cells with mTOR pathway mutations in human cortical resections.
The results suggest two possible explanations for FMCD-related seizures in humans: a] cortical dyslamination due to disrupted migration or b] increased neuronal excitability related to morphological or signaling changes. Although cortical dyslamination can cause neuronal circuitry defects such as abnormal intracortical and corticothalamic projections linked to epilepsy 16,45, epileptic seizures are traditionally characterized as the synchronous neural hyperactivity arising from imbalanced excitation and inhibition 46. While reelin signaling regulates dendrite growth 47 and ion channel compartmentalization contributing to hyperpolarization-activated cation currents 48, no epileptic phenotype has been reported in mice under- or over-expressing reelin 49. On the other hand hyperactivation of the mTOR pathway can increase neuronal excitation. Thus, the data argue in favor of mTOR pathways as making a larger contribution to epilepsy in the mTORopathies.
Mutations in PI3K-AKT pathway have long been recognized in brain tumors, with somatic mutations found in upwards of 90% of GBMs 50. Why then do persons with FMCD with the same mutation not develop tumors? One possibility is that PI3K-AKT pathway transformation is cell-cycle dependent. However, most neuroblasts are actively dividing during periods of migration, so this is unlikely to create a barrier to transformation. Another possibility is that activating mutations are not sufficient for cellular transformation in neural cells, and data certainly support cooperativity among signaling pathways such as p53 and Rb in the induction of high-grade astrocytomas in adults 50,51. In fact, selective activation of mTORC1 in a specific context produces microcephaly due to massive programmed cell death 52.
Several human syndromes such as Cowden, Lhermitte-Duclos, CLOVES, Proteus and megalencephaly syndromes (OMIM#158350, #612918, #176920 #603387, respectively) demonstrate hamartomatous non-cancerous lesions or increased organ size across the body plan, along with mutations predicted to hyperactivate the mTOR pathway 7,53,54. Mouse models in neural progenitors demonstrate roles for the mTOR pathway in determining cellular morphology and size 35,41. A picture of segmental overgrowth due to activating mutations is emerging 55, and in vivo models provide useful tools to study 10,56. Our finding that activating mutations in the PI3K-AKT pathway recapitulate FMCD in a mammalian model sets the stage for further mechanistic studies, and development of therapies for these catastrophic disorders.
ONLINE METHODS
Animals
Timed-pregnant females (C57/BL6 or CD-1) were obtained by overnight breeding with males (C57/BL6). For timed-pregnancy, noon after mating was considered embryonic day (E) 0.5. Animals were obtained from Charles River, Harlan Laboratories or Jackson Laboratory. Animal care, maintenance, and experimental procedures followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Institutional Animal Care and Use Committee standards at University of California San Diego, The Rockefeller University and The Scripps Research Institute.
DNA Constructs and siRNA
The empty vectors pCAG-IRES-GFP (pCIG) has been used to clone human AKT3 or PIK3CA. Variants (p.E17K, p.K177M, p.S472A, p.S472E, p.E17K;S472A of AKT3 or PIK3CA p.E545K) were generated with Quick change II site-directed mutagenesis kit (Agilent). PstI-digested fragment of pCIG-AKT3 p.K177M was subcloned into PstI-digested pCIG-AKT3 p.E17K to generate AKT3 p.E17K; p.K177M. AKT3 p.E17K linked to RFP via 2A was cloned into pBOB/Switch vector to generated pBOB/Switch-AKT3 p.E17K. For Foxg1-GFP fusion protein, Foxg1 constructs (wildtype, p.T271A or p.T271E) was cloned into pCAG-GFP vector. All clones were confirmed by sequencing analysis. pBOB/Switch vector and Foxg1 construct were kindly provided by Dr. Inder Verma and Dr. Gordon Fishell respectively. In Reln knockdown experiments, two siRNA constructs (Silencer® Pre-Designed siRNA; Life Technologies; cat. no. AM16708; ID 151443 and ID 151445) were tested separately and obtained similar result.
Chemicals
BrdU (Sigma) were maternally administrated (100 mg per gram-body-weight) by intraperitoneal injection. BrdU was dissolved in saline with NaOH (7 mM). Rapamycin (LC Labs) stock was prepared in ethanol (50 mg/ml), and diluted with vehicle (4% Tween 80; 4% polyethylene glycol 400) before injection.
In utero electroporation
In utero electroporation was performed as described previously 57 with modifications as follows. Endotoxin-free plasmids (0.5–1 μg) plus 0.5% Fast Green (Sigma) were injected into one lateral hemisphere of E14.5 embryos. Electroporation was performed by placing the anode on the side of DNA injection and the cathode on the other side of the head to target cortical progenitors. Four pulses of 45 V for 50 ms with 500 ms interval were used. Upon isolation of brains, contralateral hemisphere was also examined for possible anatomical changes.
hNPC culture and neurosphere migration assay
iPSC derived from human primary dermal fibroblasts (ATCC) were differentiated into neural progenitor cells (hNPCs) by standard protocol. Briefly, three micrograms of expression plasmid mixtures (OCT3/4, SOX2, KLF4, L-MYC, LIN28 and p53 shRNA) were electroporated to fibroblasts, negative for mycoplasma contamination. After 7 days, cells were re-plated onto irradiated CF-1 mouse embryonic fibroblasts (MEF) feeder layer. The culture medium was replaced the next day with hESC/iPSC medium (DMEM:F12 supplemented with 20% KOSR and 20 ng/ml bFGF, 1X nonessential amino acids, 110 μM 2-mercaptoethanol). Healthy colonies were selected for further cultivation and evaluation. After 3 passages, iPSCs were transferred to MEFs free plates and cultured with mTeSr medium (Stem Cells Technologies). For the generation of human neural progenitors cells, embryoid bodies (EBs) were formed by mechanical dissociation of cell clusters and plated in suspension in differentiation medium (DMEM F12, 1X N2, 1 μM Dorsomorphin (Tocris), 2 μM A8301 (Tocris)) and kept shaking at 95 rpm for 7 days. EBs were then plated onto Matrigel coated dishes and cultured with NBF medium (DMEM/F12 supplement with 0.5X B-27, 0.5X N-2 and 20 ng/ml bFGF). After 5–7 days, rosettes were collected and dissociated with Accutase (Millipore), and hNPCs were plated onto poly-ornithine- and laminin-coated plates with NBF medium. Medium was replaced every other day. hNPCs were characterized by immunocytochemistry for the expression of neural progenitor cell markers (SOX2, PAX6, Nestin) but for the absence of differentiated neuronal (MAP2) or glial markers (GFAP). RNA-seq allowed for expression characterization of these cells showing fragments per kilobase of transcript sequence per million mapped fragments (FPKM) values 58,59 for TUBB3, DCX, SOX2, PAX6, and NES of 296.98 ± 8.35, 1.38 ± 0.11, 90.60 ± 2.23, 1.20 ± 0.12 and 532.20 ± 1.89, respectively. For neuronal differentiation, bFGF was withdrawn from NBF (differentiation medium). For lentiviral transduction, pBOB/Switch lentiviral vectors were co-transfected with packaging plasmids into HEK293T cells using Lipofectamine LTX (Life Technologies). After 24 h, transfected cells were cultured in NBF media and viral particle-containing media was harvested twice at 48 h and 72 h. hNPCs were cultured in virus-containing NBF media for 24 h. Transduction efficiency was monitored by RFP expression for 48 h followed by FACS. The AKT3 overexpression and downstream activation was confirmed by western blot. For adenoviral Cre induction, 200 μl of adenovirus-containing media (~108 CFU/ml) was added to cultures. After 8hr, cells were cultured in fresh NBF media and monitored for Cre recombination by GFP expression for 72 h followed by fluorescence-assisted cell sorting (FACS) of RFP+GFP− cells. For neurosphere migration assay, sorted hNPCs were seeded to non-coated plates containing NBF media with gentle agitation to generate neurosphere. Neurospheres with similar diameter were then cultured in differentiation medium for two days on poly-ornithine- and laminin-coated plated followed by immunostaining.
Immunohistochemistry and staining
For frozen embedding, tissues were fixed in 4% paraformaldehyde (PFA, Sigma) for overnight at 4°C with gentle rotation, immersed in 30% sucrose for cryoprotection then embedded in OCT. Tissue sections were permeabilized with 0.1% Triton X-100 in PBS (PBT), blocked with Cas block (Life Technologies) and immunostained with antibodies against phospho-S6 (1:250; Cell Signaling; cat. no 5364), MAP2 (1:500; Millipore; cat. no. MAB3418), reelin (1:200; Abcam; cat. no. ab78540), CTIP2 (1:500; Abcam; cat. no. ab81465), SATB2 (1:500; Abcam; ab51502) and FOXG1 (1:200, Abcam; cat. no. ab18259). For BrdU (1:50; BD Biosciences; cat. no. 347580) antibody staining, tissues were treated with HCl (2 N) before blocking. Paraffin embedded tissues were sectioned, rehydrated and retrieved for antigen (citrated buffer, pH 6.0) before immunostaining for GFP (1:200; Rockland; cat. no. 600-106-215) followed by detection by DAB (Vector Labs). Hematoxylin (Fisher) counter staining was performed according to the manufacturer’s protocol.
Quantitative real-time PCR (qRT-PCR) and chromatin immunoprecipitation
Total RNA was extracted with PureLink RNA extraction kit or Picopure RNA extraction kit (Life Technologies) followed by cDNA synthesis with SuperScript (Life Technologies) and ran for qRT-PCR with CFX96 (Bio-Rad) in triplicate. Primer sequences are: GAPDH, forward, 5′-GGA GCG AGA TCC CTC CAA AAT-3′, reverse, 5′-GGC TGT TGT CAT ACT TCT CAT GG-3′ and RELN, forward, 5′-TGA GAG CCA GCC TAC AGG A-3′, reverse, 5′-TCG TTC CAC ATT CTG TAC CAA-3′. Chromatin immunoprecipitation of hNPCs was performed with IgG or FOXG1 antibodies (Abcam; cat. no. ab18259) followed by qRT-PCR. Primer sequences are: RELN-R1- forward, 5′-CCG ATT TCA GGG CCA TTG GTC-3′, RELN-R1-reverse, 5′-GCT TCT GGG CTC TTC AGG CAG-3′, RELN-R2-forward, 5′-TGG CAG TTA AGC CTC TGG GG-3′, RELN-R2-reverse, 5′-CCA AGG GGA GAC CTC TGT AAT CAC-3′, RELN-R3-forward, 5′-GGG GAC TTC CTG GGA ATT GAG G-3′, RELN-R3-reverse, 5′-GTC TGG CCA TGC TTG ATG CC-3′, RELN-R4-forward, 5′-CCC TAA TAT CCC ACC TTT CTC AGT GC-3′, RELN-R4-reverse, 5′-CAT GAG AGC AAA CCC AAT AGC AGC-3′, RELN-R4-forward, 5′-GGC TTA CTG ATC TTT AGG GTC TAT TTT TGT-3′ and RELN-R4-reverse, 5′-AAT GGT AAT AAA AGG CCA CAT GAG AGC-3′.
Mutation burden analysis using single–base pair extension
Mutation burdens were analyzed with the Sequenom MassARRAY platform as described previously 1. Online MassARRAY Designer software (https://mysequenom.com) was used to design the primers for PCR and iPLEX single-base extension analysis. PCR and subsequent single–base pair extension reactions were performed with iPLEX Gold (Sequenom) according to the manufacturer’s protocol. Allele-specific differences in mass between extension products were detected by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry and were analyzed by MassARRAY TyperAnalyzer software.
Laser-captured microdissection (LCM)
LCM was performed with MMI CellCut Laser Microdissection system. Brain tissues were sectioned (30 μm) onto membrane slides (MMI) followed by pS6 immunostaining and hematoxylin counter staining. Membrane with no tissue (as a negative control) or 2 to 20 nuclei from pS6+ or pS6− cells were cut and collected to isolation caps (MMI) followed by DNA extraction. For the mutation burden analysis, genomic region covering AKT3 c.49G>A was amplified by nested PCR followed by capillary sequencing and single-base pair extension analysis. Minimum of 7 independent samples for each condition were tested.
Imaging and quantifications
An Olympus IX51with QImaing Retiga-2000R camera was used for fluorescence imaging and a Keyence BZ-9000 for color imaging. Higher magnification images were acquired using an Olympus FV1000 confocal microscope. Analysis of images was performed with ImageJ. All images and figures were edited and created in Photoshop CS4 or Adobe Illustrator CS4.
RNA sequencing, GO enrichment and FMCD network analysis
RNA was extracted and purified using Purelink RNA kit (Life Technologies). Mature mRNA was capture by TruSeq Stranded mRNA kit (Illumina) with standard protocol. Single-end 100 nucleotide reads were generated with Illumina HiSeq 2500 to an expected ~30 million reads per sample. RNA sequencing data have been deposited in the NCBI Sequence Read Archive with BioProject accession code SRP063581. Reads were aligned to the 1000 Genomes Project’s version of GRCh37 using standard TopHat2 (http://ccb.jhu.edu/software/tophat/) v2.0.11 with single-end read options and allowing for intron-spanning reads as defined by transcripts in Illumina’s iGenomes NCBI build 37.2. Differential expression on a gene-based level was tested for with cuffdiff 60 v2.1.1 using default option, after exclusion of one AKT3E17K replicate. Genes reported as significantly differentially expressed between a pair of conditions were determined to have been so based on cuffdiff’s threshold of a 0.05 false-discovery rate corrected P-value. Gene Ontology terms for the specified sets of genes were tested for enrichment with DAVID’s functional annotation tool (http://david.abcc.ncifcrf.gov/). Cytoscape v3.1.1 with ReactomeFI plugin (http://www.cytoscape.org/) was used for functional FMCD network analysis.
EEG surgery
Mice were implanted under isoflurane anesthesia (1%–2%), with a standard set of stainless-steel screw electrodes for chronic electroencephalographic (EEG) recordings. Anesthetized animals were placed in a stereotaxic apparatus, their head hair was shaved off and the incision site was prepared with ethanol and betadine. The EEG was recorded from two bilateral electrodes inserted in the frontal bone (AP = 1.0; ML ± 1.5), and two bilateral electrodes implanted in the parietal bone over the hippocampus (AP = −2.06; ML = 1.5), whereas a fifth electrode was placed over the cerebellum and served to ground the mouse to reduce signal artifacts. Insulated leads from these electrodes were then soldered to a miniconnector that was cemented to the skull with dental acrylic. Wounds were sutured closed, topical antibiotic ointment was applied and mice were injected subcutaneously with the analgesic flunixin. After surgery, mice were housed in individual cages and allowed a 1-week recovery before recordings. The ambient temperature was maintained at 25ºC ± 1.0, and a 12 h light/dark cycle was maintained throughout the recovery period and the subsequent experiment period. Food and water were available ad libitum. Once recordings were done, animals were anesthetized and their brains were dissected for further analysis.
EEG Analysis
EEG signals were amplified in a Grass Model 7D polygraph (Quincy, MA), filtered in a frequency range of 0.3 to 100 Hz and sampled at 256 Hz. The EEG signals are displayed on a computer monitor and stored with a resolution of 128 Hz in the hard drive of a computer for the off-line analysis of the vigilance states and spectral analysis, using software supplied by Kissei Comptec (Irvine, CA). The polygraphic results were analyzed semiautomatically by 15-second epochs. Epochs with artifacts in the polygraph records were discarded. Spontaneous bursts (1–3 second burst of spontaneous 300–500 mV activity) were quantified in number and duration.
Statistical analysis
Two-tailed unpaired Student’s t-test was used to calculate P-values comparing two means. G-test of goodness-of-fit was used to calculate P-values comparing two mean ratios. A P-value less than 0.05 was considered statistically significant. No animals were excluded from the analysis. When variances between groups were found to be different, a Welch’s t-test was used. No statistical method was used to predetermine sample size. For in utero electroporation, experiments were designed to include both littermate and non-littermate controls with the exception of Figure 1h–j where control and experimental groups were non-littermate. Because the animal groups were defined by electroporated DNA or RNA, no randomization was used. For Figure 1h–j, the investigators were blinded to group allocation during the experiment and when assessing the outcome. For Figure 6g, upsteam5000.fa.gz and tfbsConsSites.txt.gz were downloaded from the UCSC Genome Browser database. These files contain the upsteam promoter sequence of all annotated genes in the human genome and binding sites for a number of transcription factors. The two data sets were intersected based on whether there was any overlap between the 5,000 bases upstream of each gene and the FOX-binding sites to decide for each gene whether its promoter region was associated with FOX. A given gene set is tested for enrichment of FOX-binding sites in its promoter by first counting the number of genes in the set whose 5′ UTRs overlap FOX-binding sites and as a null distribution counting the number of genes that overlap for randomly selected without replacement subsets of the matched number of genes in the human genome (10,000 iterations). By this process an empirical P-value is calculated for the significance of the enrichment for the gene list specified.
Supplementary Material
Acknowledgments
We thank N. Cai, M. Huynh, T. Chirwa, K. Um, J. Silhavy and U. Yang for technical expertise. This work was supported by the US National Institutes of Health R01NS083823 (J.G.G. and G.W.M), Simons Foundation for Autism Research (275275, J.G.G), the Howard Hughes Medical Institute (J.G.G.), the 2014 NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (22892, S.T.B.), the Human Frontier Science Program Long Term Fellowship (S.K.K). We thank the UCSD Neuroscience Microscopy Core P30 NS047101 for imaging support, K. Jepsen from UCSD IGM Core Facility, UCSD Human Embryonic Stem Cell Core Facility, A. Roberts from Scripps Research Institute Animal Core Facility, I. Verma (Salk Institute) for pBOB/Switch vector, A. Acharya (University of Texas Southwestern Medical Center) for Cre-expressing adenovirus, G. Fishell (New York University) for Foxg1 plasmid, P. Mischel, I. Martin-Valencia, T. Curran and T. Park for discussions.
Footnotes
ACCESSION CODE
RNA sequencing data have been deposited in the NCBI Sequence Read Archive with BioProject accession code SRP063581.
AUTHOR CONTRIBUTIONS
S.T.B. and J.G.G. designed experiments, analyzed data and wrote the manuscript. B.C. performed bioinformatics analysis of RNA-seq results. S.T.B. performed experiments. E-.J.Y. performed FOXG1 chromatin immunoprecipitation. S-.K.K. helped with in utero electroporation. A.G-.G., A. E. S., S.K., H-.C.K., S.S. and G.W.M. contributed key reagents and advice. J.G.G. conceived and supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Lee JH, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronica E, Becker AJ, Spreafico R. Malformations of cortical development. Brain Pathol. 2012;22:380–401. doi: 10.1111/j.1750-3639.2012.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumcke I, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52:158–174. doi: 10.1111/j.1528-1167.2010.02777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor DC, Falconer MA, Bruton CJ, Corsellis JA. Focal dysplasia of the cerebral cortex in epilepsy. J Neurol Neurosurg Psychiatry. 1971;34:369–387. doi: 10.1136/jnnp.34.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riviere JB, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poduri A, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakashima M, et al. Somatic mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann Neurol. 2015 doi: 10.1002/ana.24444. [DOI] [PubMed] [Google Scholar]
- 10.Lim JS, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21:395–400. doi: 10.1038/nm.3824. [DOI] [PubMed] [Google Scholar]
- 11.Evrony GD, et al. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai X, et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell reports. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conti V, et al. Focal dysplasia of the cerebral cortex and infantile spasms associated with somatic 1q21.1-q44 duplication including the AKT3 gene. Clin Genet. 2014 doi: 10.1111/cge.12476. [DOI] [PubMed] [Google Scholar]
- 14.Blumcke I, et al. Malformations of cortical development and epilepsies: neuropathological findings with emphasis on focal cortical dysplasia. Epileptic Disord. 2009;11:181–193. doi: 10.1684/epd.2009.0261. [DOI] [PubMed] [Google Scholar]
- 15.Hadjivassiliou G, et al. The application of cortical layer markers in the evaluation of cortical dysplasias in epilepsy. Acta Neuropathol. 2010;120:517–528. doi: 10.1007/s00401-010-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cepeda C, et al. Epileptogenesis in pediatric cortical dysplasia: the dysmature cerebral developmental hypothesis. Epilepsy Behav. 2006;9:219–235. doi: 10.1016/j.yebeh.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tegenge MA, Rockel TD, Fritsche E, Bicker G. Nitric oxide stimulates human neural progenitor cell migration via cGMP-mediated signal transduction. Cell Mol Life Sci. 2011;68:2089–2099. doi: 10.1007/s00018-010-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams JR, et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 2011;71:2706–2717. doi: 10.1158/0008-5472.CAN-10-0738. [DOI] [PubMed] [Google Scholar]
- 20.D’Arcangelo G, et al. Reelin is a secreted glycoprotein recognized by the CR-50 monoclonal antibody. J Neurosci. 1997;17:23–31. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jossin Y, Cooper JA. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat Neurosci. 2011;14:697–703. doi: 10.1038/nn.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto-Torii K, et al. Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron. 2008;60:273–284. doi: 10.1016/j.neuron.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekine K, et al. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin alpha5beta1. Neuron. 2012;76:353–369. doi: 10.1016/j.neuron.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong SE, et al. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 26.Kubo K, et al. Ectopic Reelin induces neuronal aggregation with a normal birthdate-dependent “inside-out” alignment in the developing neocortex. J Neurosci. 2010;30:10953–10966. doi: 10.1523/JNEUROSCI.0486-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine K, Kubo KI, Nakajima K. How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex? Neurosci Res. 2014 doi: 10.1016/j.neures.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 29.Xuan S, et al. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 30.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 31.Regad T, Roth M, Bredenkamp N, Illing N, Papalopulu N. The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat Cell Biol. 2007;9:531–540. doi: 10.1038/ncb1573. [DOI] [PubMed] [Google Scholar]
- 32.Fauser S, et al. Long-term seizure outcome in 211 patients with focal cortical dysplasia. Epilepsia. 2015;56:66–76. doi: 10.1111/epi.12876. [DOI] [PubMed] [Google Scholar]
- 33.Gutmann DH. Tumor suppressor genes as negative growth regulators in development and differentiation. Int J Dev Biol. 1995;39:895–908. [PubMed] [Google Scholar]
- 34.Kwon CH, et al. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 35.Backman SA, et al. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat Genet. 2001;29:396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- 36.Feliciano DM, Su T, Lopez J, Platel JC, Bordey A. Single-cell Tsc1 knockout during corticogenesis generates tuber-like lesions and reduces seizure threshold in mice. J Clin Invest. 2011;121:1596–1607. doi: 10.1172/JCI44909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bateup HS, et al. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78:510–522. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8:1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- 39.Fraser MM, et al. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 2004;64:7773–7779. doi: 10.1158/0008-5472.CAN-04-2487. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Curr Opin Neurobiol. 2012;22:873–879. doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai PT, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ariani F, et al. FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet. 2008;83:89–93. doi: 10.1016/j.ajhg.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Persico AM, et al. Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol Psychiatry. 2001;6:150–159. doi: 10.1038/sj.mp.4000850. [DOI] [PubMed] [Google Scholar]
- 45.Crunelli V, Leresche N. Childhood absence epilepsy: genes, channels, neurons and networks. Nat Rev Neurosci. 2002;3:371–382. doi: 10.1038/nrn811. [DOI] [PubMed] [Google Scholar]
- 46.Truccolo W, et al. Single-neuron dynamics in human focal epilepsy. Nat Neurosci. 2011;14:635–641. doi: 10.1038/nn.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuki T, et al. Reelin and stk25 have opposing roles in neuronal polarization and dendritic Golgi deployment. Cell. 2010;143:826–836. doi: 10.1016/j.cell.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kupferman JV, et al. Reelin signaling specifies the molecular identity of the pyramidal neuron distal dendritic compartment. Cell. 2014;158:1335–1347. doi: 10.1016/j.cell.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pujadas L, et al. Reelin regulates postnatal neurogenesis and enhances spine hypertrophy and long-term potentiation. J Neurosci. 2010;30:4636–4649. doi: 10.1523/JNEUROSCI.5284-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chow LM, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19:305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurek KC, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindhurst MJ, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keppler-Noreuil KM, et al. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am J Med Genet A. 2014;164:1713–1733. doi: 10.1002/ajmg.a.36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LoTurco JJ, Bai J. The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 2006;29:407–413. doi: 10.1016/j.tins.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Baek ST, et al. Off-Target Effect of doublecortin Family shRNA on Neuronal Migration Associated with Endogenous MicroRNA Dysregulation. Neuron. 2014;82:1255–1262. doi: 10.1016/j.neuron.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarnat HB. Clinical neuropathology practice guide 5-2013: markers of neuronal maturation. Clin Neuropathol. 2013;32:340–369. doi: 10.5414/NP300638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.