Abstract
More than half of human genes use alternative cleavage and polyadenylation to generate alternative 3′UTR isoforms. Most efforts have focused on transcriptome-wide mapping of alternative 3′UTRs and on the question of how 3′UTR isoform ratios may be regulated. However, it is still less clear why alternative 3′UTRs have evolved and what biological roles they play. This review summarizes our current knowledge of the functional roles of alternative 3′UTRs, including mRNA localization, mRNA stability, and translational efficiency. Recent work suggests that alternative 3′UTRs may also enable the formation of protein-protein interactions to regulate protein localization or to diversify protein functions. These recent findings open up an exciting research direction for the investigation of new biological roles of alternative 3′UTRs.
Keywords: 3′UTR, alternative polyadenylation, non-coding RNA, post-transcriptional gene regulation, multi-functionality, protein-protein interactions, diversification of protein functions, protein localization, RNA-binding protein, RNA granule, protein abundance
Evolution of 3′UTR length and function
A few years ago, it was found that a large fraction of genes use alternative cleavage and polyadenylation to generate alternative 3′ untranslated regions (UTRs) [1–5]. Initially, the majority of efforts concentrated on establishing sequencing protocols to map alternative 3′UTRs transcriptome-wide [6–13], and elucidating how alternative 3′UTR ratios are regulated [14–24], two topics that have been summarized in several recent reviews [25–28]. Here, the functional roles of alternative 3′UTRs are discussed and reasons for why they have evolved are suggested.
It is largely unknown how biological complexity of organisms is achieved. Most cellular processes are carried out by proteins through interactions with other proteins [29, 30]. Therefore, when asked what makes humans different from worms, it was initially surprising that the number of protein-encoding genes and the coding region length have remained fairly constant during evolution from worms to humans [31–34]. Intuitively, higher protein diversity would enable more complex biological functions. However, higher protein diversity within a constrained space also increases the number of nonfunctional interactions [35, 36]. Therefore, protein concentration and diversity has to be limited to avoid overcrowding. When only a fixed number of proteins are available, biological complexity can still be accomplished through compartmentalization to avoid coexistence of too many protein types, by the strengthening of specific interactions through cooperativity [35], and by enabling multi-functionality of existing proteins.
Genome size, and, thus, the non-coding part of the genome, has dramatically increased during evolution from worms to humans. The expansion in non-coding sequences includes the 3′UTRs of mRNAs. The number of genes that produce alternative 3′UTRs has doubled and 3′UTR length has increased from a median of 140 nucleotides (nt) in worms to 1,200 nt in humans and even to 2,300 nt, when examining genes that generate alternative 3′UTRs [6, 13]. This suggests that more complex organisms have increased post-transcriptional gene regulation mediated by 3′UTR elements. 3′UTRs are well known to control mRNA stability, translational efficiency, and mRNA localization [2, 5, 37–42]. In addition, 3′UTRs were recently shown to mediate protein-protein interactions [43]. By facilitating alternative protein complex formation, alternative 3′UTRs can diversify protein functions. This may increase biological complexity by implementing multi-functionality of existing proteins.
Single and multi-UTR genes represent two distinct classes of genes
3′ end sequencing methods revealed that at least half of human genes generate alternative 3′UTR isoforms [9, 12, 13]. Transcriptome-wide analyses of alternative 3′UTRs across several normal human tissues and cell lines showed the presence of two classes of genes. One class produces mRNAs with only one 3′UTR, whereas the other class generates alternative 3′UTR isoforms [13]. Single and multi-UTR genes differ in their genomic architecture, their functions, and their mode of regulation of tissue-specific expression (Table 1).
Table 1.
Differences between single and multi-UTR genes
| Single-UTR | Multi-UTR | |
|---|---|---|
|
| ||
| 3′UTR length (nt) | 625 | 2,323 |
| Transcription unit length (bp) | 20,629 | 40,519 |
| N expressed in 1 tissue | 1,637 (23.7%) | 630 (11.1%) |
| N expressed in 2–5 tissues | 2,414 (34.9%) | 1,854 (32.6%) |
| N expressed in 6–7 tissues | 2,861 (41.4%) | 3189 (56.2%) |
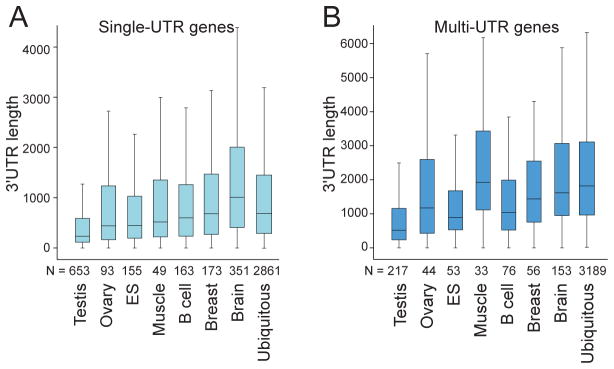
The median 3′UTR length of single-UTR genes is ~600 nt, but is ~2,300 nt for multi-UTR genes. The increase in 3′UTR length is associated with longer transcription units, which are about twice as long in multi-UTR genes (Table 1). The tissue-specific expression pattern of the two classes of genes also differs significantly. Whereas the majority of genes expressed in only one tissue has single 3′UTRs, more than half of ubiquitously transcribed genes are multi-UTR genes (Table 1) [13]. Furthermore, 3′UTR length is tissue-dependent: single or multi-UTR genes specifically expressed in testis or liver have much shorter 3′UTRs than genes only expressed in the brain (Fig. 1) [44–46]. However, ubiquitously transcribed multi-UTR genes have the longest 3′UTRs. They are even longer than the 3′UTRs of brain-specific genes and are about three-times longer than the 3′UTRs of ubiquitously expressed single-UTR genes (Fig. 1). This suggests that ubiquitously expressed genes that generate alternative 3′UTRs are especially prone to regulation by elements in their 3′UTRs. The importance of their 3′UTR-based regulation is further supported by a higher sequence conservation of multi-UTR genes with longer 3′UTRs [13].
Figure 1. 3′UTR length of ubiquitously transcribed or tissue-restricted genes.
A. Single-UTR genes. Here, ubiquitously expressed genes are defined to be expressed in at least six out of seven human tissues, including testis, ovary, embryonic stem (ES) cells, B cells, muscle, breast and brain. N, Number of genes in each category. B. Multi-UTR genes. As in (A).
What is the function of widely transcribed genes that generate alternative 3′UTRs? Whereas ubiquitously transcribed single-UTR genes are enriched in genes associated with classical housekeeping functions, such as ribosome biogenesis, translation, and energy metabolism, ubiquitous multi-UTR genes are abundant in genes with regulatory functions. They include transcription factors, RNA-binding proteins (RBPs), kinases, phosphatases, and proteins involved in RNA or protein transport [13]. Thus, widely expressed single or multi-UTR genes encode vastly different classes of proteins. Together with the finding that alternative 3′UTR isoform abundance is highly tissue- and cell type-specific [9, 13], widely expressed regulatory factors may use elements located in their alternative 3′UTRs to achieve tissue-specific expression or function [13, 43]. Thus, complex multi-cellular organisms may use elaborate post-transcriptional regulation to diversify protein functions.
3′UTRs regulate localization, stability, and translation of their cognate mRNAs
mRNAs contain cis-elements in their 3′UTRs that affect the biology of the mRNA that contains the regulatory elements. The 3′UTR elements, which are bound by RBPs, can control the subcellular localization, the half-life, or the rate of translation of an mRNA. RBPs mediate the different 3′UTR functions through interaction with diverse effector proteins.
Regulation of mRNA localization
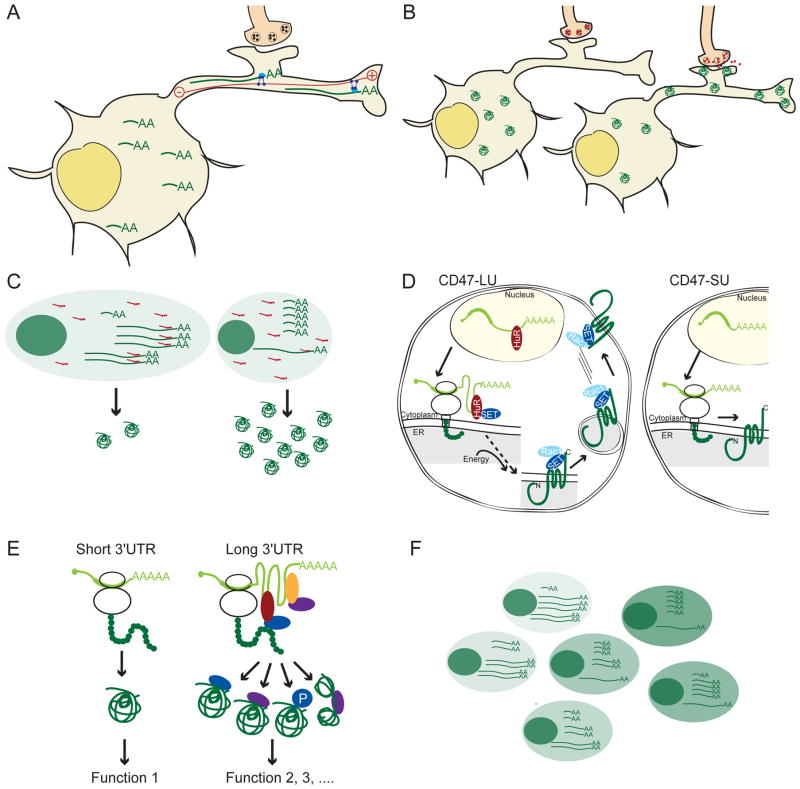
If an RBP that is bound to 3′UTR elements also interacts with a motor protein, it will localize the mRNA to different subcellular compartments [47–49]. Asymmetrically localized mRNAs are used to establish cell polarity, direct asymmetric cell division, or sequester protein activity [37]. In the case of alternative 3′UTRs of BDNF, it was shown that only the long 3′UTR isoform – which contains the localization elements – is localized to dendrites, where it regulates memory formation [38]. BDNF is a neurotrophin that plays roles in synaptogenesis and activity-dependent forms of synaptic plasticity [50]. Even modest alterations in BDNF levels are associated with behavioral changes in humans and mice, such as abnormal feeding behavior, alterations in episodic memory and susceptibility to anxiety and depression. Transcription of BDNF is regulated by alternative promoters, but, regardless of the promoter, the BDNF gene generates alternative 3′UTRs. The short 3′UTR isoform localizes BDNF mRNA and protein to the soma of neurons, but the long 3′UTR is necessary for localization of BDNF to the dendrites (Fig. 2A, Key Figure) [38]. Mice that lack the long 3′UTR of BDNF have altered dendritic spine morphology and decreased plasticity of dendritic synapses. In summary, mice lacking the long 3′UTR isoform of BDNF had impaired long-term potentiation of hippocampal neurons [38]. This study demonstrates that differential localization of BDNF 3′UTR isoforms is required for differential functions of BDNF in the soma and dendrites of neurons.
Figure 2. Diverse biological roles of alternative 3′UTRs.
A. Regulation of mRNA localization. The long 3′UTR contains a cis-element that is recognized by an RBP (light blue). Interaction with a molecular motor (dark blue) enables directional movement along actin fibers (red) and results in localization of the long 3′UTR isoform to the dendrites of neurons. The short 3′UTR isoforms lack the cis-acting element and remain localized to the soma. AA indicates a poly(A) tail. B. Activity-dependent regulation of translation of alternative 3′UTR isoforms. In the resting state (left) the short 3′UTR isoform of BDNF is translated which results in BDNF protein expression in the soma. After membrane depolarization by synaptic activation (right) the long 3′UTR isoform is translated which results in BDNF protein expression in dendrites and synapses. C. Cell-type specific regulation of protein abundance through interplay of alternative 3′UTRs with miRNAs. Presence of the miRNA and predominant expression of long 3′UTR isoforms in one cell type results in low protein output, whereas predominant expression of short 3′UTR isoforms results in escape from miRNA regulation and leads to high protein output. AA indicates a poly(A) tail. D. Alternative 3′UTRs use the scaffold function to regulate 3′UTR-dependent membrane protein localization. HuR-mediated recruitment of SET by the long 3′UTR facilitates formation of a protein complex containing SET/RAC1/CD47, which enables plasma membrane localization. E. Alternative 3′UTRs mediate protein-protein interactions to regulate protein function. Long 3′UTRs bind RBPs, which may be able to recruit effector proteins of diverse functions. Effector proteins may act as chaperones to achieve alternative protein folds or as enzymes that add alternative post-translational modifications. Or, by mediating protein-protein interactions, they may enable the formation of alternative protein complexes. F. Phenotypic diversity of single cells through variability in alternative 3′UTR isoform expression. Each cell of a clonal population expresses varying amounts of alternative 3′UTR isoforms which may contribute to differences in protein expression and phenotypic diversity among the population. AA indicates a poly(A) tail.
Regulation of translation
If an RBP that is bound to 3′UTR elements interacts with translation initiation factors, it can regulate translation of the mRNA. RBPs that mediate translational repression include Pumilio, Maskin, TIAR, and Musashi-1 [51–53]. As a consequence, alternative 3′UTR isoforms that either include or exclude the cis-elements can be translated with different efficiencies. This was shown to be the case for the alternative 3′UTR isoforms of BDNF whose translation rates are regulated in a temporal manner. It was shown that the short 3′UTR isoform of BDNF was translated in unstimulated hippocampal neurons. Neuronal activation of these neurons led to a switch in polysome association of the alternative 3′UTR isoforms and resulted in exclusive translation of the long 3′UTR isoform (Fig. 2B) [39]. This suggests that the short 3′UTR isoform is responsible for the generation of basal BDNF levels, whereas the long 3′UTR regulates activity-dependent production of BDNF. Neuronal activation also leads to transcriptional upregulation of BDNF, but the switch in isoform translation occurs first and enables an immediate increase in BDNF protein levels [39].
Another example how alternative 3′UTRs regulate translation was shown for polo, which encodes Polo-like kinase in Drosophila [54]. High levels of Polo are required for proliferation of abdominal epidermis precursor cells during development. It was demonstrated that the long 3′UTR isoform of polo was translated with much higher efficiency than the short 3′UTR isoform. Furthermore, deletion of the proximal polyadenylation signal of polo had no phenotypic effect, whereas deletion of the distal polyadenylation signal, which abrogated the function of the long 3′UTR isoform, led to death of the flies during development. This result indicated that the higher translation rates of the long 3′UTR isoform were required for the massive proliferation of epidermis precursor cells [54].
Regulation of mRNA stability
If an RBP that is bound to 3′UTR elements recruits deadenylation or decapping factors, it will destabilize the mRNA and, thus, affect its half-life. The best-studied cis-elements that control mRNA stability are microRNA (miRNA) binding sites and AU-rich elements [40, 55–58]. Thus, the binding of RBPs, such as AUF1/hnRNPD, TTP, or KSRP, that bind to AU-rich elements or the miRNA-mediated recruitment of Ago results in mRNA destabilization and short mRNA half-lives. Many proteins whose levels need to be tightly controlled are regulated at the level of mRNA stability, including oncogenes, cytokines, cell cycle regulators, and signaling proteins [57, 59–62].
If these genes generate alternative 3′UTRs, the alternative 3′UTR isoforms can control protein abundance levels, which was shown for oncogenes [5]. The shorter 3′UTRs of several oncogenes were more stable than their corresponding long 3′UTR isoforms and produced up to 40-fold more protein [5]. The genes that were chosen for investigation in this study had an enrichment of destabilizing elements in their long 3′UTRs and, thus, had a high potential for differential regulation of mRNA stability by alternative 3′UTRs [5]. Indeed, the difference in protein output generated either by the long or short 3′UTR isoforms was sufficient for a phenotypic difference, as was shown for CCND2 or IGF2BP1 (IMP-1) [5]. IGF2BP1 is an oncogene that when expressed with a short 3′UTR, generated IMP-1 protein amounts that were sufficient for oncogenic transformation of fibroblasts in soft agar or in mice [5, 63]. In contrast, the expression of IGF2BP1 under the same promoter, but with its long 3′UTR, had no transforming abilities. Thus, expression of shorter 3′UTR isoforms of genes whose long 3′UTR isoforms are enriched in negative regulatory elements resulted in increased protein expression. This study also showed that oncogenes can be activated through a change in 3′UTR isoform ratios [5].
This finding led to the prediction that shorter 3′UTRs that escape regulation by 3′UTR elements would be more stable and produce more protein, since the majority of 3′UTR elements known at the time, such as miRNA binding sites, AU- and GU-rich elements, had negative effects on gene expression [40, 55, 56, 64]. When this prediction was tested in studies carried out in yeast, mammalian cell lines or primary T cells [22, 65–67], either no correlation [65, 67] or only a weak correlation between shorter 3′UTRs and increased mRNA stability was found [22, 66]. Meanwhile, more extensive studies on the effects of regulatory sequences had shown that many 3′UTR elements have positive effects on gene expression [68–70]. Recent data even show that similar numbers of activating and repressive elements are present across all 3′UTRs [69]. These findings confirm the results of transcriptome-wide studies on mRNA stability rates and emphasize that, overall, shorter 3′UTRs are not more stable [22, 65–67].
In contrast, the earlier observations that shorter 3′UTRs are more stable was also confirmed in transcriptome-wide studies, in which the analysis focused on long 3′UTRs enriched in negative regulatory elements [5, 65, 66]. Long 3′UTRs that contained predominantly destabilizing elements, such as the Puf binding motif, AU-rich elements, or C-rich elements, showed increased decay rates in NIH3T3 cells or yeast [65, 66]. In summary, transcriptome-wide studies performed under non-stressed, steady-state conditions found that up to 35% of alternative 3′UTR isoforms differed in their mRNA stability rates, with the shorter 3′UTRs being stable slightly more often [66, 68].
Interplay of alternative 3′UTRs with miRNA-mediated regulation contributes to cell type-specific gene expression
Transcriptome-wide analyses of alternative 3′UTR isoforms revealed that the regions that vary between alternative isoforms are enriched in conserved binding sites for RBPs or miRNAs [13, 71]. Together with the finding that 3′UTR isoform ratios are highly cell-type specific [9, 13], it suggests that cell type-specific gene regulation can be partially accomplished by the interplay of alternative 3′UTR isoform expression and the presence of miRNA binding sites in distal 3′UTRs. A cell type-specific increase in usage of proximal polyadenylation sites can thus be used to avoid regulation by elements located in distal 3′UTRs. Indeed, Pax-3 mRNA, which is an important regulator of myogenesis, is targeted by miR-206 for degradation [72]. However, in a subset of muscle stem cells, high levels of Pax-3 as well as miR-206 are co-expressed. In these cells, high Pax-3 expression is accomplished by the expression of Pax-3 transcripts with shorter 3′UTRs that do not contain the binding site for miR-206. This shows that, under most conditions, Pax-3 is regulated by miR-206, but, in some cell types, expression of the shorter 3′UTR isoform leads to escape from miRNA regulation to enable Pax-3 expression despite expression of the miRNA (Fig. 2C) [72]. Avoidance of miRNA-mediated regulation as a means for cell type-specific regulation was confirmed by a transcriptome-wide study on cell lines that showed that approximately 10% of predicted miRNA target sites were affected by expression of alternative 3′UTRs [73].
The interplay of miRNA expression and alternative 3′UTRs in the regulation of tissue-specific gene regulation is further supported by the finding that ubiquitously transcribed genes that produce only one 3′UTR isoform are enriched in miRNA binding sites of tissue-specific miRNAs [13]. For these genes, the tissue-specificity of miRNA/target interaction is accomplished by the tissue-specific transcription of the miRNAs. In contrast, ubiquitously transcribed genes that generate alternative 3′UTRs are enriched in miRNA binding sites of ubiquitously expressed miRNAs. Thus, for this class of genes the tissue-specificity of miRNA/target interaction is accomplished by the tissue-specific expression of alternative 3′UTRs [13].
3′UTRs act as scaffolds and mediate protein-protein interactions
The biological functions of 3′UTRs were thought to only affect the properties of the mRNAs that carried the 3′UTR elements. However, a new function of 3′UTRs was discovered that does not alter the fate of the mRNA, but, instead, affects the newly made protein. It was shown that 3′UTRs facilitate the formation of protein-protein interactions. They do so by acting as scaffolds to recruit proteins to the site of translation, which enables the formation of protein complexes with the nascent peptide chain [43]. Protein complex formation can then determine membrane protein localization or protein functions.
The scaffold function of 3′UTRs was predominantly studied for the CD47 gene, which generates alternative 3′UTR isoforms and encodes a membrane protein. It was found that CD47 protein generated by the short CD47 3′UTR isoform (CD47-SU) was predominantly retained in the endoplasmic reticulum, whereas CD47 protein generated by the long 3′UTR isoform (CD47-LU) efficiently localized to the plasma membrane. CD47-SU and CD47-LU have an identical amino acid sequence and the difference in protein localization was shown to be independent of mRNA localization, suggesting that the 3′UTRs regulate protein localization [43].
With respect to the mechanism (Fig. 2D), it was demonstrated that the long CD47 3′UTR binds the RBP HuR, which recruits the effector protein SET [74] to the site of translation. During translation of CD47, SET is transferred from the mRNA to the nascent protein, resulting in a protein complex between SET and CD47-LU [43]. The formation of the protein complex depended on the presence of the long 3′UTR of CD47 because CD47-SU did not interact with SET. SET also binds to RAC1 and active RAC1 translocated SET/CD47-LU to the plasma membrane [43, 75]. As a result, CD47-LU efficiently localized to the plasma membrane whereas CD47-SU, which does not have binding sites for HuR and thus, does not recruit SET, was retained in the endoplasmic reticulum [43]. Intriguingly, this mechanism of 3′UTR-dependent membrane protein localization was not restricted to the regulation of CD47, but resulted in efficient plasma membrane localization of additional candidates. It was speculated that this localization mechanism may be especially important for membrane proteins with several transmembrane domains, such as G-protein coupled receptors, which are known to be difficult to express in heterologous systems [76]. These results suggest that inclusion of the endogenous 3′UTR or of a 3′UTR that is competent for SET recruitment may increase surface localization of these proteins, which are difficult to express.
In addition to regulating protein localization, the scaffold function of 3′UTRs was also shown to determine protein function, at least for CD47. CD47-LU interacts with SET and RAC1, which resulted in co-localization with RAC1 at the plasma membrane, RAC1 hyperactivation, lamellipodia formation, and cell migration [43]. In contrast, CD47-SU did not interact with SET and RAC1 and lacked all RAC1-mediated functions, despite also being localized to the plasma membrane like CD47-LU [43]. These findings demonstrate that proteins with identical amino acid sequence and that localize to the same cellular compartment, but are generated by alternative 3′UTRs, can carry out alternative functions due to the generation of alternative protein complexes. Thus, alternative 3′UTRs can diversify protein functions without changing the amino acid sequence.
It is currently unknown how widespread is the scaffold function of 3′UTRs. Many RBPs other than HuR have domains that mediate protein-protein interactions [77–81]. Thus, it is likely that HuR is not the only RBP able to recruit effector proteins to the site of translation to direct the functions of nascent proteins. It is also likely that the scaffold function of 3′UTRs is not restricted to the regulation of membrane proteins but also affects cytosolic or nuclear proteins. It can be speculated that 3′UTR-recruited effector proteins may have diverse functions. They may act as chaperones and result in alternative protein folds, or they may help with the formation of multi-protein complexes by facilitating protein-protein interactions (Fig. 2E). Furthermore, it can be imagined that they act as enzymes that add alternative post-translational modifications to the nascent protein, which was shown to be the case for CEBPB [82].
Finally, it can be speculated that the scaffold function may play a role in miRNA-mediated gene regulation. All human AGO genes have long 3′UTRs and use ApA [13]. miRNAs loaded into AGO are known to bind to 3′UTRs and repress gene expression [40, 58]. However, miRNA effects are often subtle, therefore, it is possible that AGO may act similarly to other RBPs and recruit diverse effector proteins to nascent peptide chains in order to change the function of the nascent proteins.
RNA editing and alternative 3′UTRs contribute to phenotypic diversity
RNA editing changes the sequence of the RNA without changing the DNA. Thus, mRNA editing contributes to individual variation in gene expression and results in the diversification of the transcriptome within a tissue- or cell type. Interestingly, regardless of the type of RNA editing – A to I by ADAR [83], C to U by APOBEC1 [84], mRNA pseudouridilation [85, 86], or m6A modifications [87, 88] – all modifications show enrichment in 3′UTRs. m6A modifications were thought to be enriched surrounding stop codons [87, 88]. However, a more detailed study revealed that m6A modifications mark the beginning of the terminal exon and, thus, may be involved in determining the end of a transcript [89]. An important feature of RNA modifications is that not all transcripts are modified, which suggests a role for RNA editing in diversification of the transcriptome.
Diversification of the transcriptome can also be achieved by the expression of alternative 3′UTRs. Several 3′ end sequencing studies showed that the number of 3′UTR isoforms generated by a gene depends on the stringency of the cut-offs used for calling the presence of an isoform [6, 12, 13]. When only robustly expressed, alternative 3′UTR isoforms were considered, the fraction of human genes that generated alternative 3′UTRs was about 50% [13]. However, if minor isoforms were also included, up to 79% of mouse or human genes produced alternative 3′UTRs and generated, on average, four isoforms [12, 13]. Extensive alternative 3′UTR isoform diversity was also seen in clonal populations, such as cell lines or exponentially growing yeast [12, 13, 90]. Furthermore, the mapping of 3′UTR isoform expression in single cells from mouse embryonic stem cells and neural stem cells revealed that although the developmental state globally determined isoform expression, single cells from the same state often differed in the choice of isoforms [91]. Genes with a higher variability in isoform expression were often moderately expressed, which suggests that many of these isoforms are expressed at less than one copy per cell. As a result, different transcriptomes may be expressed in different single cells (Fig. 2F) [91]. Taken together, 3′UTRs use RNA editing or alternative cleavage and polyadenylation to diversify the transcriptome and, thus, contribute to epigenetic gene regulation.
Concluding remarks
3′UTR-mediated gene regulation seems to have expanded during evolution and appears to be an integral part of the functional diversity seen in higher organisms. The multi-functionality of proteins facilitated by alternative 3′UTRs may contribute to increased biological complexity. In order to fully understand the functional roles of 3′UTRs, it will be important to determine all the RBPs that bind to specific 3′UTRs (see Outstanding questions). Two similar methods that identify the RBPs associated with long non-coding RNAs were recently established and identified the proteins bound to Xist [92, 93]. These methods can also be applied to identify RBPs bound to specific 3′UTRs.
Outstanding questions.
What are all the RBPs that bind to a specific 3′UTR and how does the set of bound proteins differ in various cellular contexts? Methods that were established to identify the composition and dynamics of proteins bound to long non-coding RNAs can be applied to identify the ensemble of proteins bound to specific 3′UTRs. The comparison of the 3′UTR-bound proteins across cell types will help to elucidate the contribution of 3′UTRs in the regulation of cell-type specific gene expression and function.
What are the protein interaction partners of 3′UTR-bound RBPs? Ultimately, the protein interaction partners of RBPs determine the functional outcomes of 3′UTRs. Therefore, the identification of protein interaction partners of RBPs will be necessary to address the diverse biological roles of 3′UTRs.
What determines the specific function of an RBP that acts on a single mRNA? As was illustrated for HuR, an RBP can accomplish diverse functions in gene regulation. It will be important to understand what regulates the specific function of an RBP in the context of a single gene. Post-translational modifications, protein interaction partners and RNA granule composition have the potential to be regulators of specific RBP functions.
The scaffold function of 3′UTRs seems to diversify protein functions. Is the scaffold function of 3′UTRs also used by non-membrane proteins to alter protein function? Do all 3′UTRs mediate protein-protein interactions?
RBPs are highly interactive at the RNA and protein levels [80, 81, 94]. In addition to identifying the RBPs that bind to specific 3′UTRs, it will be important to determine the protein interaction partners of these RBPs. As was illustrated above, the interaction partners of the RBPs are ultimately responsible for the different functions that are mediated by each 3′UTR. If the interaction partner is a motor protein, the mRNA will localize to different subcellular compartments, whereas if the interaction partner is a deadenylase or a translation initiation factor, the stability or the translation rate of the mRNA may change [48, 58].
One challenge will be to dissect the different functions that can be carried out by a single RBP. This is illustrated for HuR. HuR can regulate alternative polyadenylation [95, 96], stabilize transcripts [97], increase translation [98], or regulate membrane protein localization [43]. It will be important to understand how a specific function of an RBP is selected to act on a single mRNA. One possibility is that post-translational modifications or protein interaction partners determine context-specific functions of RBPs [99, 100].
The recent identification of approximately 800 RBPs in a cell [80, 81], with the majority of them binding to 3′UTRs hints at a myriad of possible interactions that ultimately determine the individual fate of each mRNA. In addition to well-known RNA-binding motifs, RBPs are also strongly enriched in intrinsically disordered regions (IDRs) [80]. Proteins with IDRs, together with mRNAs or small RNAs, are the major components of RNA granules [101, 102]. Overexpression of IDRs of known RBPs resulted in liquid phase transitions, leading to the generation of RNA granules or RNA droplets because of their liquid-like behavior [101, 103, 104]. RNA granules can contain 100-fold higher concentrations of proteins with IDRs compared with the surrounding cytoplasm [105] and, thus, may facilitate many mRNA-based processes such as signaling, splicing, degradation, storage, and transport of mRNAs [105, 106]. The purification of the mRNA components of RNA granules showed that the average 3′UTR length within granules was over five-fold longer than the average 3′UTR length found in the cytoplasm [102]. Intriguingly, 3′UTR length of mRNAs within RNA granules is similar to the length of long 3′UTR isoforms, whereas single-UTR genes or mRNAs with short 3′UTRs seem to be present in the cytoplasm. These findings suggest that mRNAs with long 3′UTRs, which seem to be covered with RBPs, predominantly reside and carry out their functions within RNA granules. In order to understand the biology of 3′UTRs, it will be important to view them in the context of the bound RBPs and within diverse types of RNA granules.
Trends Box.
During animal evolution, the number of protein-encoding genes has remained fairly constant, but 3′UTR length and the fraction of genes expressing alternative 3′UTRs has increased substantially.
3′UTRs mediate protein-protein interactions. Thus, alternative 3′UTRs facilitate the formation of alternative protein complexes, which can carry out alternative protein functions. This diversifies proteome function without a change in amino acid sequence.
About 15–35% of alternative 3′UTRs have significantly different half-lives, which may contribute to the transcriptome diversity of single cells.
Translation rates of mRNAs with alternative 3′UTRs can be differentially affected by signaling. Whereas one isoform generates basal protein levels, translation of the other is induced by signaling.
Long 3′UTRs seem to be bound by many RBPs and may exert their functions within RNA granules.
Acknowledgments
I apologize to all colleagues whose work could not be cited because of space constraints. I am grateful to Eric Kallin, Binyamin Berkovits, Nikolaus Rajewsky, and Nicholas Proudfoot for critical comments on the manuscript. Funding for work on alternative 3′UTRs was obtained from NIH/NCI, the Starr Cancer Foundation, the Sidney Kimmel Cancer Foundation and from the Innovator Award of the Damon Runyon-Rachleff Cancer Foundation and the Island Outreach Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tian B, et al. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandberg R, et al. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji Z, et al. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jan CH, et al. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepard PJ, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y, et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Y, et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40:8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkon R, et al. E2F mediates enhanced alternative polyadenylation in proliferation. Genome biology. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoque M, et al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lianoglou S, et al. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenal M, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 17.de Klerk E, et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 2012;40:9089–9101. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin G, et al. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell reports. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Bava FA, et al. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature. 2013;495:121–125. doi: 10.1038/nature11901. [DOI] [PubMed] [Google Scholar]
- 21.Lackford B, et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33:878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masamha CP, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Giammartino DC, et al. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3′ UTRs. Genes Dev. 2014;28:2248–2260. doi: 10.1101/gad.245787.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batra R, et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elkon R, et al. Alternative cleavage and polyadenylation: extent, regulation and function. Nature reviews Genetics. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 26.Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends in biochemical sciences. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley DL. Coupling mRNA processing with transcription in time and space. Nature reviews Genetics. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng D, Tian B. RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Advances in experimental medicine and biology. 2014;825:97–127. doi: 10.1007/978-1-4939-1221-6_3. [DOI] [PubMed] [Google Scholar]
- 29.Han JD, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 30.Nooren IM, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 32.Hillier LW, et al. Genomics in C. elegans: so many genes, such a little worm. Genome Res. 2005;15:1651–1660. doi: 10.1101/gr.3729105. [DOI] [PubMed] [Google Scholar]
- 33.Ezkurdia I, et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Human molecular genetics. 2014;23:5866–5878. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spieth J, Lawson D, Davis P, Williams G, Howe K. Wormbook. 2014. Overview of gene structure in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, et al. Constraints imposed by non-functional protein-protein interactions on gene expression and proteome size. Molecular systems biology. 2008;4:210. doi: 10.1038/msb.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy ED, et al. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc Natl Acad Sci U S A. 2012;109:20461–20466. doi: 10.1073/pnas.1209312109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An JJ, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau AG, et al. Distinct 3′UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF) Proc Natl Acad Sci U S A. 2010;107:15945–15950. doi: 10.1073/pnas.1002929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Jambor H, et al. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife. 2015:4. doi: 10.7554/eLife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berkovits BD, Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sood P, et al. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smibert P, et al. Global Patterns of Tissue-Specific Alternative Polyadenylation in Drosophila. Cell reports. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulitsky I, et al. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 48.Niedner A, et al. Of social molecules: The interactive assembly of ASH1 mRNA-transport complexes in yeast. RNA biology. 2014;11:998–1009. doi: 10.4161/rna.29946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ephrussi A, et al. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 50.Greenberg ME, et al. New insights in the biology of BDNF synthesis and release: implications in CNS function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:12764–12767. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Moor CH, et al. Mechanisms of translational control by the 3′ UTR in development and differentiation. Seminars in cell & developmental biology. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Mazan-Mamczarz K, et al. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Battelli C, et al. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Molecular and cellular neurosciences. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Pinto PA, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barreau C, et al. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends in biochemical sciences. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 57.Liao B, et al. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 58.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nature reviews Genetics. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 59.Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dani C, et al. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984;81:7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiavi SC, et al. Regulation of proto-oncogene mRNA stability. Biochim Biophys Acta. 1992;1114:95–106. doi: 10.1016/0304-419x(92)90009-n. [DOI] [PubMed] [Google Scholar]
- 62.Stoecklin G, et al. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- 63.Tessier CR, et al. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 2004;64:209–214. doi: 10.1158/0008-5472.can-03-2927. [DOI] [PubMed] [Google Scholar]
- 64.Vlasova IA, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta I, et al. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Molecular systems biology. 2014;10:719. doi: 10.1002/msb.135068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spies N, et al. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013;23:2078–2090. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gruber AR, et al. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nature communications. 2014;5:5465. doi: 10.1038/ncomms6465. [DOI] [PubMed] [Google Scholar]
- 68.Geisberg JV, et al. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oikonomou P, et al. Systematic identification of regulatory elements in conserved 3′ UTRs of human transcripts. Cell reports. 2014;7:281–292. doi: 10.1016/j.celrep.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kristjansdottir K, et al. Systematic analysis of the Hmga2 3′ UTR identifies many independent regulatory sequences and a novel interaction between distal sites. RNA. 2015;21:1346–1360. doi: 10.1261/rna.051177.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pelechano V, et al. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boutet SC, et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell stem cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nam JW, et al. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2014;53:1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brennan CM, et al. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. The Journal of cell biology. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.ten Klooster JP, et al. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007;26:336–345. doi: 10.1038/sj.emboj.7601518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dunham JH, Hall RA. Enhancement of the surface expression of G protein-coupled receptors. Trends in biotechnology. 2009;27:541–545. doi: 10.1016/j.tibtech.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bandziulis RJ, et al. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 78.Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 79.Lunde BM, et al. RNA-binding proteins: modular design for efficient function. Nature reviews Molecular cell biology. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 81.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 82.Basu SK, et al. 3′UTR elements inhibit Ras-induced C/EBPbeta post-translational activation and senescence in tumour cells. EMBO J. 2011;30:3714–3728. doi: 10.1038/emboj.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peng Z, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 84.Rosenberg BR, et al. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol. 2011;18:230–236. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 88.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ke S, et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015;29:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pelechano V, et al. Genome-wide polyadenylation site mapping. Methods in enzymology. 2012;513:271–296. doi: 10.1016/B978-0-12-391938-0.00012-4. [DOI] [PubMed] [Google Scholar]
- 91.Velten L, et al. Single-cell polyadenylation site mapping reveals 3′ isoform choice variability. Molecular systems biology. 2015;11:812. doi: 10.15252/msb.20156198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim PM, et al. The role of disorder in interaction networks: a structural analysis. Molecular systems biology. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hilgers V, et al. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 2012;26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oktaba K, et al. ELAV Links Paused Pol II to Alternative Polyadenylation in the Drosophila Nervous System. Mol Cell. 2015;57:341–348. doi: 10.1016/j.molcel.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mazan-Mamczarz K, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Current opinion in genetics & development. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 101.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han TW, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 103.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]